Genetic homogeneity and historical expansions of the slipper lobster, Scyllarides brasiliensis, in the south-west Atlantic
Ghennie T. Rodríguez-Rey A , Antonio M. Solé-Cava A and Cristiano Lazoski A BA Laboratório de Biodiversidade Molecular, Departamento de Genética, Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, 21941-590, Brazil.
B Corresponding author. Email: lazoski@acd.ufrj.br
Marine and Freshwater Research 65(1) 59-69 https://doi.org/10.1071/MF12359
Submitted: 25 December 2012 Accepted: 18 June 2013 Published: 20 September 2013
Abstract
Management strategies for fisheries species require understanding their connectivity and population dynamics. The Brazilian slipper lobster, Scyllarides brasiliensis, is one of the most commercially important slipper lobster species in South America. We investigated, for the first time, the population genetic structure and evolutionary history of this species. Analyses of sequences of the cytochrome oxidase I gene (COI) and the control region (CR) did not reveal any significant genetic structure of S. brasiliensis (N = 202) along 2700 km of the Atlantic coast (COI: ΦST = 0.0004, ΦCT = 0–0.005, P > 0.05; CR: ΦST = 0.004, ΦCT = 0–0.029, P > 0.05). The genetic homogeneity found suggests high levels of gene flow along the area that are possibly related to the high dispersal potential of the planktonic larvae of the species. Furthermore, the data indicate that demographic and geographical expansions of this slipper lobster population have occurred during the late and middle Pleistocene, which could be related to the fluctuating environmental conditions of that period.
Additional keywords: fisheries, genetic structure, phylogeography, Scyllaridae.
Introduction
Slipper lobsters (family Scyllaridae) have received little scientific and fisheries management attention because of their lower economic importance compared with that of clawed (family Nephropidae) or spiny (family Palinuridae) lobsters (Spanier and Lavalli 2007). However, slipper lobster catches have increased over the last 10 years as a consequence of the overfishing of the Palinuridae (Groeneveld et al. 2006; Phillips and Melville-Smith 2006; Spanier and Lavalli 2007). This led to an initial increase in the role of slipper lobster fisheries in the economies of areas where they are fished (Australia, Brazil, Galapagos Islands, Hawaii, India, and the Mediterranean), but poor fishery management subsequently resulted in a decrease of their relative abundance (Spanier and Lavalli 2006, 2007; Oliveira et al. 2008; Duarte et al. 2010, 2011). Within the Scyllaridae, three genera, Ibacus (subfamily Ibacinae), Scyllarides (subfamily Arctidinae) and Thenus (subfamily Theninae) have significant fisheries because of their large body size (Spanier and Lavalli 2006). Within Scyllarides, the Brazilian slipper lobster, S. brasiliensis Rathbun, 1906, is one of the main species caught in South America, especially in the North-east region (Santos and Freitas 2002). S. brasiliensis can be found at 20–40 m depth in the western Atlantic, along the Brazilian coast, from 2°S (Maranhão State) to 30°S (Santa Catarina State) (Dall’Occo et al. 2007). The species has also been reported in Dominica, in the West Indies (Holthuis 1991).
A key factor in fisheries management is delimiting stocks (Ward 2002). The structuring and connectivity of populations is determined largely by the dispersal of individuals. The environment in which marine organisms live allows for a variety of mechanisms for dispersal among populations (Cowen and Sponaugle 2009). For most benthic invertebrates that are sessile or with limited mobility, gene exchange between populations occurs mainly during the pelagic larval phase (Thorpe et al. 2000) and depends on the biological characteristics of larvae and their interaction with ecological (food availability, species interactions) and physical (past and present currents, and climatic features) processes (Palumbi 2003; Shanks et al. 2003). Usually, species with a short planktonic larval duration tend to present higher levels of population structuring than those with long planktonic larval durations (Palumbi 2003). Nevertheless, there are exceptions. Strong population structuring can also be found in species with long planktonic larval duration, since dispersal may be affected by larval behaviour or oceanographic mechanisms that can result in high degrees of larval retention, or high selection against recruits in new environments (Palumbi 2003; Taylor and Hellberg 2003; Palero et al. 2008).
Achelata lobsters (spiny, slipper, and coral lobsters) have a larval phase (phyllosoma) specially adapted for long dispersal (Palero et al. 2008). This dispersal ability is among the greatest found in crustaceans; in some species of Scyllarides, the phyllosome larvae can live in the plankton for up to 9 months (Coutures 2000; Booth et al. 2005; Sekiguchi et al. 2007). Accordingly, for some of the lobster species of Jasus, Panulirus, Palinurus and Scyllarides, no genetic differentiation is usually observed, even between populations distributed over large oceanographic distances (e.g. Tolley et al. 2005; Naro-Maciel et al. 2011; Kennington et al. 2013) (Table 1). However, there are exceptions in which some genetic heterogeneity has been reported (e.g. Brasher et al. 1992; Palero et al. 2008; Babbucci et al. 2010) (Table 1). In Jasus verreauxi, for example, heterogeneity among southern Australia and New Zealand populations was observed, probably due to a fall in larval survivorship while crossing the Tasman Sea (Brasher et al. 1992). In Palinurus delagoae and P. elephas, low genetic differentiation was found, possibly caused by the retention of some of the larvae by mesoscale oceanic processes such as eddies (Gopal et al. 2006; Palero et al. 2008; Babbucci et al. 2010). Similarly, strong differentiation was found between the Caribbean and south-west Atlantic populations of Panulirus argus (Diniz et al. 2005), but that diversity was later attributed to the presence of cryptic species (Tourinho et al. 2012). Also, in Panulirus penicillatus, the heterogeneity found between the eastern Pacific and the central to western Pacific was probably due to the presence of cryptic subspecies (Chow et al. 2011).

|
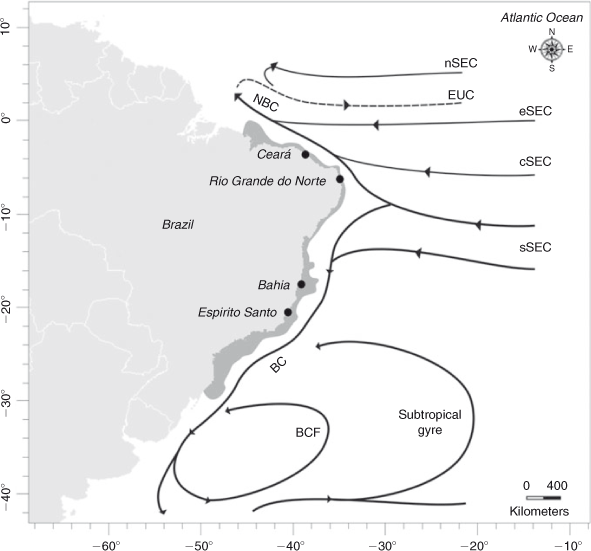
Circulation in the southern Atlantic is complex, and offers an interesting setting to study the influence of current systems on the larval transport of marine species. In that area, the surface circulation pattern shows some dependence on predominant wind and sea depth (Stramma and England 1999). The South-Equatorial Current (SEC) flows westward across the Atlantic until it reaches the Brazilian coast, at which point (9–15°S) it splits into the Northern Brazilian Current, which flows north to Guyana and the Caribbean Sea, and the Brazil Current, which flows south along the Brazilian coast (Cirano et al. 2006). The South-Equatorial Current has four main branches, the south (sSEC), the central (cSEC), the north (nSEC), and the equatorial (eSEC). The Brazil Current originates from the sSEC, has several associated eddies, and flows to the region of the Subtropical Convergence (~35°S), where it breaks into two branches: one turning north, forming a recirculation cell by the Brazil Current Front, and the second continuing southward, associating with the South Atlantic subtropical gyre (Cirano et al. 2006) (Fig. 1). The intensity and bifurcation of the South-Equatorial Current could have an effect in the population structure of marine species living in this area, acting as a barrier to gene flow between populations of either side of this current. For example, genetic heterogeneity between populations north and south of the South-Equatorial Current has been observed in Micropogonias furnieri and Holacanthus ciliares (Puchnick-Legat and Levy 2006; Affonso and Galetti Jr 2007). Moreover, the presence of eddies and fronts could cause larval retention, affecting the dispersal potential of the species, as was observed in Panulirus delagoae on the south-eastern coast of Africa (Gopal et al. 2006).
|
To date, no studies have been performed on the population genetic structure of Scyllarides brasiliensis, one of the most commercially important slipper lobster species in South America. The aim of our study was to estimate the population structure and demographic history of S. brasiliensis along 2700 km of the south-west Atlantic, including samples from both sides of the South-Equatorial Current. For that, we used two mitochondrial markers: the cytochrome c oxidase I gene and the control region, which have been shown to be useful for population genetics studies in crustaceans.
Materials and methods
Sampling
Samples of Scyllarides brasiliensis were collected at four sites along 2700 km of the Brazilian coast: Ceará (03°43′S, 38°32′W; N = 34), Rio Grande do Norte (05°47′S, 35°12′W; N = 21), Bahia (17°31′S, 39°11′W; N = 75) and Espírito Santo (21°02′S, 40°49′W; N = 73) (Fig. 1). Slipper lobsters were identified morphologically (Holthuis 1991) and one pereiopod or muscle tissue from each lobster was removed and preserved in absolute ethanol.
DNA extraction, amplification and sequencing
Total genomic DNA was obtained by salt extraction (Lysis buffer: 50 mm TRIS-HCl, 50 mm EDTA, 1% SDS, 50 mm NaCl; Proteinase K 20 mg mL–1), followed by precipitation in isopropanol (modified from Miller et al. 1988). The 5′ end of the cytochrome c oxidase subunit I (COI) gene was amplified using the universal primers LCO1490 and HCO2198 (Folmer et al. 1994). The control region (CR) of the mtDNA genome was amplified using the specific internal primers RCSBE_F (ATA GCA AGA ATC AAA CTT AT) and RCSBE_R (TCA GGC ATC ATA TTT ATC). These primers were developed from sequences initially obtained using the primers CRLF and CRLR that were originally designed to amplify the CR of Panulirus argus (Diniz et al. 2005). Amplification reactions included ~10–50 ng of genomic DNA, 1 U of GoTaq Flexi DNA polymerase (Promega), 3 μL of Green GoTaq Flexi Buffer (5X), 0.2 mm of dNTPs, 2.5 mm of MgCl2, 0.3 μm of each primer and 4 μg of Bovine Serum Albumin (BSA) in a final volume of 15 μL. Reactions were carried out with an initial denaturation step of 2 min at 95°C, followed by 35 cycles consisting of a denaturation step of 1 min at 92°C, an annealing step of 40 s at 46°C (LCO1490/HCO2198 and RCSBE_F/RCSBE_R), or at 40°C (CRLF/CRLR), and an extension step of 50 s at 72°C; and a final extension step of 5 min at 72°C. Forward and reverse sequences of purified PCR products were obtained using an ABI 3500 automated DNA sequencer (Applied Biosystems)(annealing temperature 64°C). Preliminary analyses indicated coamplification of multiple sequences when using universal primers for COI. This prompted us to design specific sequencing primers COISB_F (GAA CTA GGA CAG CCT GGG AGG TTG A) and COISB_R (GAG GTG TTG AGA TTA CGG TCA GT). CR sequences were obtained using the same primer pairs used for PCR amplification.
Intraspecific variability and population genetic differentiation
The sequences obtained for each gene were edited in SEQMAN II 4.0 (DNAstar Inc.). Due to the different mutation rates between the COI gene and the CR, analyses were carried out separately for each region. Sequences were aligned using the CLUSTALW (Thompson et al. 1994) algorithm implemented in MEGA 5.0 (Tamura et al. 2011) and checked manually for misalignments. Gaps were not observed in the COI sequences. For CR analysis, each gap region in the alignment was conservatively replaced, without violating assumptions of independence of characters, by a single transition, regardless of gap size. All haplotype sequences were deposited in GenBank (Accession Numbers COI: JX896692 to JX896755; CR: JX896756 to JX896954).
Standard genetic diversity indices such as nucleotide (π) and haplotype (h) diversities and their variances were estimated for each locality using DNASP 5 (Librado and Rozas 2009). The number of haplotypes, polymorphic sites, transitions, and transversions were obtained using ARLEQUIN 3.11 (Excoffier et al. 2005). Pairwise genetic divergences between populations were estimated using FST statistics, and population structure was examined through an analysis of molecular variance (AMOVA) using ARLEQUIN. The statistical significance of estimates was assessed by 10 000 permutations. Sequential Bonferroni (Rice 1989) and False Discovery Rate (Benjamini and Hochberg 1995) corrections were used to adjust significance levels to account for multiple simultaneous tests. False Discovery Rate estimates were performed using the Excel spread sheet designed by Pike (2011). The genealogical relationships among haplotypes were assessed through a parsimony haplotype network constructed using a median-joining algorithm as implemented in the software NETWORK 4.5.1.6 (Bandelt et al. 1999). Due to the high diversity observed in the control region of this species (see Results), the network for that region was built by considering only the transversions. This approach aims to minimise the effect of homoplasies due to mutational saturation of transition sites, and has been used successfully in previous studies (Bowen and Grant 1997; Ball et al. 2007). The network for the COI gene was built by taking both types of nucleotide substitutions into account.
Neutrality tests and demographic analyses
Tajima’s D (Tajima 1989) and Fu’s Fs (Fu 1997), as implemented in ARLEQUIN, were used to test for mutation-drift equilibrium deviation in the overall sample. Most variation at the molecular level of neutral or nearly neutral sequences is the result of the equilibrium between genetic drift and mutation. When a large number of low-frequency haplotypes accumulate in a population, deviation from neutrality can be explained as resulting from selection or from demographic changes (Fu and Li 1993; Tajima 1996). Thus, putative temporal changes in population size of the overall sample were investigated through R2 tests (Ramos-Onsins and Rozas 2002), Bayesian Skyline Plots (Drummond et al. 2005) and mismatch distribution analyses (Rogers and Harpending 1992). R2 statistics and their confidence intervals were estimated by coalescence simulations using DNASP.
Bayesian Skyline Analyses were done in BEAST 1.6.2 (Drummond and Rambaut 2007). The posterior distributions of gene genealogies and population parameters were used to generate credibility intervals [highest posterior densities (HPD)] that represent both phylogenetic and coalescent uncertainty (Drummond et al. 2005). The nucleotide substitution models were identified through the Bayesian Information Criterion implemented in MEGA. For the COI gene, the HKY85 model and a substitution rate of 2.3% million year–1 (Schubart et al. 1998) were employed. For the CR, the substitution GTR+G+I model, with a substitution rate of 19% Myear–1 (McMillen-Jackson and Bert 2003), was used. The analyses assumed a strict molecular clock model, and the number of groups was set to six. For each marker, three independent MCMC analyses of 100 million generations were run by sampling every 1000th generation, with the first 10% of each run discarded as burn-in. The analyses for each gene were combined using LOGCOMBINER 1.6.2 (distributed with BEAST) and the results were summarised in piecewise-constant Bayesian Skyline Plots using TRACER 1.5 (Rambaut and Drummond 2007).
The parameters and approximate confidence intervals for the mismatch distributions were estimated using the parametric bootstrap approach implemented in ARLEQUIN under models of demographic and spatial expansion. Population growth has a strong effect on the frequency distribution of pairwise differences among all haplotypes (Rogers and Harpending 1992). Thus, populations that are stable or that experienced a sudden reduction in size present multimodal mismatch distributions, whereas the distribution appears unimodal (approximately Poisson) in populations that have passed through recent demographic or spatial expansions (possibly following a bottleneck) (Rogers and Harpending 1992; Bunje and Wirth 2008). The validity of the expansion models was evaluated using the sum of square deviations (SSD) between the observed and expected mismatch distributions and by the raggedness index (r) (Harpending 1994).
Finally, time since expansion started (t), was estimated as t = τ/2u (Rogers and Harpending 1992), where τ is the age of the demographic change in mutational units estimated in the mismatch distribution, and u is the mutation rate for the whole haplotype (Harpending 1994). The value of u was calculated as u = 2μk, where μ is the mutation rate per nucleotide and k is the length of the sequence.
Results
Intraspecific variability and population genetic differentiation
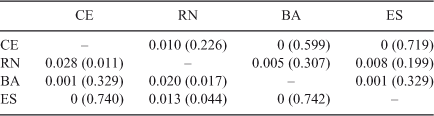
After alignment, a segment of 537 bp of the COI gene was obtained from each of the 202 S. brasiliensis specimens analysed. Fifty-three sites were polymorphic (56 substitutions, 43 of which were transitions). Three of these yielded non-synonymous changes. The percentage of A+T found (57.8%) was similar to that reported for lobsters of the genera Jasus (59.5%) and Panulirus (55–57%) (Ovenden et al. 1997; Tourinho et al. 2012). Of the 64 haplotypes identified, 50 were private (unique to a single locality). Seven of the 14 shared haplotypes showed a wide geographical distribution. The most common haplotype was found in 78 (38.6%) individuals from all localities. The other haplotypes differed little from the common one, resulting in a low nucleotide diversity (overall π = 0.004; range = 0.001–0.004) and moderate to high haplotype diversity (overall h = 0.840; range = 0.429–0.923) (Table 2).

|
For the control region, an aligned segment of 739 bp was obtained from each of the 201 S. brasiliensis individuals analysed. The alignment revealed 260 (35.4%) polymorphic sites with 315 substitutions, 247 of which were transitions. The percentage of A+T found (70.5%) was similar to that reported for the same region in lobsters of the genera Panulirus (73.2%) and Palinurus (75.4%) (Diniz et al. 2005; Babbucci et al. 2010). Levels of sequence variation (overall h = 0.999; range = 0.988–1.000), and nucleotide diversities (overall π = 0.026; range = 0.026–0.028) were very high (Table 2).
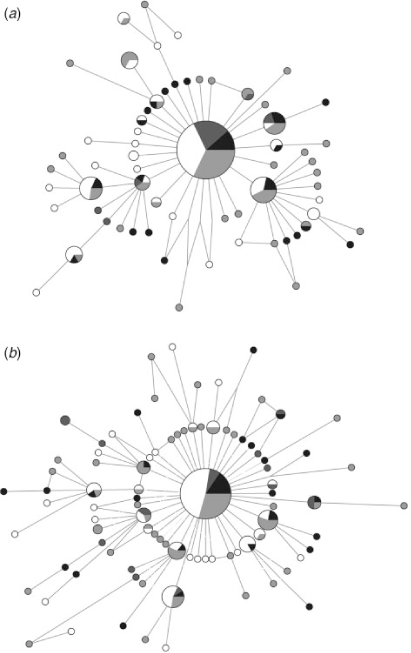
No population differentiation was observed using any of the analytical methods with either marker. The overall ΦST (COI: 0.0004; CR: 0.004) and the pairwise FST values (COI: 0–0.010; CR: 0–0.028) were low and not significant after sequential Bonferroni and False Discovery Rate corrections (P > 0.05) (Table 3). Similarly, the analyses of molecular variance showed that nearly 100% of the variation occurred within localities in all scenarios of population structure simulated (COI: ΦCT = 0.001–0.005, P > 0.05; CR: ΦCT = 0.001–0.029, P > 0.05). Finally, the parsimony median-joining networks were also consistent with very little population differentiation among groups of the four geographic areas. The networks showed a star-like shape, in which most of the unique haplotypes were closely related to the common central haplotype (Fig. 2).
|
|
Neutrality tests and demographic inferences
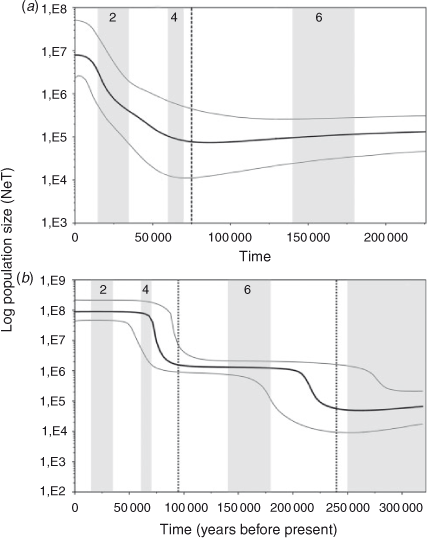
The Tajima’s D (COI: –2.334, P < 0.001; CR: –1.770, P < 0.005) and Fu’s Fs (COI: –27.315, P < 0.001; CR: –23.720, P < 0.001) tests indicate a significant deviation from neutrality for both markers in the overall sample. The deviation is related to a large number of rare haplotypes and can be explained by either selection or demographic factors. Considering the likely neutrality of the variation in the CR and in COI (only three amino acid substitutions), the significant negative values of D and Fs observed in S. brasiliensis are more probably due to population expansion (Aris-Brosou and Excoffier 1996). Significant values were obtained for the R2 statistics (COI: 0.082, P < 0.001; CR: 0.161, P < 0.001). The Bayesian Skyline Plots detected a clear increase in effective population size (Fig. 3). Similarly, the mismatch distributions were compatible with models of demographic and spatial expansion (P(SSD) and P(r) >0.05 for COI and CR under both models) (Fig. 4).
|
The age of the demographic expansion (τ) in mutational units estimated in the mismatch distributions was 2.178 (95% CI = 1.123–3.076) for the COI gene and 20.084 (95% CI = 18.770–20.305) for the control region. For spatial expansion, τ was 2.071 (95% CI = 1.924–2.841) for the COI gene and 20.085 (95% CI = 17.850–20.252) for the control region. The demographic and spatial expansions started at ~88 000 (95% CI = 45 000–124 000) years ago for the COI gene, similar to 71 000 (95% CI = 66 000–72 000) years ago estimated for the control region. These estimates were supported by the Bayesian Skyline Plots, which indicate that the overall population has experienced a period of relatively constant population size, followed by an evident expansion that started ~75 000 years ago (95% HPD = 65 000–100 000) for the COI gene and 95 000 years ago (95% HPD = 85 000–115 000) for the control region (Fig. 3). Furthermore, an older expansion event around 240 000 years ago (95% HPD = 230 000–290 000), was also hypothesised for the control region (Fig. 3).
Discussion
This study is the first to investigate the genetic population structure of a Scyllarides species in the west Atlantic. S. brasiliensis presented very high levels of genetic variation, homogeneously distributed along 2700 km of the South American coast. Bayesian reconstructions of historical demography revealed two events of population expansion: an older one, which started ~240 000 years ago, and a younger expansion that started ~95–70 000 years ago.
Molecular diversity and population genetic differentiation
The high genetic variability observed in S. brasiliensis (COI: h = 0.840, π = 0.004; CR: h = 0.999, π = 0.026) seems to be typical of decapods, since similar results have been reported for lobsters and shrimp using the same mitochondrial markers (e.g. McMillen-Jackson and Bert 2004; Naro-Maciel et al. 2011).
Analyses of the two mitochondrial markers did not reveal any significant genetic structure for S. brasiliensis along the South American coast, suggesting extensive gene flow among localities. The lack of differentiation between populations of S. brasiliensis could be explained on the basis of biological traits that determine the dispersal potential. Population homogeneity is generally expected in marine species with high fecundities and long planktonic phases (Palumbi 2003; Cowen and Sponaugle 2009). In the south-west Atlantic slipper lobster, Scyllarides deceptor, adult females can spawn up to ~190 000 eggs (Oliveira et al. 2008), and in some Scyllarides species the phyllosome larvae can live in the plankton for up to 9 months (Booth et al. 2005). Panmixia has recently been reported for the slipper lobster Scyllarides latus across 3300 km in the north-eastern Atlantic and Mediterranean Sea (Faria et al. 2013). Likewise, high levels of genetic homogeneity have been observed between geographically distant populations of other lobster species. In Panulirus japonicus, for example, no significant genetic differentiation was found (COI sequences) across ~2200 km of the eastern coast of Japan (Inoue et al. 2007). Similarly, no significant differences were observed for the control region of mtDNA among populations of Panulirus inflatus along over 2000 km of the Pacific coast of Mexico (García-Rodríguez and Perez-Enriquez 2008) or among Palinurus gilchristi populations along ~800 km of the southern coast of South Africa (Tolley et al. 2005). Other examples of genetic homogeneity in lobster populations can be found in the literature (Table 1). In most cases, ocean currents and the long larval phase are considered the primary bases for the similarities, and panmixia has been partially attributed to mixing during the planktonic phase (e.g. Tolley et al. 2005; García-Rodríguez and Perez-Enriquez 2006).
Similarly high levels of population homogeneity have also been reported in several fish species and invertebrates with planktonic larval phases along the south-west Atlantic coast (e.g. with Lutjanus purpureus: Gomes et al. 2012; Scomberomorus cavalla: Santa-Brígida et al. 2007; Ocyurus chrysurus: Vasconcellos et al. 2008; Farfantepenaeus brasiliensis and Litopenaeus schmitti: Gusmão et al. 2005; Ucides cordatus: Oliveira-Neto et al. 2007; Cardisoma guanhumi: Oliveira-Neto et al. 2008).
The lack of population structuring observed in S. brasiliensis contrasts with the restricted gene flow reported for Micropogonias furnieri and Holacanthus ciliares from the same area (Puchnick-Legat and Levy 2006; Affonso and Galetti Jr 2007), indicating that the bifurcation of the South-Equatorial Current can have different effects on the genetic structure of marine species, not acting as a barrier to gene flow between localities for S. brasiliensis (Fig. 1). This different phylogeographic pattern could be explained by ecological and biological features of the species. For example, the eggs and larvae of Micropogonias furnieri are subject to estuarine retention, decreasing their dispersal potential (Puchnick-Legat and Levy 2006). The presence of eddies and fronts in the south-west Atlantic also seems not to affect the retention of S. brasiliensis larvae, as observed in Palinurus elephas and P. delagoae (Gopal et al. 2006; Palero et al. 2008; Babbucci et al. 2010). Climate phenomena and oceanographic processes that vary seasonally and yearly are clearly important for larval drift, and partly explain the genetic homogenisation of lobster populations (García-Rodríguez and Perez-Enriquez 2008). For example, the intensification of mesoscale ocean processes associated with ‘El Niño’ events seems to promote larval transport and, hence, connectivity between lobster populations in the tropical Atlantic (Rudorff et al. 2009). Larval dispersal patterns and larval development for many lobster species, including S. brasiliensis, remain poorly understood, but the circulation and the oceanographic processes in the southern Atlantic coupled with the long larval period of slipper lobsters could be an important mechanism for dispersal and larval mixing, allowing gene flow among S. brasiliensis localities along the South American coast.
Demographic history
The lack of population structuring observed in S. brasiliensis could be a result of contemporary gene flow, but it may also reflect historical processes. The star-like haplotype networks obtained (Fig. 2) indicate that a population expansion occurred during the recent history of the species, where the common and widespread haplotype is probably the ancestral condition from which the rare haplotypes were recently derived by point mutations (Slatkin and Hudsont 1991). This result has also been found for other lobsters (e.g. Gopal et al. 2006; García-Rodríguez and Perez-Enriquez 2008; Babbucci et al. 2010).
During the Pleistocene (~2.6 million to 10 000 years ago), climatic fluctuations of the glacial–interglacial cycles (Imbrie et al. 1992) influenced the demographic history and distribution of several marine species (e.g. Lavery et al. 1996; Maggs et al. 2008; Babbucci et al. 2010; Fernández et al. 2011). These climatic oscillations produced changes in sea levels, temperatures, current patterns, upwelling intensity, and coastal habitats (Rohling et al. 1998; Lambeck et al. 2002). The advance and retreat of the ice sheets through multiple glacial cycles, the most recent of which was the last glacial maximum (LGM) ~23 000–18 000 years ago, had a major impact on the present-day species distribution (Provan and Bennett 2008).
The mismatch analysis suggests that both demographic and spatial expansion of S. brasiliensis occurred at ~95 000–70 000 years ago during an interglacial episode of Late Pleistocene related to the marine isotope Stage (MIS) 5 (Selivanov 1992; Gibbard and Cohen 2009). Additionally, the Bayesian Skyline Plot for the control region detected that this population appears to have experienced an older demographic expansion followed by a longer period of relatively constant population size (Fig. 3). The onset of this older expansion event occurred ~240 000 years ago during an interglacial episode in the Middle Pleistocene related to MIS 7 (Selivanov 1992; Gibbard and Cohen 2009). The more recent main glaciations (MIS 2, 4 and 6) appear to have had little impact on population size, given that the population showed nearly constant growth or continuous exponential growth during those periods (Fig. 3). Moreover, given its long population history, pre-LGM coalescence times indicate that S. brasiliensis was not affected by LGM. This trend was also observed in some species of the Mollusca, Arthropoda, Echinodermata and Chordata (Cárdenas et al. 2009; Marko et al. 2010; Ibañez et al. 2012). Due to the uncertainty in mutation rates and previous calculation errors in applying the formula of Rogers and Harpending (t = τ/2u) (Schenekar and Weiss 2011), it is difficult to compare the date of demographic events between lobster studies. However, it is feasible that the demographic expansions for S. brasiliensis and Panulirus argus, both distributed in the west Atlantic, occurred probably during the same period, and that expansions of Palinurus elephas, P. delagoe, P. elephas, P. gilchristi, and Panulirus inflatus were more recent, since the τ value reported for P. argus was similar to that found for S. brasiliensis, but larger than those reported for other species (Table 4).

|
Palaeontological and palynological records, and a large body of biogeographical data, suggest that most of the biota, particularly temperate species, persisted through glacial periods in lower-latitude refugia where climatic conditions were less extreme. They then recolonised other areas when the climate became warmer (Hewitt 1999; Provan and Bennett 2008). Populations inside the refugia thus have a longer demographic history with higher levels of genetic diversity than populations in areas that were recolonised from those refugia (Maggs et al. 2008; Provan and Bennett 2008). This scenario of ‘recolonisation from different refugial areas’ is not apparent for S. brasiliensis given the genetic homogeneity observed along the South America coast.
The similarity between the times estimated for expansion events (demographic and spatial expansion) and the genetic homogeneity suggests that demographic expansion may have occurred simultaneously along the complete range of distribution and was thus associated with a geographical (spatial) expansion. A similar demographic pattern was also observed in the gastropod Concholepas concholepas (Cárdenas et al. 2009) and in the fish Larimichthys polyactis (Wu et al. 2012). The long-distance dispersal during expansion helps preserve high levels of diversity (Fayard et al. 2009). It is possible that the high dispersal potential of planktonic larvae helps this preservation. The patterns that shape the genetic structure of species are complex; nevertheless, the relationship between expansion events and interglacial periods during the late and middle Pleistocene suggests that this species is able to increase its distribution and population size during periods of suitable climate (‘habitat tracking’: Provan and Bennett 2008). The lack of population structure, coupled with recent population expansion, has been observed in several marine crustaceans (e.g. McMillen-Jackson and Bert 2003, 2004; García-Rodríguez and Perez-Enriquez 2008). Nonetheless, further studies of comparative phylogeography and demography are necessary to clarify how the glacial–interglacial cycles during the Pleistocene influenced the demographic history of species in the south-west Atlantic.
The high levels of genetic variation observed indicate that the recent strong exploitation of S. brasiliensis has not had time to produce any detectable effects on the genetic make-up of its population. Furthermore, the genetic homogeneity observed along 2700 km of south-west Atlantic coast suggests extensive gene flow promoted by the high dispersal potential of its larvae. This discovery has important implications for the conservation of this slipper lobster, since it indicates that the whole Brazilian coast could be treated as a single management unit for fishery purposes.
Acknowledgements
The authors thank C. Schrago by invaluable support during the first stages in the Bayesian Skyline Analysis, B. A. Rodríguez for the supporting calculations in the GFAM computer cluster and D. Almeida, F. Azevedo, E. Araujo, L. Barbosa, P. Coelho Filho, P. Cruz, H. Cunha, M.C.O. Matos, N. Leite Jr., S. Lima, F. Magalhães, R. Monteiro, P. Paiva, A. Reis, S. Ribeiro, E. Rios, and E. Routledge for help with sample collections. The authors thank the two anonymous reviewers for providing valuable comments and suggestions. Financial support for this work was provided by Brazilian Ministries for Fisheries and Aquaculture (MPA) and Science, Technology and Innovation (MCTI), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). This work is part of the thesis in Genetics of the first author at the Federal University of Rio de Janeiro (UFRJ).
References
Affonso, P. R. A. M., and Galetti Jr, P. M. (2007). Genetic diversity of three ornamental reef fishes (Families Pomacanthidae and Chaetodontidae) from the Brazilian coast. Brazilian Journal of Biology 67, 925–933.| Genetic diversity of three ornamental reef fishes (Families Pomacanthidae and Chaetodontidae) from the Brazilian coast.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1c7msFWjsg%3D%3D&md5=09b1fde16e76f2eeedc21996cfb68dc5CAS |
Aris-Brosou, S., and Excoffier, L. (1996). The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism. Molecular Biology and Evolution 13, 494–504.
| The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28Xht1Kgurs%3D&md5=9e05f93bd6ef8370942fb05b8fa3dfcdCAS | 8742638PubMed |
Babbucci, M., Buccoli, S., Cau, A., Cannas, R., Goñi, R., Díaz, D., Marcato, S., Zane, L., and Patarnello, T. (2010). Population structure, demographic history, and selective processes: contrasting evidences from mitochondrial and nuclear markers in the European spiny lobster Palinurus elephas (Fabricius, 1787). Molecular Phylogenetics and Evolution 56, 1040–1050.
| Population structure, demographic history, and selective processes: contrasting evidences from mitochondrial and nuclear markers in the European spiny lobster Palinurus elephas (Fabricius, 1787).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXos1CjsrY%3D&md5=409c9c4e82e89be246063d5f5f2da850CAS | 20510378PubMed |
Ball, A. O., Beal, M. G., Chapman, R. W., and Sedberry, G. R. (2007). Population structure of red porgy, Pagrus pagrus, in the Atlantic Ocean. Marine Biology 150, 1321–1332.
| Population structure of red porgy, Pagrus pagrus, in the Atlantic Ocean.Crossref | GoogleScholarGoogle Scholar |
Bandelt, H.-J., Forster, P., and Röhl, A. (1999). Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution 16, 37–48.
| Median-joining networks for inferring intraspecific phylogenies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXjvVGltA%3D%3D&md5=23b8b0385ad557269cf17606563da0c6CAS | 10331250PubMed |
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. Series B. Methodological 57, 289–300.
Booth, J. D., Webber, R., Sekiguchi, H., and Coutures, E. (2005). Diverse larval recruitment strategies within the Scyllaridae. New Zealand Journal of Marine and Freshwater Research 39, 581–592.
| Diverse larval recruitment strategies within the Scyllaridae.Crossref | GoogleScholarGoogle Scholar |
Bowen, B. W., and Grant, W. S. (1997). Phylogeography of the sardines (Sardinops spp.): assessing biogeographic models and population histories in temperate upwelling zones. Evolution 51, 1601–1610.
| Phylogeography of the sardines (Sardinops spp.): assessing biogeographic models and population histories in temperate upwelling zones.Crossref | GoogleScholarGoogle Scholar |
Brasher, D. J., Ovenden, J. R., Booth, J. D., and White, R. W. G. (1992). Genetic subdivision of Australian and New Zealand populations of Jasus verreauxi (Decapoda: Palinuridae) – preliminary evidence from the mitochondrial genome. New Zealand Journal of Marine and Freshwater Research 26, 53–58.
| Genetic subdivision of Australian and New Zealand populations of Jasus verreauxi (Decapoda: Palinuridae) – preliminary evidence from the mitochondrial genome.Crossref | GoogleScholarGoogle Scholar |
Bunje, P. M. E., and Wirth, T. (2008). Inferring patterns of migration. In ‘Bioinformatics. Volume I: Data, Sequence Analysis, and Evolution’. (Ed. J. M. Keith.) pp. 485–506. (Humana Press: Totowa.)
Cárdenas, L., Castilla, J. C., and Viard, F. (2009). A phylogeographical analysis across three biogeographical provinces of the south-eastern Pacific: the case of the marine gastropod Concholepas concholepas. Journal of Biogeography 36, 969–981.
| A phylogeographical analysis across three biogeographical provinces of the south-eastern Pacific: the case of the marine gastropod Concholepas concholepas.Crossref | GoogleScholarGoogle Scholar |
Chow, S., Jeffs, A., Miyake, Y., Konishi, K., Okazaki, M., Suzuki, N., Abdullah, M. F., Imai, H., Wakabayasi, T., and Sakai, M. (2011). Genetic isolation between the western and eastern Pacific populations of pronghorn spiny lobster Panulirus penicillatus. PLoS ONE 6, e29280.
| Genetic isolation between the western and eastern Pacific populations of pronghorn spiny lobster Panulirus penicillatus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xis1ajsQ%3D%3D&md5=082b931650e10ecbe4c8dcd3b674b44eCAS | 22195038PubMed |
Cirano, M., Mata, M. M., Campos, E. J. D., and Deir, N. F. R. (2006). A circulação oceânica de larga-escala na região oeste do Atlântico Sul com base no modelo de circulação global OCCAM. Revista Brasileira de Geofísica 24, 209–230.
| A circulação oceânica de larga-escala na região oeste do Atlântico Sul com base no modelo de circulação global OCCAM.Crossref | GoogleScholarGoogle Scholar |
Coutures, E. (2000). Distribution of phyllosoma larvae of Scyllaridae and Palinuridae (Decapoda: Palinuridea) in the south-western lagoon of New Caledonia. Marine and Freshwater Research 51, 363–369.
| Distribution of phyllosoma larvae of Scyllaridae and Palinuridae (Decapoda: Palinuridea) in the south-western lagoon of New Caledonia.Crossref | GoogleScholarGoogle Scholar |
Cowen, R. K., and Sponaugle, S. (2009). Larval dispersal and marine population connectivity. Annual Review of Marine Science 1, 443–466.
| Larval dispersal and marine population connectivity.Crossref | GoogleScholarGoogle Scholar | 21141044PubMed |
Dall’Occo, P. L., Bento, R. T., and Melo, G. A. (2007). Range extensions for lobsters off the Brazilian coast (Crustacea, Decapoda, Palinura, Astacidea). Biociências (Porto Alegre) 15, 47–52.
Diniz, F. M., Maclean, N., Ogawa, M., Cintra, I. H. A., and Bentzen, P. (2005). The hypervariable domain of the mitochondrial control region in Atlantic spiny lobsters and its potential as a marker for investigating phylogeographic structuring. Marine Biotechnology (New York, N.Y.) 7, 462–473.
| The hypervariable domain of the mitochondrial control region in Atlantic spiny lobsters and its potential as a marker for investigating phylogeographic structuring.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtFGhtLvL&md5=27469d0d8714bfada5a385328ec740aaCAS |
Drummond, A. J., and Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology 7, 214.
| BEAST: Bayesian evolutionary analysis by sampling trees.Crossref | GoogleScholarGoogle Scholar | 17996036PubMed |
Drummond, A. J., Rambaut, A., Shapiro, B., and Pybus, O. G. (2005). Bayesian coalescent inference of past population dynamics from molecular sequences. Molecular Biology and Evolution 22, 1185–1192.
| Bayesian coalescent inference of past population dynamics from molecular sequences.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjvVKitb4%3D&md5=663738ef319a5c64bf57cb828b08ac98CAS | 15703244PubMed |
Duarte, L. F. A., Severino-Rodrigues, E., and Gasalla, M. A. (2010). Slipper lobster (Crustacea, Decapoda, Scyllaridae) fisheries off the southeastern coast of Brazil: I. Exploitation patterns between 23°00′ and 29°65′S. Fisheries Research 102, 141–151.
| Slipper lobster (Crustacea, Decapoda, Scyllaridae) fisheries off the southeastern coast of Brazil: I. Exploitation patterns between 23°00′ and 29°65′S.Crossref | GoogleScholarGoogle Scholar |
Duarte, L. F. A., Severino-Rodrigues, E., and Gasalla, M. A. (2011). Contextualização da pesca mundial de lagostas e características de comercialização de Scyllarides spp. e Panulirus spp. na baixada santista, estado de São Paulo, Brasil. Boletim do Instituto de Pesca, São Paulo 37, 235–246.
Excoffier, L., Laval, G., and Schneider, S. (2005). Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1, 47–50.
| 1:CAS:528:DC%2BD28XjsFSltg%3D%3D&md5=a34b11451ded86925196f3b5d358655bCAS |
Faria, J., Froufe, E., Tuya, F., Alexandrino, P., and Pérez-Losada, M. (2013). Panmixia in the endangered slipper lobster Scyllarides latus from the northeastern Atlantic and western Mediterranean. Journal of Crustacean Biology , .
| Panmixia in the endangered slipper lobster Scyllarides latus from the northeastern Atlantic and western Mediterranean.Crossref | GoogleScholarGoogle Scholar |
Fayard, J., Klein, E. K., and Lefèvre, F. (2009). Long distance dispersal and the fate of a gene from the colonization front. Journal of Evolutionary Biology 22, 2171–2182.
| Long distance dispersal and the fate of a gene from the colonization front.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3c%2FitFOquw%3D%3D&md5=8d75469a897068581a2b9d3db87fb3afCAS | 20069723PubMed |
Fernández, P. J., Alonso, M. A., Sabadin, D. E., Arauz, P. A., and Iudica, C. M. (2011). Phylogeography of weakfish Cynoscion guatucupa (Perciformes: Sciaenidae) from the southwestern Atlantic. Scientia Marina 75, 701–706.
| Phylogeography of weakfish Cynoscion guatucupa (Perciformes: Sciaenidae) from the southwestern Atlantic.Crossref | GoogleScholarGoogle Scholar |
Folmer, O., Black, M., Hoeh, W., Lutz, R., and Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3, 294–299.
| 1:CAS:528:DyaK2MXjt12gtLs%3D&md5=0b1ac07b325bba0e3d8d3405393cf5dfCAS | 7881515PubMed |
Fu, Y. X. (1997). Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147, 915–925.
| 1:STN:280:DyaK2svns1egtQ%3D%3D&md5=6ac6772257390f76d3baecac5ab69118CAS | 9335623PubMed |
Fu, Y. X., and Li, W. H. (1993). Statistical tests of neutrality of mutations. Genetics 133, 693–709.
| 1:STN:280:DyaK3s3gt1Crsw%3D%3D&md5=77600cec2f9da0e6f23d25fb7de6cfb3CAS | 8454210PubMed |
García-Rodríguez, F. J., and Perez-Enriquez, R. (2006). Genetic differentiation of the California spiny lobster Panulirus interruptus (Randall, 1840) along the west coast of the Baja California Peninsula, Mexico. Marine Biology 148, 621–629.
| Genetic differentiation of the California spiny lobster Panulirus interruptus (Randall, 1840) along the west coast of the Baja California Peninsula, Mexico.Crossref | GoogleScholarGoogle Scholar |
García-Rodríguez, F. J., and Perez-Enriquez, R. (2008). Lack of genetic differentiation of blue spiny lobster Panulirus inflatus along the Pacific coast of Mexico inferred from mtDNA sequences. Marine Ecology Progress Series 361, 203–212.
| Lack of genetic differentiation of blue spiny lobster Panulirus inflatus along the Pacific coast of Mexico inferred from mtDNA sequences.Crossref | GoogleScholarGoogle Scholar |
Gibbard, P. L., and Cohen, K. M. (2009). Global chronostratigraphical correlation table for the last 2.7 million years, v. 2009. Subcommission on Quaternary Stratigraphy, International Commission on Stratigraphy, Cambridge.
Gomes, G., Sampaio, I., and Schneider, H. (2012). Population structure of Lutjanus purpureus (Lutjanidae – Perciformes) on the Brazilian coast: further existence evidence of a single species of red snapper in the western Atlantic. Annals of Brazilian Academy of Science 84, 979–999.
| Population structure of Lutjanus purpureus (Lutjanidae – Perciformes) on the Brazilian coast: further existence evidence of a single species of red snapper in the western Atlantic.Crossref | GoogleScholarGoogle Scholar |
Gopal, K. A., Tolley, K., Groeneveld, J., and Matthee, C. A. (2006). Mitochondrial DNA variation in spiny lobster Palinurus delagoae suggests genetically structured populations in the southwestern Indian Ocean. Marine Ecology Progress Series 319, 191–198.
| Mitochondrial DNA variation in spiny lobster Palinurus delagoae suggests genetically structured populations in the southwestern Indian Ocean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xht1enu7%2FP&md5=f404498db629ff0267f5d0de6e9f94caCAS |
Groeneveld, J. C., Goñi, R., and Latrouite, D. (2006). Palinurus species. In ‘Lobsters: Biology, Management, Aquaculture and Fisheries’. (Ed. B. F. Phillips.) pp. 385–411. (Blackwell Publishing: Oxford.)
Gusmão, J., Lazoski, C., and Solé-Cava, A. M. (2005). Population genetic structure of Brazilian shrimp species (Farfantepenaeus sp., F. brasiliensis, F. paulensis and Litopenaeus schmitti: Decapoda: Penaeidae). Genetics and Molecular Biology 28, 165–171.
| Population genetic structure of Brazilian shrimp species (Farfantepenaeus sp., F. brasiliensis, F. paulensis and Litopenaeus schmitti: Decapoda: Penaeidae).Crossref | GoogleScholarGoogle Scholar |
Harpending, H. (1994). Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biology 66, 591–600.
| 1:STN:280:DyaK2cznsV2lsQ%3D%3D&md5=1df4fab78b933fe90f30a16706472957CAS | 8088750PubMed |
Hewitt, G. M. (1999). Post-glacial re-colonization of European biota. Biological Journal of the Linnean Society. Linnean Society of London 68, 87–112.
| Post-glacial re-colonization of European biota.Crossref | GoogleScholarGoogle Scholar |
Holthuis, L. B. (1991). FAO species catalogue. Vol. 13. Marine lobsters of the world. An annotated and illustrated catalogue of species of interest to fisheries known to date. FAO Fisheries Synopsis No. 125, Rome.
Ibañez, C., Argüelles, J., Yamashiro, C., Adasme, L., Céspedes, R., and Poulin, E. (2012). Spatial genetic structure and demographic inference of the Patagonian squid Doryteuthis gahi in the south-eastern Pacific Ocean. Journal of the Marine Biological Association of the United Kingdom 92, 197–203.
Imbrie, J., Boyle, E. A., Clemens, S. C., Duffy, A., Howard, W. R., Kukla, G., Kutzbach, J., Martinson, D. G., McIntyre, A., Mix, A. C., Molfino, B., Morley, J. J., Peterson, L. C., Pisias, N. G., Prell, W. L., Raytoo, M. E., Shackletons, N. J., and Toggweiler, J. R. (1992). On the structure and origin of major glaciation cycles. 1. Linear responses to Milankovitch forcing. Paleoceanography 7, 701–738.
| On the structure and origin of major glaciation cycles. 1. Linear responses to Milankovitch forcing.Crossref | GoogleScholarGoogle Scholar |
Inoue, N., Watanabe, H., Kojima, S., and Sekiguchi, H. (2007). Population structure of Japanese spiny lobster Panulirus japonicus inferred by nucleotide sequence analysis of mitochondrial COI gene. Fisheries Science 73, 550–556.
| Population structure of Japanese spiny lobster Panulirus japonicus inferred by nucleotide sequence analysis of mitochondrial COI gene.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXntlGitrs%3D&md5=10dc89c11d9c5c3f5ac583a9e0e962d2CAS |
Kennington, W. J., Cadee, S. A., Berry, O., Groth, D. M., Johnson, M. S., and Melville-Smith, R. (2013). Maintenance of genetic variation and panmixia in the commercially exploited western rock lobster (Panulirus cygnus). Conservation Genetics 14, 115–124.
| Maintenance of genetic variation and panmixia in the commercially exploited western rock lobster (Panulirus cygnus).Crossref | GoogleScholarGoogle Scholar |
Lambeck, K., Esat, T. M., and Potter, E.-K. (2002). Links between climate and sea levels for the past three million years. Nature 419, 199–206.
| Links between climate and sea levels for the past three million years.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XmvV2qtrc%3D&md5=bd6a91fcc3ed181749edd26fce181011CAS | 12226674PubMed |
Lavery, S., Moritz, C., and Fielder, D. R. (1996). Genetic patterns suggest exponential population growth in a declining species. Molecular Biology and Evolution 13, 1106–1113.
| Genetic patterns suggest exponential population growth in a declining species.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28Xmt1ektr0%3D&md5=b3c1ab6bef4a858f2a3fb4c1325927fdCAS |
Librado, P., and Rozas, J. (2009). DnaSP v. 5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452.
| DnaSP v. 5: a software for comprehensive analysis of DNA polymorphism data.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXmtFeqtr8%3D&md5=91dff07328478a12a2acd6e1fcd3f58aCAS | 19346325PubMed |
Maggs, C. A., Castilho, R., Foltz, D., Henzler, C., Jolly, M. T., Kellt, J., Olsen, J., Perez, K. E., Stam, W., Väinölä, R., Viard, F., and Wares, J. (2008). Evaluating signatures of glacial refugia for north Atlantic benthic marine taxa. Ecology 89, S108–S122.
| Evaluating signatures of glacial refugia for north Atlantic benthic marine taxa.Crossref | GoogleScholarGoogle Scholar | 19097488PubMed |
Marko, P. B., Hoffman, J. M., Emme, S. A., McGovern, T. M., Keever, C. C., and Cox, L. N. (2010). The ‘Expansion–Contraction’ model of Pleistocene biogeography: rocky shores suffer a sea change? Molecular Ecology 19, 146–169.
| The ‘Expansion–Contraction’ model of Pleistocene biogeography: rocky shores suffer a sea change?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXitVSrsLY%3D&md5=6704d812f4513231e34a47ecc93fabc1CAS | 20092033PubMed |
Matthee, C. A., Cockcroft, A. C., Gopal, K., and von der Heyden, S. (2007). Mitochondrial DNA variation of the west-coast rock lobster, Jasus lalandii: marked genetic diversity differences among sampling sites. Marine and Freshwater Research 58, 1130–1135.
| Mitochondrial DNA variation of the west-coast rock lobster, Jasus lalandii: marked genetic diversity differences among sampling sites.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhsVantLbL&md5=50e0538bf2de18c835711ed4e060bc33CAS |
McMillen-Jackson, A. L., and Bert, T. M. (2003). Disparate patterns of population genetic structure and population history in two sympatric penaeid species in the southeastern United States. Molecular Ecology 12, 2895–2905.
| Disparate patterns of population genetic structure and population history in two sympatric penaeid species in the southeastern United States.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD3srltFCitw%3D%3D&md5=ea1ac3ebcc68b65cfd012f400c3e31c2CAS | 14629371PubMed |
McMillen-Jackson, A. L., and Bert, T. M. (2004). Genetic diversity in the mtDNA control region and population structure in the pink shrimp Farfantepenaeus duorarum. Journal of Crustacean Biology 24, 101–109.
| Genetic diversity in the mtDNA control region and population structure in the pink shrimp Farfantepenaeus duorarum.Crossref | GoogleScholarGoogle Scholar |
Miller, S. A., Dykes, D. D., and Polesky, H. F. (1988). A simple salting procedure for extracting DNA from human nucleated cells. Nucleic Acids Research 16, 1215.
| A simple salting procedure for extracting DNA from human nucleated cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1cXhsVKlsrs%3D&md5=b78997f8e44c3b4acff61935a03570a7CAS | 3344216PubMed |
Naro-Maciel, E., Reid, B., Holmes, K. E., Brumbaugh, D. R., Martin, M., and DeSalle, R. (2011). Mitochondrial DNA sequence variation in spiny lobsters: population expansion, panmixia, and divergence. Marine Biology 158, 2027–2041.
| Mitochondrial DNA sequence variation in spiny lobsters: population expansion, panmixia, and divergence.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtVOrtLjM&md5=565a9a3857b31f42b7cc294a7b2e6215CAS |
Oliveira, G., Freire, A. S., and Bertoul, P. R. K. (2008). Reproductive biology of the slipper lobster Scyllarides deceptor (Decapoda: Scyllaridae) along the southern Brazilian coast. Journal of the Marine Biological Association of the United Kingdom 88, 1433–1440.
| Reproductive biology of the slipper lobster Scyllarides deceptor (Decapoda: Scyllaridae) along the southern Brazilian coast.Crossref | GoogleScholarGoogle Scholar |
Oliveira-Neto, J. F., Pie, M. R., Boeger, W. A., Ostrensky, A., and Bagiio, R. A. (2007). Population genetics and evolutionary demography of Ucides cordatus (Decapoda: Ocypodidae). Marine Ecology (Berlin) 28, 460–469.
| Population genetics and evolutionary demography of Ucides cordatus (Decapoda: Ocypodidae).Crossref | GoogleScholarGoogle Scholar |
Oliveira-Neto, J. F., Pie, M. R., Chammas, M. A., Ostrensky, A., and Boeger, W. A. (2008). Phylogeography of the blue land crab, Cardisoma guanhumi (Decapoda: Gecarcinidae) along the Brazilian coast. Journal of the Marine Biological Association of the United Kingdom 88, 1417–1423.
| Phylogeography of the blue land crab, Cardisoma guanhumi (Decapoda: Gecarcinidae) along the Brazilian coast.Crossref | GoogleScholarGoogle Scholar |
Ovenden, J. R., Brasher, D. J., and White, R. W. G. (1992). Mitochondrial DNA analyses of the red rock lobster Jasus edwardsii supports an apparent absence of population subdivision throughout Australasia. Marine Biology 112, 319–326.
| Mitochondrial DNA analyses of the red rock lobster Jasus edwardsii supports an apparent absence of population subdivision throughout Australasia.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK38XhsVyltrY%3D&md5=67a8b034355c599a0e7ca8aa359aed98CAS |
Ovenden, J. R., Booth, J. D., and Smolenski, A. J. (1997). Mitochondrial DNA phylogeny of red and green rock lobsters (genus Jasus). Marine and Freshwater Research 48, 1131–1136.
| Mitochondrial DNA phylogeny of red and green rock lobsters (genus Jasus).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXisV2gsrc%3D&md5=c402727d6aa7d0ec9f9a9805eb5ef69eCAS |
Palero, F., Abelló, P., Macpherson, E., Gristina, M., and Pascual, M. (2008). Phylogeography of the European spiny lobster (Palinurus elephas): influence of current oceanographical features and historical processes. Molecular Phylogenetics and Evolution 48, 708–717.
| Phylogeography of the European spiny lobster (Palinurus elephas): influence of current oceanographical features and historical processes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXptVOhsr8%3D&md5=11ed49ead31fadd5f6fa9ccfe86c4081CAS | 18515152PubMed |
Palumbi, S. R. (2003). Population genetics, demographic connectivity, and the design of marine reserves. Ecological Applications 13, S146–S158.
| Population genetics, demographic connectivity, and the design of marine reserves.Crossref | GoogleScholarGoogle Scholar |
Perez-Enriquez, R., Vega, A., Avila, S., and Sandoval, J. L. (2001). Population genetics of red spiny lobster (Panulirus interruptus) along the Baja California Peninsula, Mexico. Marine and Freshwater Research 52, 1541–1549.
| Population genetics of red spiny lobster (Panulirus interruptus) along the Baja California Peninsula, Mexico.Crossref | GoogleScholarGoogle Scholar |
Peterson, R. G., and Stramma, L. (1991). Upper-level circulation in the south Atlantic Ocean. Progress in Oceanography 26, 1–73.
| Upper-level circulation in the south Atlantic Ocean.Crossref | GoogleScholarGoogle Scholar |
Phillips, B. F., and Melville-Smith, R. (2006). Panulirus species. In ‘Lobsters: Biology, Management, Aquaculture and Fisheries’. (Ed. B. F. Phillips.) pp. 359–384. (Blackwell Publishing: Oxford.)
Pike, N. (2011). Using false discovery rates for multiple comparisons in ecology and evolution. Methods in Ecology and Evolution 2, 278–282.
| Using false discovery rates for multiple comparisons in ecology and evolution.Crossref | GoogleScholarGoogle Scholar |
Provan, J., and Bennett, K. D. (2008). Phylogeographic insights into cryptic glacial refugia. Trends in Ecology & Evolution 23, 564–571.
| Phylogeographic insights into cryptic glacial refugia.Crossref | GoogleScholarGoogle Scholar |
Puchnick-Legat, A., and Levy, J. A. (2006). Genetic structure of Brazilian populations of white mouth croaker Micropogonias furnieri (Perciformes: Sciaenidae). Brazilian Archives of Biology and Technology 49, 429–439.
| Genetic structure of Brazilian populations of white mouth croaker Micropogonias furnieri (Perciformes: Sciaenidae).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xpt1yitbk%3D&md5=cd1d61776242e4652f5f0f522ad3ade6CAS |
Rambaut, A., and Drummond, A. J. (2007). Tracer v1.4. Available at: http://beast.bio.ed.ac.uk/Tracer [accessed 20 January 2012].
Ramos-Onsins, S. E., and Rozas, J. (2002). Statistical properties of new neutrality tests against population growth. Molecular Biology and Evolution 19, 2092–2100.
| Statistical properties of new neutrality tests against population growth.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xps12hsrc%3D&md5=2fd18f760576aeae23e663e063184e7cCAS | 12446801PubMed |
Rice, W. R. (1989). Analyzing tables of statistical tests. Evolution 43, 223–225.
| Analyzing tables of statistical tests.Crossref | GoogleScholarGoogle Scholar |
Rogers, A. R., and Harpending, H. (1992). Population growth makes waves in the distribution of pairwise genetic differences. Molecular Biology and Evolution 9, 552–569.
| 1:STN:280:DyaK383mtFeitA%3D%3D&md5=bbc3f6487ab7cb488a43ab54fd335ce4CAS | 1316531PubMed |
Rohling, E. J., Fenton, M., Jorissen, F. J., Bertrand, P., Ganssen, G., and Caulet, J. P. (1998). Magnitudes of sea-level lowstands of the past 500,000 years. Nature 394, 162–165.
| Magnitudes of sea-level lowstands of the past 500,000 years.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXksFymsr0%3D&md5=912145eeae68cea6593dd380cc0e67adCAS |
Rudorff, C. A. G., Lorenzzetti, J. A., Gherardi, D. F. M., and Lins-Oliveira, J. E. (2009). Application of remote sensing to the study of the pelagic spiny lobster larval transport in the tropical Atlantic. Brazilian Journal of Oceanography 57, 7–16.
| Application of remote sensing to the study of the pelagic spiny lobster larval transport in the tropical Atlantic.Crossref | GoogleScholarGoogle Scholar |
Santa-Brígida, E., Cunha, D. B., Rego, P. S., Sampaio, I., Schneider, H., and Vallinoto, M. (2007). Population analysis of Scomberomorus cavalla (Cuvier, 1829) (Perciformes, Scombridae) from the northern and northeastern coast of Brazil. Brazilian Journal of Biology 67, 919–924.
| Population analysis of Scomberomorus cavalla (Cuvier, 1829) (Perciformes, Scombridae) from the northern and northeastern coast of Brazil.Crossref | GoogleScholarGoogle Scholar |
Santos, M. C. F., and Freitas, A. E. T. S. (2002). Estudo sobre a lagosta sapata Scyllarides brasiliensis Rathbun, 1906 (Crustacea: Decapoda: Scyllaridae) no litoral dos estados de Pernambuco e Alagoas, Brasil. Boletim Técnico Científico do CEPENE 10, 123–143.
Schenekar, T., and Weiss, S. (2011). High rate of calculation errors in mismatch distribution analysis results in numerous false inferences of biological importance. Heredity 107, 511–512.
| High rate of calculation errors in mismatch distribution analysis results in numerous false inferences of biological importance.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC3MbpslGgug%3D%3D&md5=a5463df82f0b2755e91086e941f6af4bCAS | 21731052PubMed |
Schubart, C. D., Diesel, R., and Hedges, B. (1998). Rapid evolution to terrestrial life in Jamaican crabs. Nature 393, 363–365.
| Rapid evolution to terrestrial life in Jamaican crabs.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXjsFKis78%3D&md5=aa0f66fcf190d4cb26386ea0f01acc17CAS |
Sekiguchi, H., Booth, J. D., and Webber, R. (2007). Early life histories of slipper lobsters. In ‘The Biology and Fisheries of the Slipper Lobster’. (Eds K. L. Lavalli and E. Spanier.) pp. 69–90. (CRC Press: Florida.)
Selivanov, A. O. (1992). Spatial-temporal analysis of large Pleistocene sea-level fluctuations: marine terrace data. Journal of Coastal Research 8, 408–418.
Shanks, A. L., Grantham, B. A., and Carr, M. H. (2003). Propagule dispersal distance and the size and spacing of marine reserves. Ecological Applications 13, S159–S169.
| Propagule dispersal distance and the size and spacing of marine reserves.Crossref | GoogleScholarGoogle Scholar |
Silberman, J. D., Sarver, S. K., and Walsh, P. J. (1994). Mitochondrial DNA variation and population structure in the spiny lobster Panulirus argus. Marine Biology 120, 601–608.
| Mitochondrial DNA variation and population structure in the spiny lobster Panulirus argus.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXjsVGju7k%3D&md5=abb36ef5790de5344a51d05e113fe5f7CAS |
Slatkin, M., and Hudsont, R. R. (1991). Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129, 555–562.
| 1:CAS:528:DyaK38Xhs12mtr8%3D&md5=a8d3e0ed519be8e6f1a942a0bc8066a2CAS | 1743491PubMed |
Spanier, E., and Lavalli, K. L. (2006). Scyllarides species. In ‘Lobsters: Biology, Management, Aquaculture and Fisheries’. (Ed. B. F. Phillips.) pp. 462–496. (Blackwell Publishing: Oxford.)
Spanier, E., and Lavalli, K. L. (2007). Slipper lobster fisheries – present status and future perspectives. In ‘The Biology and Fisheries of the Slipper Lobster’. (Eds K. L. Lavalli and E. Spanier.) pp. 377–391. (CRC Press: Florida.)
Stramma, L., and England, M. (1999). On the water masses and mean circulation of the south Atlantic Ocean. Journal of Geophysical Research 104, 20863–20883.
| On the water masses and mean circulation of the south Atlantic Ocean.Crossref | GoogleScholarGoogle Scholar |
Tajima, M. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595.
| 1:CAS:528:DyaK3cXhslentA%3D%3D&md5=3195a8d501393c0ff6c382d33172974cCAS |
Tajima, M. (1996). The amount of DNA polymorphism maintained in a finite population when the neutral mutation rate varies among sites. Genetics 143, 1457–1465.
| 1:STN:280:DyaK28vgtV2guw%3D%3D&md5=5e3ed3674c974de27eb4b23798c2b7e3CAS |
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28, 2731–2739.
| MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1eiu73K&md5=4feabfd2916eb73d10bf98384eecf75bCAS | 21546353PubMed |
Taylor, M., and Hellberg, M. (2003). Genetic evidence for local retention of pelagic larvae in a Caribbean reef fish. Science 299, 107–109.
| Genetic evidence for local retention of pelagic larvae in a Caribbean reef fish.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XpvVensL0%3D&md5=2357789114ab50162fe4df90034e3862CAS | 12511651PubMed |
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680.
| CLUSTAL-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXitlSgu74%3D&md5=9d02f84fd1683b4c4ca4923b268e3185CAS | 7984417PubMed |
Thompson, A. P., Hanley, J. R., and Johnson, M. S. (1996). Genetic structure of the western rock lobster, Panulirus cygnus, with the benefit of hindsight. Marine and Freshwater Research 47, 889–896.
| Genetic structure of the western rock lobster, Panulirus cygnus, with the benefit of hindsight.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XntF2lurk%3D&md5=da674c962636923dddc49d9a2d69b136CAS |
Thorpe, J. P., Solé-Cava, A. M., and Watts, P. C. (2000). Exploited marine invertebrates: genetics and fisheries. Hydrobiologia 420, 165–184.
| Exploited marine invertebrates: genetics and fisheries.Crossref | GoogleScholarGoogle Scholar |
Tolley, K. A., Groeneveld, J. C., Gopal, K., and Matthee, C. A. (2005). Mitochondrial DNA panmixia in spiny lobster Palinurus gilchristi suggests a population expansion. Marine Ecology Progress Series 297, 225–231.
| Mitochondrial DNA panmixia in spiny lobster Palinurus gilchristi suggests a population expansion.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtFeltrrM&md5=1ca0a6f9f465dc25443958d43af3e6adCAS |
Tourinho, J. L., Solé-Cava, A. M., and Lazoski, C. (2012). Cryptic species within the commercially most important lobster in the tropical Atlantic, the spiny lobster Panulirus argus. Marine Biology 159, 1897–1906.
| Cryptic species within the commercially most important lobster in the tropical Atlantic, the spiny lobster Panulirus argus.Crossref | GoogleScholarGoogle Scholar |
Vasconcellos, A., Vianna, P., Paiva, P. C., Schama, R., and Solé-Cava, A. M. (2008). Genetic and morphometric differences between yellowtail snapper (Ocyurus chrysurus, Lutjanidae) populations of the tropical west Atlantic. Genetics and Molecular Biology 31, 308–316.
| Genetic and morphometric differences between yellowtail snapper (Ocyurus chrysurus, Lutjanidae) populations of the tropical west Atlantic.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXptlWrs78%3D&md5=1a870f508a053a48f84bed03c2f2354cCAS |
von der Heyden, S., Groeneveld, J. C., and Matthee, C. A. (2007). Long current to nowhere? Genetic connectivity of Jasus tristani populations in the southern Atlantic Ocean. African Journal of Marine Science 29, 491–497.
| Long current to nowhere? Genetic connectivity of Jasus tristani populations in the southern Atlantic Ocean.Crossref | GoogleScholarGoogle Scholar |
Ward, R. D. (2002). Genetics of fish populations. In ‘Handbook of Fish Biology and Fisheries’. (Eds P. J. B. Hart and J. D. Reynolds.) pp. 200–224. (Blackwell Publishing: Oxford.)
Wu, R., Liu, S., Zhuang, Z., Su, Y., and Tang, Q. (2012). Population genetic structure and demographic history of small yellow croaker, Larimichthys polyactis (Bleeker, 1877), from coastal waters of China. African Journal of Biotechnology 11, 12500–12509.
| 1:CAS:528:DC%2BC38XhtFOlsrnJ&md5=61b4c6d767d827b859e631701fdf0846CAS |



