One Health: the global challenge of Clostridium difficile infection
Su-Chen Lim A , Thomas V Riley A B C D and Daniel R Knight B EA School of Medical and Health Sciences, Edith Cowan University, Joondalup, WA, Australia
B Medical, Molecular and Forensic Sciences, Murdoch University, Murdoch, WA, Australia
C School of Biomedical Sciences, The University of Western Australia, Nedlands, WA, Australia
D PathWest Laboratory Medicine, Department of Microbiology, Nedlands, WA, Australia
E Corresponding author. Tel: +61 8 6457 4092, Email: daniel.knight@murdoch.edu.au
Microbiology Australia 41(1) 23-27 https://doi.org/10.1071/MA20007
Published: 28 February 2020
The One Health concept recognises that the health of humans is interconnected to the health of animals and the environment. It encourages multidisciplinary communication and collaboration with the aim of enhancing surveillance and research and developing integrative policy frameworks. Clostridium difficile (also known as Clostridioides difficile) infection (CDI) has long been viewed as a hospital-associated (HA) enteric disease mainly linked to the use of broad-spectrum antimicrobials that cause dysbiosis in the gut and loss of ‘colonisation resistance’. However, since the early 2000s, the rate of community-associated CDI (CA-CDI) has increased to ~15% in Europe, ~30% in Australia and ~40% in the USA in populations often without obvious risk factors. Since the 1990s, it has become apparent that food animals are now a major reservoir and amplification host for C. difficile, including lineages of clinical importance. Cephalosporin antimicrobials, to which C. difficile is intrinsically resistant, were licensed for animal use in North America in 1990. By the second decade of the 21st century, there were reports of C. difficile contamination of food and the environment in general. Using whole-genome sequencing (WGS) and high-resolution typing, C. difficile isolates from humans, animals, food and the environment were proven to be genetically closely related and, in some cases, indistinguishable. This suggests possible zoonoses and/or anthroponoses, with contaminated food and the environment acting as the conduit for transmission between animals and humans. This paper summarises the key evidence that demonstrates the One Health importance of C. difficile.
The role of asymptomatic carriers in the spread of C. difficile
In 2013, the landmark study of Eyre et al.1 provided the first compelling evidence that asymptomatic carriers, and possibly other unknown sources external to the healthcare setting, were playing a major role in C. difficile transmission. The study, conducted in Oxfordshire in the United Kingdom used WGS and core-genome single nucleotide variant (cgSNV) analysis to examine C. difficile strains from 957 hospital- and community-identified CDI cases collected between 2008 and 20111. The authors found only 333 isolates (35%) had a clonal relationship (≤2 SNVs difference) with isolates of the same PCR ribotype (RT) from other CDI cases, and 428 isolates (45%) were genetically distinct with >10 SNVs difference in their core-genomes. Of the 333 cases with evidence of clonal transmission, only 126 (38%) had close hospital contact with another CDI patient and 120 (36%) had no plausible epidemiological link to a patient in the hospital system or community.
In early 2019, Sheth et al.2 and Halstead et al.3 presented more evidence for asymptomatic carriers playing a part in the dissemination of C. difficile in hospitals. In both studies, asymptomatic patients were screened for C. difficile on admission and isolates were compared to isolates from symptomatic CDI patients using Multi-Locus Variable Number Tandem Repeat Analysis (MLVA) or cgSNV analysis. Of the 10–15% asymptomatic patients that tested positive for C. difficile, >80% were colonised by toxigenic C. difficile strains capable of causing disease2,3. These studies revealed C. difficile transmission from asymptomatic patients to previously C. difficile-negative patients2, and clustering of asymptomatic patients with symptomatic CDI patients3, supporting the idea that asymptomatic carriers are spreading C. difficile. Furthermore, in a separate study by Gonzalez-Orta et al.4, 27% of HA-CDI cases in Cleveland, USA, were infected with strains that the patients were previously colonised with on admission. This suggests that they were not true HA cases and that C. difficile was likely acquired in the community, with disease manifesting only after admission to hospital. With continuous importation of C. difficile into the hospital setting via asymptomatic carriers, community reservoirs are undoubtedly playing a much bigger role in the transmission of CDI than previously thought and the incidence of CA-CDI might have been grossly underestimated using the current guidelines5.
Community reservoirs
To date, C. difficile has been isolated from diverse array of sources/reservoirs including food animals (pigs, cattle, sheep and poultry), meat (veal, beef, pork, lamb, chicken and turkey), seafood (clams, salmon, shrimp and mussels), vegetables (lettuce, pea sprouts, ginger, carrots, potatoes and salad), the household environment (toilets, floors, bathroom sinks and soles of shoes) and the natural environment (rivers, lakes and soil)6. In summary, food animals, retail food and the environment are important reservoirs of C. difficile. The average prevalence of C. difficile in neonatal animals is always high, ranging from ∼20% in calves to ∼70% in piglets6. C. difficile prevalence as high as 42% in retail meat has been reported in the USA7; however, European studies reported a much lower figure of ∼3%8, possibly related to differences in slaughtering practices. Meanwhile, the prevalence of C. difficile in natural environments such as soil and water averages ∼30%6. The most common strains identified in these studies are C. difficile RT 014, belonging to multi-locus sequence types (MLSTs [STs]) 2, 13 and 49, and ST11 RTs 078, 126, 127 and 033. All these strains are toxigenic, associated with human CDI6, well established in multiple animal and environmental sources and invariably resistant to numerous antimicrobials used in human and veterinary medicine9,10. This further demonstrates the relevance of C. difficile to the One Health concept, i.e. there are three independent yet convergent problems that require an integrative solution: a human health issue, an animal health issue and an environmental issue.
Long-range interspecies transmission of C. difficile
To date, there has been no incontrovertible proof of foodborne or environmental transmission of C. difficile. Such proof remains elusive given C. difficile is not a typical foodborne or enteric pathogen: (1) not all individuals exposed to C. difficile will develop symptoms (depending on the vulnerability of their gut microbiota); (2) C. difficile is ubiquitous in the environment; and (3) the usual rules for source attribution are often not obeyed11. Nevertheless, largely due to the advent of microbial genomics, there is now ample evidence that: (1) C. difficile common to humans and production animals share a recent evolutionary history; and (2) CDI has a substantial zoonotic component which results in the spillover of C. difficile into retail food and the environment. Building on their earlier work showing clonal transmission of C. difficile between a pig and a pig farmer12, Knetsch et al.13 sequenced 247 C. difficile RT 078 strains from diverse sources in 22 countries across four continents (North America, Europe, Australia and Asia). Core-genome analysis revealed extensive clustering of human and animal strains, evidence of potential bidirectional spread of C. difficile between farm animals and humans. There was limited geographical clustering with clones of C. difficile RT 078 spread across towns, countries and continents. One clonal group of RT 078 showed intercontinental transmission between an animal in Canada and humans in the United Kingdom. Another study10, this time focusing on a global population of ST11, corroborated the findings of Knetsch et al.13 revealing a globally disseminated network of C. difficile ST11 clones (of RTs 078, 126, 127, 033 and 288) with the propensity for reciprocal zoonotic and/or anthroponotic transmission. Tetracycline use in agriculture and animal husbandry is widespread and its inappropriate use in the latter is well recognised. Dingle et al.14 found tetracycline selection to be a key driver of C. difficile RT 078 evolution, with multiple independent tetM-associated clonal expansions of this lineage occurring around the year 2000. Further supporting an agricultural focus for C. difficile RT 078, the evolutionary origins of these different tetracycline resistance elements were Tn916-like elements (which are capable of inter-species transfer) from established zoonotic species including Streptococcus suis, Enterococcus faecalis and Escherichia coli.
RT 014 is the most successful C. difficile lineage worldwide. In Australia, this RT 014 is well established in humans with CDI and pigs, accounting for around 30% and 25% of isolates, respectively15–17. Knight et al.9 sequenced a contemporaneous collection of C. difficile RT 014 strains of human and porcine origin and cgSNV analysis revealed recent interspecies transmission, with 42% of human isolates having a clonal relationship with at least one animal isolate. Again, these clones were isolated months and thousands of kilometres apart across different States of Australia. Thus, it is unlikely that there was any direct contact between the animals and humans, however, it appears that C. difficile frequently moves between food animals and humans and that the zoonotic spread is not confined to any geographical region or local population. This strongly suggests an interconnected long-range zoonotic and/or anthroponotic transmission pathway involving recycled waste-products such as manure, biosolids and compost which could result in contaminated crops and/or widespread dissemination of C. difficile in the environment. Indeed, studies have shown that retail meat, vegetables, compost, public lawn, household environment and companion animals are reservoirs of clinically important and often antimicrobial-resistant (AMR) C. difficile lineages, including RT 0146. This is also in agreement with a WGS study involving 482 European hospitals which revealed no within-country clustering for RTs 078, 015, 002, 014 and 020, consistent with Europe-wide dissemination18.
Transmission cycle
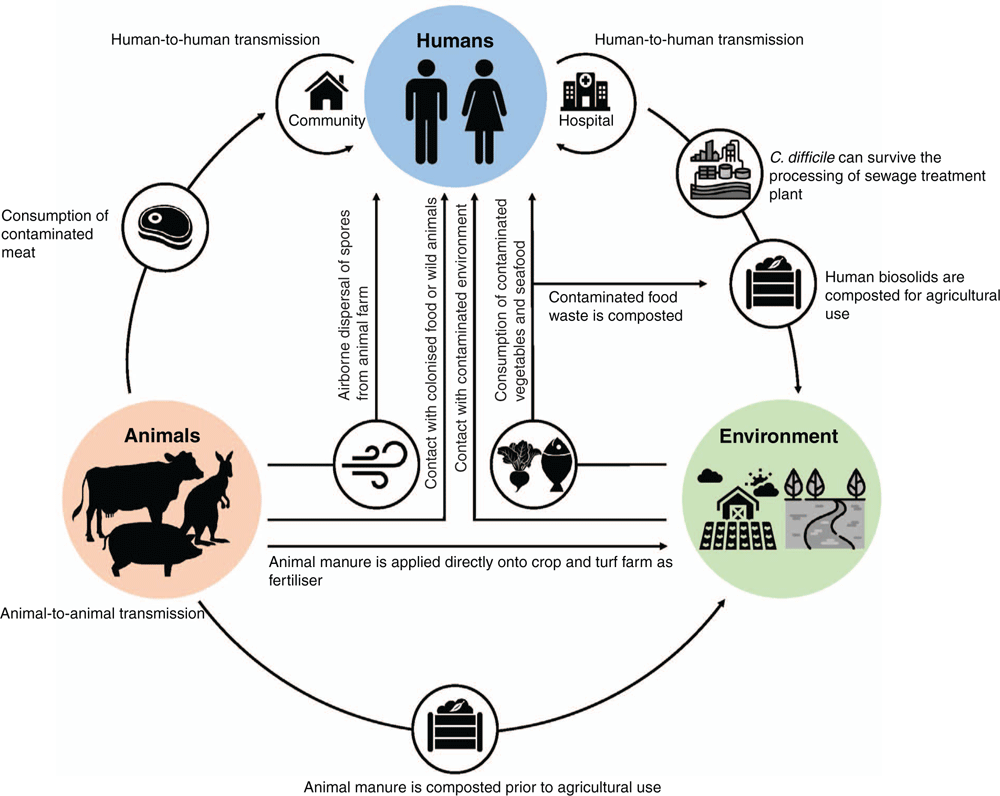
How does C. difficile spread between food animals and humans with limited geographical clustering? The principal amplification hosts of C. difficile are animals, both human and non-human. C. difficile from food animals can contaminate meat during processing at the slaughterhouse and survive up to the point of human consumption as C. difficile spores can endure the recommended cooking temperature for meat (71°C) for over 2 h19 as well as freezing, chilling and disinfection processes20,21. C. difficile spores can also disseminate in the air, in hospitals22 and animal production facilities23. Transmission by invertebrate vectors also occurs. Depending on local agricultural practices and policies, manure from food animals can either be composted or applied directly onto farmland as fertiliser resulting in contamination of the farming environment. Even if the manure is composted, complete elimination of C. difficile spores is unlikely; ∼60% of composted products such as garden mixes and mulches are contaminated with C. difficile (Lim et al. unpublished). Contaminated food waste can also be composted for use in gardening and landscaping. C. difficile can survive the process of sewage treatment24 and release of treated sewage effluent can impact rivers and marine life25. While direct zoonotic transfer of C. difficile between pig farmers and pigs has been reported12, for the general public indirect zoonotic transmission through food and the environment is more likely. With C. difficile being so widely disseminated in the community, household environments and companion animals are also being contaminated/colonised with C. difficile26, providing yet another route for CDI transmission. Figure 1 shows the major reservoirs and known transmission pathways of CDI27.
Conclusions
In summary, C. difficile is a pathogen with substantial community reservoirs and evidence of long-range interspecies transmission. This appears to be a recent (in the past 50 years) event, likely linked to anthropomorphic factors such as high-intensity animal husbandry, international travel and trade and, most critically, injudicious antimicrobial usage in farm animals. High-resolution One Health-focused surveillance of C. difficile from diverse human, animal and environmental sources will continue to be critical to the development of a better understanding of the epidemiological and genetic factors contributing to the emergence, evolution and spread of CDI. The control of CDI is currently focused on antimicrobial stewardship and infection control around CDI patients in the hospital setting. With the new knowledge of asymptomatic carriers spreading C. difficile in hospitals, early screening and isolating C. difficile carriers on hospital admission could help prevent HA-CDI as suggested in a recent Canadian study28, which saw the incidence of HA-CDI decrease significantly from 6.9 to 3.0 per 10 000 patient-days, a 62.4% reduction in expected CDI cases. However, if we are to make meaningful interventions which impact upon both human and animal health, it is imperative that we move beyond the hospital setting and foster collaborative relationships between industry, government, veterinarians, clinicians and researchers. Enhanced antimicrobial stewardship in both human and veterinary settings is crucial but a more productive approach in reducing CDI would be to minimise the environmental burden of C. difficile by reducing carriage/infection in both animals and humans with a vaccine. Despite the recent demise of the Sanofi C. difficile vaccine program29, several other contenders remain in the pipeline including Pfizer who are currently conducting a phase III trial of a vaccine based on genetically and chemically detoxified toxins A and B30. In addition, a vaccine that offers protection against both CDI and colonisation (via mucosal antibodies to reduce the adhesion of C. difficile to mucus-producing intestinal cells) is currently being tested by GSK in a phase I clinical trial31. C. difficile is already considered a critical AMR pathogen by US Centers for Disease Control and Prevention32 and should also be recognised as the most significant One Health problem in the world today.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This paper did not receive any specific funding.
References
[1] Eyre, D.W. et al. (2013) Diverse sources of C. difficile infection identified on whole-genome sequencing. N. Engl. J. Med. 369, 1195–1205.| Diverse sources of C. difficile infection identified on whole-genome sequencing.Crossref | GoogleScholarGoogle Scholar | 24066741PubMed |
[2] Sheth, P.M. et al. (2019) Evidence of transmission of Clostridium difficile in asymptomatic patients following admission screening in a tertiary care hospital. PLoS One 14, e0207138.
| Evidence of transmission of Clostridium difficile in asymptomatic patients following admission screening in a tertiary care hospital.Crossref | GoogleScholarGoogle Scholar | 31287834PubMed |
[3] Halstead, F.D. et al. (2019) Whole genome sequencing of toxigenic Clostridium difficile in asymptomatic carriers: insights into possible role in transmission. J. Hosp. Infect. 102, 125–134.
| Whole genome sequencing of toxigenic Clostridium difficile in asymptomatic carriers: insights into possible role in transmission.Crossref | GoogleScholarGoogle Scholar | 30359648PubMed |
[4] Gonzalez-Orta, M. et al. (2019) Are many patients diagnosed with healthcare-associated Clostridioides difficile infection colonized with the infecting strain on admission? Clin. Infect. Dis. 69, 1801–1804.
| Are many patients diagnosed with healthcare-associated Clostridioides difficile infection colonized with the infecting strain on admission?Crossref | GoogleScholarGoogle Scholar | 30855075PubMed |
[5] McDonald, L.C. et al. (2007) Recommendations for surveillance of Clostridium difficile-associated disease. Infect. Control Hosp. Epidemiol. 28, 140–145.
| Recommendations for surveillance of Clostridium difficile-associated disease.Crossref | GoogleScholarGoogle Scholar | 17265394PubMed |
[6] Knight, D.R. and Riley, T.V. (2019) Genomic delineation of zoonotic origins of Clostridium difficile. Front. Public Health 7, 164.
| Genomic delineation of zoonotic origins of Clostridium difficile.Crossref | GoogleScholarGoogle Scholar | 31281807PubMed |
[7] Songer, J.G. et al. (2009) Clostridium difficile in retail meat products, USA, 2007. Emerg. Infect. Dis. 15, 819–821.
| Clostridium difficile in retail meat products, USA, 2007.Crossref | GoogleScholarGoogle Scholar | 19402980PubMed |
[8] Candel-Pérez, C. et al. (2019) A review of Clostridioides [Clostridium] difficile occurrence through the food chain. Food Microbiol. 77, 118–129.
| A review of Clostridioides [Clostridium] difficile occurrence through the food chain.Crossref | GoogleScholarGoogle Scholar | 30297042PubMed |
[9] Knight, D.R. et al. (2017) Genome analysis of Clostridium difficile PCR ribotype 014 lineage in Australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission. Front. Microbiol. 7, 2138.
| Genome analysis of Clostridium difficile PCR ribotype 014 lineage in Australian pigs and humans reveals a diverse genetic repertoire and signatures of long-range interspecies transmission.Crossref | GoogleScholarGoogle Scholar | 28123380PubMed |
[10] Knight, D.R. et al. (2019) Evolutionary and genomic insights into Clostridioides difficile sequence type 11: A diverse zoonotic and antimicrobial-resistant lineage of global One Health importance. MBio 10, e00446-19.
| Evolutionary and genomic insights into Clostridioides difficile sequence type 11: A diverse zoonotic and antimicrobial-resistant lineage of global One Health importance.Crossref | GoogleScholarGoogle Scholar | 30992351PubMed |
[11] Mughini-Gras, L. et al. (2018) Source attribution of foodborne diseases: potentialities, hurdles, and future expectations. Front. Microbiol. 9, 1983.
| Source attribution of foodborne diseases: potentialities, hurdles, and future expectations.Crossref | GoogleScholarGoogle Scholar | 30233509PubMed |
[12] Knetsch, C.W. et al. (2014) Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011. Euro Surveill. 19, 20954.
| Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011.Crossref | GoogleScholarGoogle Scholar | 25411691PubMed |
[13] Knetsch, C.W. et al. (2018) Zoonotic transfer of Clostridium difficile harboring antimicrobial resistance between farm animals and humans. J. Clin. Microbiol. 56, e01384-17.
| Zoonotic transfer of Clostridium difficile harboring antimicrobial resistance between farm animals and humans.Crossref | GoogleScholarGoogle Scholar | 29237792PubMed |
[14] Dingle, K.E. et al. (2019) A role for tetracycline selection in recent evolution of agriculture-associated Clostridium difficile PCR ribotype 078. MBio 10, e02790-18.
| A role for tetracycline selection in recent evolution of agriculture-associated Clostridium difficile PCR ribotype 078.Crossref | GoogleScholarGoogle Scholar | 30862754PubMed |
[15] Collins, D.A. et al. (2017) Laboratory-based surveillance of Clostridium difficile strains circulating in the Australian healthcare setting in 2012. Pathology 49, 309–313.
| Laboratory-based surveillance of Clostridium difficile strains circulating in the Australian healthcare setting in 2012.Crossref | GoogleScholarGoogle Scholar | 28237369PubMed |
[16] Knight, D.R. et al. (2015) Surveillance for antimicrobial resistance in Australian isolates of Clostridium difficile, 2013-14. J. Antimicrob. Chemother. 70, 2992–2999.
| Surveillance for antimicrobial resistance in Australian isolates of Clostridium difficile, 2013-14.Crossref | GoogleScholarGoogle Scholar | 26221017PubMed |
[17] Knight, D.R. et al. (2015) Nationwide surveillance study of Clostridium difficile in Australian neonatal pigs shows high prevalence and heterogeneity of PCR ribotypes. Appl. Environ. Microbiol. 81, 119–123.
| Nationwide surveillance study of Clostridium difficile in Australian neonatal pigs shows high prevalence and heterogeneity of PCR ribotypes.Crossref | GoogleScholarGoogle Scholar | 25326297PubMed |
[18] Eyre, D.W. et al. (2018) Two distinct patterns of Clostridium difficile diversity across Europe indicating contrasting routes of spread. Clin. Infect. Dis. 67, 1035–1044.
| Two distinct patterns of Clostridium difficile diversity across Europe indicating contrasting routes of spread.Crossref | GoogleScholarGoogle Scholar | 29659747PubMed |
[19] Rodriguez-Palacios, A. et al. (2010) Clostridium difficile survives minimal temperature recommended for cooking ground meats. Anaerobe 16, 540–542.
| Clostridium difficile survives minimal temperature recommended for cooking ground meats.Crossref | GoogleScholarGoogle Scholar | 20488251PubMed |
[20] Deng, K. et al. (2015) Survival of Clostridium difficile spores at low temperatures. Food Microbiol. 46, 218–221.
| Survival of Clostridium difficile spores at low temperatures.Crossref | GoogleScholarGoogle Scholar | 25475288PubMed |
[21] Doan, L. et al. (2012) Clinical and cost effectiveness of eight disinfection methods for terminal disinfection of hospital isolation rooms contaminated with Clostridium difficile 027. J. Hosp. Infect. 82, 114–121.
| Clinical and cost effectiveness of eight disinfection methods for terminal disinfection of hospital isolation rooms contaminated with Clostridium difficile 027.Crossref | GoogleScholarGoogle Scholar | 22902081PubMed |
[22] Best, E.L. et al. (2010) The potential for airborne dispersal of Clostridium difficile from symptomatic patients. Clin. Infect. Dis. 50, 1450–1457.
| The potential for airborne dispersal of Clostridium difficile from symptomatic patients.Crossref | GoogleScholarGoogle Scholar | 20415567PubMed |
[23] Keessen, E.C. et al. (2011) Aerial dissemination of Clostridium difficile on a pig farm and its environment. Environ. Res. 111, 1027–1032.
| Aerial dissemination of Clostridium difficile on a pig farm and its environment.Crossref | GoogleScholarGoogle Scholar | 22014605PubMed |
[24] Romano, V. et al. (2012) Toxigenic Clostridium difficile PCR ribotypes from wastewater treatment plants in southern Switzerland. Appl. Environ. Microbiol. 78, 6643–6646.
| Toxigenic Clostridium difficile PCR ribotypes from wastewater treatment plants in southern Switzerland.Crossref | GoogleScholarGoogle Scholar | 22798376PubMed |
[25] Troiano, T. et al. (2015) Toxigenic Clostridium difficile PCR ribotypes in edible marine bivalve molluscs in Italy. Int. J. Food Microbiol. 208, 30–34.
| Toxigenic Clostridium difficile PCR ribotypes in edible marine bivalve molluscs in Italy.Crossref | GoogleScholarGoogle Scholar | 26022983PubMed |
[26] Orden, C. et al. (2018) Recreational sandboxes for children and dogs can be a source of epidemic ribotypes of Clostridium difficile. Zoonoses Public Health 65, 88–95.
| Recreational sandboxes for children and dogs can be a source of epidemic ribotypes of Clostridium difficile.Crossref | GoogleScholarGoogle Scholar | 28686001PubMed |
[27] Lim, S.C. et al. (2019) Clostridium difficile and One Health. Clin. Microbiol. Infect. , .
| Clostridium difficile and One Health.Crossref | GoogleScholarGoogle Scholar | 31682985PubMed |
[28] Longtin, Y. et al. (2016) Effect of detecting and isolating Clostridium difficile carriers at hospital admission on the incidence of C. difficile infection a Quasi-experimental controlled study. JAMA Intern. Med. 176, 796–804.
| Effect of detecting and isolating Clostridium difficile carriers at hospital admission on the incidence of C. difficile infection a Quasi-experimental controlled study.Crossref | GoogleScholarGoogle Scholar | 27111806PubMed |
[29] Sanofi (2017) Sanofi ends development of Clostridium difficile vaccine. [Press release.] https://www.sanofi.com/en/media-room/press-releases/2017/2017-12-01-22-00-00
[30] Kitchin, N. et al. (2020) A phase 2 study evaluating the safety, tolerability, and immunogenicity of two 3-dose regimens of a Clostridium difficile vaccine in healthy US adults aged 65 to 85 years. Clin. Infect. Dis. 70, ciz153.
| A phase 2 study evaluating the safety, tolerability, and immunogenicity of two 3-dose regimens of a Clostridium difficile vaccine in healthy US adults aged 65 to 85 years.Crossref | GoogleScholarGoogle Scholar | 31125055PubMed |
[31] Hong, H.A. et al. (2017) Mucosal antibodies to the C terminus of toxin A prevent colonization of Clostridium difficile. Infect. Immun. 85, e0160-16.
| Mucosal antibodies to the C terminus of toxin A prevent colonization of Clostridium difficile.Crossref | GoogleScholarGoogle Scholar | 28536258PubMed |
[32] Centers for Disease Control and Prevention (2019) Antibiotic resistance threats in the United States 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf
Biographies
Su-Chen Lim is a postdoctoral research associate in the School of Medical and Health Sciences at Edith Cowan University. She completed her PhD at The University of Western Australia in 2019, investigating sources of C. difficile in the community of One Health importance. Her current work extends on this theme to determine the role of the contaminated environment in the transmission of C. difficile.
Tom Riley holds positions in various universities in Western Australia, as well as in Pathwest Laboratory Medicine. He has had a long-standing interest in healthcare-related infections, particularly the diagnosis, pathogenesis and epidemiology of CDI, in both humans and animals.
Daniel Knight is an NHMRC Early Career Research Fellow at Murdoch University in Western Australia. He has a background in AMR and infectious disease surveillance and currently manages a genomics-based research program focusing on evolutionary and One Health aspects of C. difficile Infection.