Rotavirus surveillance informs diarrhoea disease burden in the WHO Western-Pacific region
Celeste M Donato A B C F , Sarah Thomas A , Sokoveti Covea D , Felisita T Ratu D , Aalisha Sahu Khan D , Eric Rafai D and Julie E Bines A B EA Enteric Diseases Group, Murdoch Children’s Research Institute, Parkville, Vic., Australia
B Department of Paediatrics, The University of Melbourne, Parkville, Vic., Australia
C Biomedicine Discovery Institute and Department of Microbiology, Monash University, Clayton, Vic., Australia
D Ministry of Health and Medical Services, Suva, Fiji
E Department of Gastroenterology and Clinical Nutrition, Royal Children’s Hospital, Parkville, Vic., Australia
F Email: celeste.donato@mcri.edu.au
Microbiology Australia 42(4) 161-164 https://doi.org/10.1071/MA21046
Submitted: 27 August 2021 Accepted: 13 October 2021 Published: 8 November 2021
Journal Compilation © The Authors 2021 Open Access CC BY-NC-ND, published (by CSIRO Publishing) on behalf of the ASM
Abstract
The surveillance of enteric pathogens is critical in assessing the burden of diarrhoeal disease and informing vaccine programs. Surveillance supported by the World Health Organization in Fiji, Vietnam, the Lao People’s Democratic Republic, and the Philippines previously focussed on rotavirus. There is potential to expand surveillance to encompass a variety of enteric pathogens to inform vaccine development for norovirus, enterotoxigenic Escherichia coli and Shigella.
Rotavirus: the predominant enteric pathogen in children
Group A rotaviruses remain the predominant aetiological agent associated with acute gastroenteritis in young children worldwide, estimated to have caused 128 500 deaths and 258 173 300 episodes of diarrhoea among children <5 years of age in 20161. Four group A rotavirus vaccines; Rotarix® (GlaxoSmithKline, Rixenstart, Belgium), Rotasiil® (Serum Institute of India, Pune, India), RotaTeq® (Merck & Co, Pennsylvania, USA) and Rotavac® (Bharat Biotech, Hyderabad, India) have been prequalified by the World Health Organization (WHO) for global use and have been included in the National Immunisation Programs of over 112 countries worldwide2. In the WHO designated Western-Pacific region, some countries have introduced rotavirus vaccines into their national immunisation programs (Fiji, Australia, New Zealand, Kiribati, Micronesia, Solomon Islands, Marshall Islands, Japan, Niue, and Palau), but there is limited access to rotavirus vaccines across other countries in the region3.
The genotyping of rotavirus strains underpins national and global surveillance efforts. The binomial classification is based on the outer capsid proteins VP7 and VP4, which define G and P genotypes, respectively and these proteins elicit neutralising antibodies. The most prevalent genotypes in humans are G1P[8], G2P[4], G3P[8], G9P[8] and G12P[8]4,5. Several genotypes have emerged in recent years to become globally important, highlighting the need for continued surveillance efforts. Monitoring rotavirus genotypes provides critical information regarding ongoing effectiveness of implemented vaccination programs, informs planned vaccine introduction, and can assist in outbreak investigation. Given the substantial diversity and evolution of rotavirus strains, surveillance also provides fundamental information on the changing diversity of rotavirus strains in the vaccine era and may provide insight into the emergence of vaccine-escape variants.
Rotavirus surveillance in the Western-Pacific region
Many countries in the region do not have formal rotavirus surveillance programs. The Enteric Diseases Group at the Murdoch Children’s Research Institute hosts the WHO Collaborating Centre for Child Health, the WHO Rotavirus Regional Reference Laboratory (RRL) for the Western-Pacific Region and the Australian Rotavirus Surveillance Program. The WHO Rotavirus RRL provides support for both national and reference laboratories in the Western-Pacific region in the laboratory diagnosis of rotavirus infection. The Enteric Diseases Group provides training and conducts surveillance of rotavirus genotypes in the Western-Pacific region, with a focus on Fiji, Viet Nam, and the Philippines. In 2017, rotavirus positivity among children aged <5 years hospitalised with diarrhoea was 48% in the Philippines and 43% in Viet Nam, compared with 14% in Fiji, highlighting the effectiveness of vaccine introduction in decreasing the burden of disease6. The RRL also supports rotavirus outbreak responses for the region (Solomon Islands7 and Kiribati8) and assists in surveillance activities in Papua New Guinea9. There are several limitations associated with the surveillance program as a limited number of sites in each country participate and not all children admitted to hospital with diarrhoea have a stool sample collected and sent for analysis.
Rotavirus surveillance in Fiji
Fiji has participated in the WHO Global Rotavirus Surveillance Program since 2006, monitoring the rotavirus disease burden and characterising the diversity of genotypes circulating in the population. Notably, Fiji is the only Pacific nation that participates in the WHO surveillance program. Stool samples are routinely collected from children aged <5 years hospitalised with acute, non-bloody diarrhoea. The participating hospitals service the two main islands: the Colonial War Memorial Hospital on Viti Levu and the Savusavu District Hospital on Vanua Levu. Samples are sent to the Fiji Centre for Communicable Disease Control in Suva for rotavirus antigen testing. Rotavirus positive samples are then sent to the WHO RRL for confirmation of rotavirus positivity and subsequent genotyping. In-depth descriptions of rotavirus surveillance in Fiji have been previously described10–12.
We have recently completed an analysis of longitudinal surveillance data generated between 2005 and 2018 which has allowed for a detailed description of rotavirus genotype diversity in Fiji, to characterise any changes in genotype patterns that may have occurred following vaccine introduction in 201210. This study was the first to describe rotavirus genotype patterns following vaccine introduction in a low- or middle-income country in the Western-Pacific region.
Prior to vaccine introduction, rotavirus was a leading cause of diarrhoea-related hospitalisations detected in 52% (2006) and 60% (2007) of children aged <5 years hospitalised with acute diarrhoea11. Due to this substantial burden of rotavirus disease, Fiji introduced Rotarix into the National Immunisation Program in October 2012. Vaccine coverage in eligible infants reached 85% by 2013 and 99% from 2014 onwards13. Vaccine introduction has resulted in an 82% reduction in rotavirus diarrhoea-related hospitalisations in children aged <5 years12.
Surveillance in Fiji prior to Rotarix introduction (2005–2012) revealed G1P[8], G2P[4] and G3P[8] were the predominant genotypes detected in children aged <5 years (Figure 1). Genotype dominance fluctuated annually with G3P[8] dominant in 2006 (94%) and 2009 (59%), G2P[4] dominant in 2008 (40%) and 2010 (74%), and G1P[8] dominant in 2011 (85%) and 2012 (67%) (Figure 1). No clear dominant genotype could be determined in 2005 and 2007 due to low sample numbers. Other genotypes including G2P[8], G3P[6], G8P[8], G9P[8] and G12P[4] were infrequently detected in the pre-vaccine period10 (Figure 1).
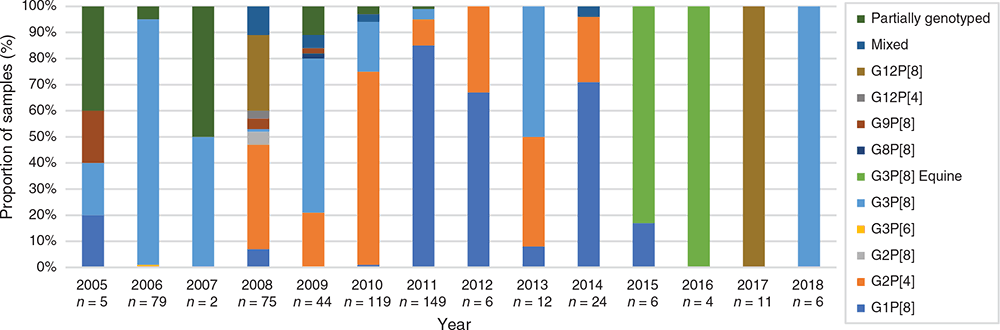
|
Due to the substantial decrease in rotavirus disease following vaccine introduction in Fiji, there was a dramatic decrease in the number of samples available for genotype analysis – reflecting the success of the vaccination program. This does not represent a bias in sample collection, rather it reflects the limited cases of rotavirus in Fiji. The limited samples available for analysis does impact the ability to make a strong inference about genotype dominance. The genotype dominance continued to fluctuate annually, with G3P[8] dominant in 2013 (50%), G1P[8] in 2014 (71%), G12P[8] in 2017 (100%) and G3P[8] in 2018 (100%) (Figure 1). Overall, the diversity of genotypes decreased following vaccine introduction with some genotypes no longer detected. Fewer genotypes were observed to co-circulate each year and one of the most interesting findings was the later absence of previously dominant genotypes. Despite accounting for a similar proportion of genotyped samples in the pre- and post-vaccine eras (28% vs 30%); G1P[8] strains were not detected after 2015 (Figs 1, 2). The identification of G2P[4] decreased from 30% to 17% following vaccine introduction and were not detected in Fiji after 2014 (Figs 1, 2). This disappearance of G1P[8] and G2P[4] in Fiji following vaccine introduction is in contrast to Australia where G1P[8] continued to be detected at reduced levels following vaccine introduction, and G2P[4] prevalence increased in regions using Rotarix14. In recent years, G2P[4] has also increased in other countries within the region, independent of whether the country had a rotavirus vaccination program or not; including the Philippines15, Cambodia and Lao PDR16,17. G3P[8] also decreased from 28% to 19% of genotyped samples following vaccine introduction (Figure 2). G3P[8] strains were not detected for four years in Fiji before re-emerging in 2018 as the dominant genotype. G3P[8] also remerged in Australia, Cambodia and Mongolia in recent years16,18,19. G12P[8] strains were also not detected in Fiji for a period of 8 years before re-emerging as the only genotype identified in 2017. G12P[8] accounted for a higher proportion of genotyped samples in the vaccine era; increasing from 6% to 17% (Figure 2). Australia and New Zealand have also reported the dominance of G12P[8] following vaccine introduction14,20,21.
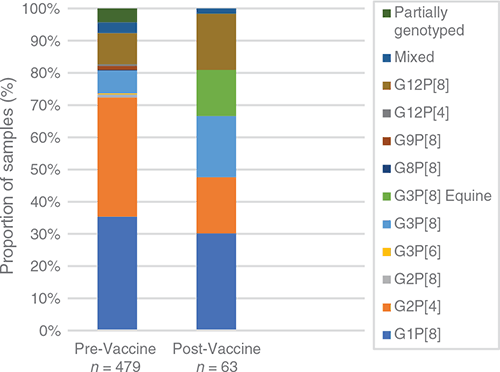
|
Another genotype that exhibited a transient circulation period in Fiji was the equine-like G3P8 variant. This variant emerged in 2013 and continues to be detected as a dominant genotype in many countries in the region regardless of vaccination22,23. The equine-like G3P[8] variant was dominant in Fiji for two consecutive years (2015–2016), accounting for 83% and 100% of samples genotyped respectively, but was not detected during surveillance in 2017 or 2018.
The sporadic detection of genotypes in the vaccine era likely represents that the vaccinated paediatric population in Fiji no longer sustains the wide transmission of rotavirus and that the intermittent emergence of genotypes is associated with the introduction of these variants from outside Fiji, followed by limited local circulation.
Expansion of enteric pathogen surveillance in the region
The Enteric Diseases Group is also part of the Global Paediatric Diarrhea Surveillance Study (GPDS) which leverage the existing Global Rotavirus Surveillance Network to screen for a broad range of enteropathogens in children aged <5 years hospitalised with diarrhoea in Fiji, Viet Nam and Lao PDR utilising quantitative PCR via TACMan Array cards. This program aims to give a greater understanding of the burden of these targeted enteropathogens, ultimately aiding vaccine development for norovirus, enterotoxigenic Escherichia coli and Shigella. Based on 2017 surveillance data, the most prevalent enteric pathogen in Viet Nam and Lao PDR continues to be rotavirus followed by norovirus GII and sapovirus, compared to Fiji where Shigella is now most prevalent, followed by rotavirus6.
Conclusions
Rotavirus surveillance provides insights into vaccine performance, informing vaccine introduction in neighbouring countries. Surveillance of other enteric pathogens is key to understand the burden of diarrhoeal disease and inform vaccine development strategies.
Conflicts of interest
Celeste Donato has served on vaccine advisory boards for GSK (2019, 2021), all payments were paid directly to an administrative fund held by Murdoch Children’s Research Institute. All other authors declare no conflicts of interest.
Declaration of funding
Rotavirus Surveillance activities of the WHO Regional Reference Laboratory and the Global Paediatric Diarrhoea Surveillance Study is funded by a grant from WHO.
Acknowledgements
We gratefully acknowledge the contributions of the personnel from each participating surveillance site, in particular the contributions of Fiona M. Russell to rotavirus surveillance efforts in Fiji, and Varja Grabovac from the World Health Organization Western Pacific Regional Office.
References
[1] Troeger, C. et al. (2018) Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. JAMA Pediatr. 172, 958–965.| Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years.Crossref | GoogleScholarGoogle Scholar | 30105384PubMed |
[2] Burke, R.M. et al. (2019) Current and new rotavirus vaccines. Curr. Opin. Infect. Dis. 32, 435–444.
| Current and new rotavirus vaccines.Crossref | GoogleScholarGoogle Scholar | 31305493PubMed |
[3] International Vaccine Access Center (2021) VIEW-hub. Johns Hopkins Bloomberg School of Public Health. https://view-hub.org (accessed 20 August 2021).
[4] Bányai, K. et al. (2012) Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine 30, A122–A130.
| Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs.Crossref | GoogleScholarGoogle Scholar | 22520121PubMed |
[5] Dóró, R. et al. (2014) Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: is there evidence of strain selection from vaccine pressure? Infect. Genet. Evol. 28, 446–461.
| Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: is there evidence of strain selection from vaccine pressure?Crossref | GoogleScholarGoogle Scholar | 25224179PubMed |
[6] Cohen, A. (2018) Global Rotavirus Surveillance Network (GRSN) and Global Pediatric Diarrhea Surveillance (GPDS) Preliminary Results. Global Rotavirus and Pediatric Diarrhea Surveillance Meeting. Cape Town, South Africa. November 2018
[7] Jones, F.K. et al. (2016) Increased rotavirus prevalence in diarrheal outbreak precipitated by localized flooding, Solomon Islands, 2014. Emerg. Infect. Dis. 22, 875–879.
| Increased rotavirus prevalence in diarrheal outbreak precipitated by localized flooding, Solomon Islands, 2014.Crossref | GoogleScholarGoogle Scholar | 27088272PubMed |
[8] Tabunga, T. et al. (2014) Response to a large rotavirus outbreak on South Tarawa, Kiribati, 2013. Western Pac. Surveill. Response J. 5, 9–14.
| Response to a large rotavirus outbreak on South Tarawa, Kiribati, 2013.Crossref | GoogleScholarGoogle Scholar | 25077032PubMed |
[9] Horwood, P.F. et al. (2012) Surveillance and molecular characterization of group A rotaviruses in Goroka, Papua New Guinea. Am. J. Trop. Med. Hyg. 87, 1145–1148.
| Surveillance and molecular characterization of group A rotaviruses in Goroka, Papua New Guinea.Crossref | GoogleScholarGoogle Scholar | 23128293PubMed |
[10] Thomas, S. et al. (2021) Genotype Diversity before and after the introduction of a rotavirus vaccine into the National Immunisation Program in Fiji. Pathogens 10, 358.
| Genotype Diversity before and after the introduction of a rotavirus vaccine into the National Immunisation Program in Fiji.Crossref | GoogleScholarGoogle Scholar | 33802966PubMed |
[11] Jenney, A. et al. (2009) The burden of hospitalised rotavirus infections in Fiji. Vaccine 27, F108–F111.
| The burden of hospitalised rotavirus infections in Fiji.Crossref | GoogleScholarGoogle Scholar | 19931707PubMed |
[12] Jenney, A.W.J. et al. (2021) The impact of the rotavirus vaccine on diarrhoea, five years following national introduction in Fiji. Lancet Reg. Health West. Pac. 6, .
| The impact of the rotavirus vaccine on diarrhoea, five years following national introduction in Fiji.Crossref | GoogleScholarGoogle Scholar |
[13] World Health Organization (2020) WHO and UNICEF Estimates of National Immunization Coverage. https://www.who.int/immunization/monitoring_surveillance/data/fji.pdf (accessed 14 January 2020).
[14] Roczo-Farkas, S. et al. (2018) The impact of rotavirus vaccines on genotype diversity: a comprehensive analysis of 2 decades of Australian surveillance data. J. Infect. Dis. 218, 546–554.
| The impact of rotavirus vaccines on genotype diversity: a comprehensive analysis of 2 decades of Australian surveillance data.Crossref | GoogleScholarGoogle Scholar | 29790933PubMed |
[15] Yamamoto, D. et al. (2017) First detection of DS-1-like G1P[8] human rotavirus strains from children with diarrhoea in the Philippines. New Microbes New Infect. 18, 54–57.
| First detection of DS-1-like G1P[8] human rotavirus strains from children with diarrhoea in the Philippines.Crossref | GoogleScholarGoogle Scholar | 28626585PubMed |
[16] Angkeabos, N. et al. (2018) Pediatric hospitalizations attributable to rotavirus gastroenteritis among Cambodian children: seven years of active surveillance, 2010–2016. Vaccine 36, 7856–7861.
| Pediatric hospitalizations attributable to rotavirus gastroenteritis among Cambodian children: seven years of active surveillance, 2010–2016.Crossref | GoogleScholarGoogle Scholar | 29588120PubMed |
[17] SoukAloun, D. (2018) Rotavirus diarrhea in hospitalized children under 5 years of age in Vientiane, Lao PDR, 2009–2015. Vaccine 36, 7878–7882.
| Rotavirus diarrhea in hospitalized children under 5 years of age in Vientiane, Lao PDR, 2009–2015.Crossref | GoogleScholarGoogle Scholar | 29739715PubMed |
[18] Samdan, A. et al. (2018) Hospital-based surveillance for rotavirus diarrhea in Ulaanbaatar, Mongolia, April 2009 through March 2016. Vaccine 36, 7883–7887.
| Hospital-based surveillance for rotavirus diarrhea in Ulaanbaatar, Mongolia, April 2009 through March 2016.Crossref | GoogleScholarGoogle Scholar | 29429811PubMed |
[19] Thomas, S. et al. (2021) Australian Rotavirus Surveillance Program: annual report, 2019. Commun. Dis. Intell. (2018) 45, .
| Australian Rotavirus Surveillance Program: annual report, 2019.Crossref | GoogleScholarGoogle Scholar |
[20] Health Intelligence Team, Health and Environment Group (2019) Rotavirus in New Zealand, 2016. https://surv.esr.cri.nz/PDF_surveillance/Rotavirus/2016Rotavirus.pdf (accessed 20 August 2021).
[21] Health Intelligence Team, Health and Environment Group (2017) Rotavirus in New Zealand, 2015. https://surv.esr.cri.nz/PDF_surveillance/Rotavirus/2015Rotavirus.pdf (accessed 20 August 2021).
[22] Cowley, D. et al. (2016) Emergence of a novel equine-like G3P[8] inter-genogroup reassortant rotavirus strain associated with gastroenteritis in Australian children. J. Gen. Virol. 97, 403–410.
| Emergence of a novel equine-like G3P[8] inter-genogroup reassortant rotavirus strain associated with gastroenteritis in Australian children.Crossref | GoogleScholarGoogle Scholar | 26588920PubMed |
[23] Tacharoenmuang, R. et al. (2020) High prevalence of equine-like G3P[8] rotavirus in children and adults with acute gastroenteritis in Thailand. J. Med. Virol. 92, 174–186.
| High prevalence of equine-like G3P[8] rotavirus in children and adults with acute gastroenteritis in Thailand.Crossref | GoogleScholarGoogle Scholar | 31498444PubMed |


