Rhizobial genetic and genomic resources for sustainable agriculture
MacLean G. Kohlmeier A * , Graham W. O’Hara
A * , Graham W. O’Hara  A , Joshua P. Ramsay
A , Joshua P. Ramsay  B and Jason J. Terpolilli
B and Jason J. Terpolilli  A
A
A
B

MacLean Kohlmeier received his PhD from the University of Manitoba in 2020 with Prof. Ivan Oresnik. He is currently a Postdoctoral Research Fellow in Microbial Genomics in the Legume Rhizobium Sciences group at Murdoch University. His research interests include gene transfer events between soil bacteria, as well as carbon metabolism and transport, and how these features affect symbiotic establishment and efficiency. |

Graham O’Hara is Director of Murdoch University’s Legume Rhizobium Sciences centre in the Food Futures Institute, and co-curator of the International Legume Inoculant Genebank (ILIG). He has worked extensively in national and international projects on applied nitrogen fixation leading to the development and dissemination of inoculant rhizobia strains. His research interest is focussed on the soil ecology, mineral nutrition, physiology and stress tolerance of rhizobia, and how this affects their performance as inoculants in the field. |

Joshua Ramsay carried out his PhD in 2004–2008 at the University of Otago with Prof. Clive Ronson. He was then a University of Cambridge Herchel Smith Postdoctoral Fellow from 2008 to 2011 and a Health Sciences Career Development Fellow at the University of Otago. Ramsay started his own lab at Curtin University in 2013. Dr Ramsay’s investigations explore the effects of mobile genetic elements in both health related and agricultural contexts. In 2018, Dr Ramsay was awarded an ARC Future Fellowship and he is currently an Associate Professor in the Curtin Medical School and Curtin Health Innovation Research Institute. |

Jason Terpolilli is Research Director of Murdoch University’s Legume Rhizobium Sciences centre in the Food Futures Institute, and co-curator of ILIG. He works closely with colleagues across Australia on several projects aimed at increasing the application of effective nitrogen-fixing rhizobia for grain and pasture legumes. His main areas of research interest are in rhizobia genetics and biochemistry and in applying genomics to develop robust approaches to studying the evolution of these organisms and their effect on farming systems in the field. |
Abstract
Rhizobia are a diverse group of α- and β-proteobacteria that boost soil fertility by forming a nitrogen-fixing symbiosis with legumes, which is why legumes are grown in rotation with cereals in agriculture. Rhizobia that naturally populate Australian soils are largely incompatible with exotic agricultural legumes, therefore, compatible strains have been imported from all over the world for use as inoculants. An amalgamated collection of these strains, called the International Legume Inoculant Genebank (ILIG), has been established at Murdoch University, to provide a centralised strain storage facility and support rhizobial research and inoculant development (see http://ilig.murdoch.edu.au). The ILIG contains 11,558 strains representing 96 bacterial species from 778 legume species collected from >1200 locations across 100 countries. New and sometimes inefficient rhizobia evolve in the field following legume inoculation, through horizontal symbiosis gene transfer from inoculants to soil bacteria. To provide a benchmark to monitor and assess the impact of this evolution, all commercial Australian inoculant strains were genome sequenced and these data made available (PRJNA783123, see https://www.ncbi.nlm.nih.gov/bioproject/PRJNA783123/). These data, and the further sequencing of the >11,000 historical strains in the ILIG, will increase our understanding of rhizobial evolution and diversity and provide the backbone for efforts to safeguard Australia’s legume inoculation program.
Keywords: horizontal gene transfer, inoculant, legume, rhizobia, sequencing, symbiosis.
The nitrogen-fixing symbiosis between diverse soil saprophytic bacteria known as rhizobia (or root nodule bacteria) and legumes is responsible for providing a substantial proportion of the biosphere’s available nitrogen.1 This symbiosis is established when rhizobia infect legume roots, differentiating into their nitrogen-fixing (or bacteroid) form, within plant-derived root nodules.2 Bacteroids reduce dinitrogen (N2) gas into ammonia (NH3), secreting this source of fixed nitrogen to the host plant, in exchange for a supply of carbon from the legume. When the plant senesces, some of this fixed nitrogen makes its way into the soil, providing a source of bioavailable nitrogen for subsequent plants. For this reason, legumes are often grown in rotation with cereals in agriculture.3–5 Symbiotic nitrogen fixation through grain legumes (e.g. chickpea, lupin, peas) and pasture legumes (e.g. clover, lucerne, serradella) is estimated to save the Australian agricultural sector A$4 billion in synthetic fertiliser costs annually, and reduce the CO2 footprint associated with fertiliser production and application.6
Although bacteria in the genus Rhizobium are well known as legume symbionts, this capability is much more phylogenetically diverse, with 15 genera of α-proteobacteria (including Rhizobium) and 3 genera of β-proteobacteria engaging in legume symbioses.7 Close to 100 species of legumes are cultivated across Australia, and, although only a small proportion of these are widely grown, their requirement for a compatible symbiotic partner can be quite specific.8,9 To maximise the agricultural benefits of symbiotic nitrogen fixation, legumes are often inoculated with elite strains of rhizobia that specifically nodulate the target legume and are highly efficient nitrogen fixers10–12 (Fig. 1). This is of primary importance in Australia, as our soils tend to lack rhizobia that nodulate and efficiently fix nitrogen with the exotic legumes we use in agriculture.
Benefits of inoculation with Rhizobium sp. strain WSM4643. (a) Field plot comparison of uninoculated vetch (Vicia sativa) (left plot) and inoculated plants (right plot). (b) Comparison of nodulation and plant growth responses in uninoculated field pea (Piscum sativum) (left) with inoculated plant (right). Photos courtesy of Dr Ron Yates (Department of Primary Industries and Regional Development).

Since the 1950s in Australia, thousands of strains of rhizobia have been collected from legumes and soils from all over the world by various University and government agencies and stored in small, dispersed collections. To preserve and develop this genetically diverse resource critical for inoculum development, the International Legume Inoculant Genebank (ILIG) was established in 2020 at Murdoch University in Western Australia, through the financial support of the Grains Research and Development Corporation (GRDC) of Australia. The ILIG has amalgamated strain collections from all over Australia into one facility, which currently houses 11,558 strains of rhizobia representing 96 different bacterial species, isolated from 778 species of legumes that were collected from more than 1200 locations throughout 100 countries. The ILIG has an online catalogue (see http://ilig.murdoch.edu.au) to browse available information on strains within the collection and request strains of interest for research purposes. Critically, as the majority of the strains in the ILIG were collected prior to the genome sequencing era, much of their genetic potential remains unknown.
Rhizobia show a remarkable diversity in genome size and complexity. In comparison to enteric bacteria, rhizobial genomes are relatively large, ranging in size from 6 Mb to more than 10 Mb, and they can be structurally complex, consisting of a chromosome with or without multiple additional plasmids.2 These plasmids can be very large, such as in Sinorhizobium meliloti 1021, which carries 1.3- and 1.7-Mb plasmids, along with its 3.7-Mb chromosome.13–15 Genes essential to the establishment and maintenance of symbiosis (i.e. nod or nodulation, and nif and fix or nitrogen fixation) are typically found on plasmids (e.g. Rhizobium and Sinorhizobium) or chromosomally on symbiosis islands (e.g. Mesorhizobium and Bradyrhizobium) and are generally considered to be part of the accessory genome.16
For symbiotic Mesorhizobium spp., chromosomal symbiosis genes are encoded on mobile integrative and conjugative elements (ICEs). These symbiosis ICEs (previously referred to as symbiosis islands) were discovered when nodule isolates from the pasture legume Lotus corniculatus (birds foot trefoil) were found to have evolved through the acquisition of a 508-kb region from an introduced inoculant strain in New Zealand.17 Subsequent work showed that the symbiosis ICE excises from the chromosome of a donor cell, transferring by conjugation to a recipient, and integrating into the recipient chromosome in a site-specific manner.18 ICE transfer is a complex process that is epigenetically activated and controlled by quorum sensing.19,20 Horizontal transfer of related symbiosis ICEs has been documented for Mesorhizobium in Australian soils, including inoculants used for the pasture legume Biserrula pelecinus21 and grain legume Cicer arietinum (chickpea).22 These studies showed that some symbiosis ICEs are tripartite in structure, whereby the ICE exists as three separate regions of DNA dispersed across the chromosome. These three regions recombine into a singular circular DNA molecule immediately prior to conjugal transfer.23,24 Although Australian soils seem to lack native Mesorhizobium strains compatible with introduced legumes, they do harbour an abundance of non-symbiotic Mesorhizobium spp. capable of acquiring symbiosis ICEs and gaining the ability to nodulate legumes.25 Critically, however, ICE transfer recipients are not always effective symbionts,21,26,27 so transfer events have the potential to create strains less efficient at nitrogen fixation that can out-compete the inoculant strain for nodulation, leading to a reduced benefit of legume inoculation.
Accurately following and assessing the impact of symbiosis gene transfer in agriculture is only possible with knowledge of the inoculant genome sequence, which has not been historically available for the majority of commercially available strains of rhizobia in Australia. To close this gap, a genome sequencing pipeline was established to provide closed genomes of all commercial inoculant strains. Long-read assemblies were first created using the Oxford Nanopore Technologies MinION Mk1B, which were then polished with short-read sequences obtained from the same DNA using an Illumina NextSeq. 500 (Fig. 2). The commercial legume inoculants were sourced from the 2019 Mother Culture maintained by the Australian Inoculants Research Group (AIRG), which supplies manufacturers with a standardised and quality-controlled source of rhizobia for the inoculant industry. Sequencing from this source, rather than historical collections of these inoculant strains, ensures that the acquired genomic data for each strain directly relates to the germplasm supplied to growers.
Genome sequencing pipeline. The pipeline consists of six steps: (1) isolation of genomic DNA, (2) sequencing with MinION Mk1B, (3) base calling or raw data conversion to sequence data, (4) read assembly, (5) incorporation of short-read sequence for polishing and error correction, and (6) gene assignment and annotation. Created with BioRender.com.
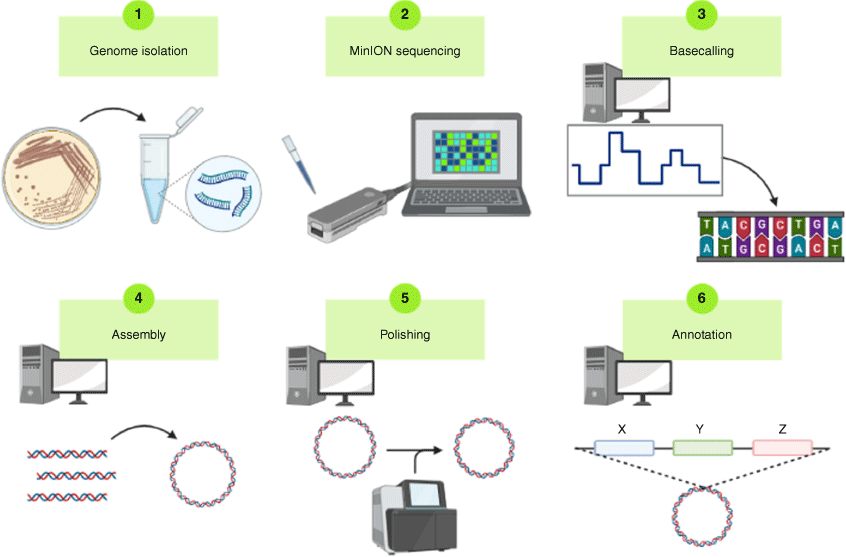
With a breadth of strains spanning five genera, the diversity of strains used as inoculants in Australia complicates genome sequencing efforts, in that many have very different growth requirements and phenotypic characteristics.28 Depending on the genus of the inoculant, different media types and extraction techniques were required for growth and isolation of genomic DNA. For many strains, thorough washing of the samples was necessary to limit carryover of cell lysate contaminants that could inhibit the sequencing reaction.
Closed genomes were completed for 38 strains, which are now available online under National Center for Biotechnology Information (NCBI) BioProject accession number PRJNA783123 (see https://www.ncbi.nlm.nih.gov/bioproject/PRJNA783123/). These genome sequences have in the first instance been used to assign more appropriate taxonomy to these strains, which is a critical first step in building a clear picture of the genetic diversity of the Australian inoculant cohort. However, many strains could not be given a designation at the species level due to a lack of closely related type strains. This reflects both the inherently novel biology of many of the inoculant organisms and the paucity of accurate genomic information currently available for many species of rhizobia. More sequencing of diverse rhizobia isolates from a range of representative legumes and biogeographical locations is therefore needed to address this deficit. Nevertheless, most inoculants showed genome architectures that were consistent with other members of their respective genera, although Rhizobium sp. SRDI969 was unusual in that it carried chromosomally encoded symbiosis genes, notable because Rhizobium spp., unlike Bradyrhizobium spp. or Mesorhizobium spp., typically carry symbiosis genes on plasmids.29 Rhizobium sp. SRDI969 is new strain for faba bean (Vicia faba), which, alongside the new pea (Pisum sativum), lentil (Lens culinaris) and vetch (Vicia spp.) strain Rhizobium sp. WSM4643, is being released for the 2024 growing season as inoculants in Australia for these important grain legumes. This marks the first time in inoculant history that novel strains are to be released to commerce alongside their complete genome sequence. This will allow more-accurate and longer-term monitoring of inoculant efficacy and should become standard practice to also ensure the catalogue of the genetic material released as inoculants remains up to date.
This ILIG strain collection and inoculant genome sequences are invaluable resources for plant and microbe research in Australia. The ILIG collection, with its thousands of diverse root-nodule bacteria collected from around the world, will serve as the backbone for future research and development of novel inoculants for the benefit of Australian agriculture, and act as a secure repository for new isolates. However, with no genomic data on most of the historical strains in the collection, the full scope of the genetic potential of this collection is unclear. Future work will sequence the historical germplasm in the ILIG. These data will help in the development of rapid and accurate methods to monitor inoculation success, as well as identify and follow the impact of inoculant symbiosis gene transfer events on nitrogen fixation. It will also provide us with a better understanding of the dual saprophytic and symbiotic lifestyle of rhizobia and the evolutionary forces that have shaped this fascinatingly diverse group of bacteria.
Data availability
Genome sequence data of Australian commercial inoculant strains sequenced in this project has been deposited in NCBI under BioProject accession number PRJNA783123 (see https://www.ncbi.nlm.nih.gov/bioproject/PRJNA783123/).
Declaration of funding
The establishment and maintenance of the International Legume Inoculant Genebank (ILIG) at Murdoch University was supported by funding from the Grains Research and Development Corporation of Australia (Grant UMU1810-001RTX). J. P. Ramsay is the recipient of an Australian Research Council Future Fellowship (Project ID FT170100235) funded by the Australian Government.
References
1 Fowler D et al. (2013) The global nitrogen cycle in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 368, 20130164.
| Crossref | Google Scholar | PubMed |
2 Poole P et al. (2018) Rhizobia: from saprophytes to endosymbionts. Nat Rev Microbiol 16, 291-303.
| Crossref | Google Scholar | PubMed |
3 Lötjönen S, Ollikainen M (2017) Does crop rotation with legumes provide an efficient means to reduce nutrient loads and GHG emissions? Rev Agric Food Environ Stud 98, 283-312.
| Crossref | Google Scholar |
4 Smith CJ, Chalk PM (2020) Grain legumes in crop rotations under low and variable rainfall: are observed short-term N benefits sustainable? Plant Soil 453, 271-279.
| Crossref | Google Scholar |
5 Xing H et al. (2017) Incorporating grain legumes in cereal-based cropping systems to improve profitability in southern New South Wales, Australia. Agric Syst 154, 112-123.
| Crossref | Google Scholar |
8 Wang D et al. (2012) Symbiosis specificity in the legume – rhizobial mutualism. Cell Microbiol 14, 334-342.
| Crossref | Google Scholar | PubMed |
9 Allito BB et al. (2021) Legume-rhizobium specificity effect on nodulation, biomass production and partitioning of faba bean (Vicia faba L.). Sci Rep 11, 3678.
| Crossref | Google Scholar | PubMed |
10 Corbin E et al. (1977) Nodulation studies on chickpea (Cicer arietinum). Aust J Exp Agric 17, 126-134.
| Crossref | Google Scholar |
11 Howieson JG et al. (1988) Selection for acid tolerance in Rhizobium meliloti. Plant Soil 105, 179-188.
| Crossref | Google Scholar |
12 Howieson J et al. (1995) Biserrula pelecinus L.—a legume pasture species with potential for acid, duplex soils which is nodulated by unique root-nodule bacteria. Aust J Agric Res 46, 997-1009.
| Crossref | Google Scholar |
13 Finan TM et al. (2001) The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc Natl Acad Sci USA 98, 9889-9894.
| Crossref | Google Scholar | PubMed |
14 Galibert F et al. (2001) The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293, 668-672.
| Crossref | Google Scholar | PubMed |
15 Capela D et al. (2001) Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc Natl Acad Sci USA 98, 9877-9882.
| Crossref | Google Scholar | PubMed |
16 Remigi P et al. (2016) Symbiosis within symbiosis: evolving nitrogen-fixing legume symbionts. Trends Microbiol 24, 63-75.
| Crossref | Google Scholar | PubMed |
17 Sullivan JT et al. (1995) Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc Natl Acad Sci USA 92, 8985-8989.
| Crossref | Google Scholar | PubMed |
18 Ramsay JP et al. (2006) Excision and transfer of the Mesorhizobium loti R7A symbiosis island requires an integrase IntS, a novel recombination directionality factor RdfS, and a putative relaxase RlxS. Mol Microbiol 62, 723-734.
| Crossref | Google Scholar | PubMed |
19 Ramsay JP et al. (2022) An epigenetic switch activates bacterial quorum sensing and horizontal transfer of an integrative and conjugative element. Nucleic Acids Res 50, 975-988.
| Crossref | Google Scholar | PubMed |
20 Jowsey WJ et al. (2023) DUF2285 is a novel helix-turn-helix domain variant that orchestrates both activation and antiactivation of conjugative element transfer in proteobacteria. Nucleic Acids Res 51, 6841-6856.
| Crossref | Google Scholar | PubMed |
21 Nandasena KG et al. (2007) In situ lateral transfer of symbiosis islands results in rapid evolution of diverse competitive strains of mesorhizobia suboptimal in symbiotic nitrogen fixation on the pasture legume Biserrula pelecinus L. Environ Microbiol 9, 2496-2511.
| Crossref | Google Scholar | PubMed |
22 Hill Y et al. (2021) Evolution of diverse effective N2-fixing microsymbionts of Cicer arietinum following horizontal transfer of the Mesorhizobium ciceri CC1192 symbiosis integrative and conjugative element. Appl Environ Microbiol 87, e02558-20.
| Crossref | Google Scholar | PubMed |
23 Haskett TL et al. (2016) Assembly and transfer of tripartite integrative and conjugative genetic elements. Proc Natl Acad Sci USA 113, 12268-12273.
| Crossref | Google Scholar | PubMed |
24 Haskett TL et al. (2018) Sequential induction of three recombination directionality factors directs assembly of tripartite integrative and conjugative elements. PLoS Genet 14, e1007292.
| Crossref | Google Scholar | PubMed |
25 Colombi E et al. (2023) Population genomics of Australian indigenous Mesorhizobium reveals diverse nonsymbiotic genospecies capable of nitrogen-fixing symbioses following horizontal gene transfer. Microb Genom 9, 000918.
| Crossref | Google Scholar | PubMed |
26 Nandasena KG et al. (2006) Rapid in situ evolution of nodulating strains for Biserrula pelecinus L. through lateral transfer of a symbiosis island from the original mesorhizobial inoculant. Appl Environ Microbiol 72, 7365-7367.
| Crossref | Google Scholar | PubMed |
27 Elias NV, Herridge DF (2015) Naturalised populations of mesorhizobia in chickpea (Cicer arietinum L.) cropping soils: effects on nodule occupancy and productivity of commercial chickpea. Plant Soil 387, 233-249.
| Crossref | Google Scholar |
28 Howieson JG, Dilworth MJ (eds) (2016) Working with rhizobia. ACIAR Monograph Number 173. Australian Centre for International Agricultural Research, Canberra, ACT, Australia. https://www.aciar.gov.au/sites/default/files/legacy/aciar_mn_173_web-updated_31_may_2016.pdf
29 Kohlmeier MG et al. (2023) Complete genome sequence of Rhizobium leguminosarum bv. viciae SRDI969, an acid-tolerant, efficient N2-fixing microsymbiont of Vicia faba. Microbiol Resour Announc 12, e00489-00423.
| Crossref | Google Scholar | PubMed |
 MacLean Kohlmeier received his PhD from the University of Manitoba in 2020 with Prof. Ivan Oresnik. He is currently a Postdoctoral Research Fellow in Microbial Genomics in the Legume Rhizobium Sciences group at Murdoch University. His research interests include gene transfer events between soil bacteria, as well as carbon metabolism and transport, and how these features affect symbiotic establishment and efficiency. |
 Graham O’Hara is Director of Murdoch University’s Legume Rhizobium Sciences centre in the Food Futures Institute, and co-curator of the International Legume Inoculant Genebank (ILIG). He has worked extensively in national and international projects on applied nitrogen fixation leading to the development and dissemination of inoculant rhizobia strains. His research interest is focussed on the soil ecology, mineral nutrition, physiology and stress tolerance of rhizobia, and how this affects their performance as inoculants in the field. |
 Joshua Ramsay carried out his PhD in 2004–2008 at the University of Otago with Prof. Clive Ronson. He was then a University of Cambridge Herchel Smith Postdoctoral Fellow from 2008 to 2011 and a Health Sciences Career Development Fellow at the University of Otago. Ramsay started his own lab at Curtin University in 2013. Dr Ramsay’s investigations explore the effects of mobile genetic elements in both health related and agricultural contexts. In 2018, Dr Ramsay was awarded an ARC Future Fellowship and he is currently an Associate Professor in the Curtin Medical School and Curtin Health Innovation Research Institute. |
 Jason Terpolilli is Research Director of Murdoch University’s Legume Rhizobium Sciences centre in the Food Futures Institute, and co-curator of ILIG. He works closely with colleagues across Australia on several projects aimed at increasing the application of effective nitrogen-fixing rhizobia for grain and pasture legumes. His main areas of research interest are in rhizobia genetics and biochemistry and in applying genomics to develop robust approaches to studying the evolution of these organisms and their effect on farming systems in the field. |


