Skerman and beyond: 2019 status of the Global Catalogue of Microorganisms
Juncai Ma A , Linhuan Wu A and İpek Kurtböke B CA World Data Center of Microorganisms, Institute of Microbiology, Chinese Academy of Sciences, No. 1–3 West Beichen Road, Chaoyang District, Beijing 100101, P. R. China
B WFCC
C University of the Sunshine Coast, School of Science and Engineering and the GeneCology Research Centre, Maroochydore DC, Qld 4558, Australia. Email: ikurtbok@usc.edu.au
Microbiology Australia 40(3) 121-124 https://doi.org/10.1071/MA19033
Published: 18 September 2019
The World Federation for Culture Collections (WFCC)-MIRCEN World Data Centre for Microorganisms (WDCM) was set up as a data centre of WFCC and UNESCO World Network of Microbiological Resources Centres (MIRCEN). The WDCM is a vehicle for networking microbial resource centres of various types of microorganisms. It also serves as an information resource for the customers of the microbial resource centres (http://www.wdcm.org/). The WDCM was established in 1966 by the late Professor V.B.D. Skerman in Australia, later moved to Japan in 1986 and since 2010 is based in China under the Directorship of Dr Juncai Ma. Current databases at the WDCM are the Culture Collections Information Worldwide (CCINFO), Global Catalogue of Microorganisms (GCM) and the WDCM Reference Strain Catalogue. In addition, Analyzer of Bio-resource citations (ABC) and Statistics on Patented Microorganisms are available (http://www.wdcm.org/databases.html). In this article the status of the GCM and its associated 10K type strain sequencing project that currently provides services to taxonomists for standard genome sequencing and annotation will be communicated.
In the 50-year history of the WDCM, the capabilities and uses of information technologies have expanded greatly, with high-throughput sequencing technology leading to an exponential increase in DNA sequence data. Microbiology and biotechnology are sciences that rely on DNA sequence data that is vital for determination of the genetic make-up and functional roles of microorganisms in nature1. Culture collections in this context have an important function as data and information repositories thus serving academia, industry and the public. As a result, the WDCM is now facilitating the application of cutting-edge information technology to improve the interoperability of microbial data, promote the access to and use of data and information, and to coordinate international cooperation between culture collections, scientists and other user communities. Curators and scientists from culture collections not only share data but also design and implement the generated data platforms that meet the changing requirements of microbiologists.
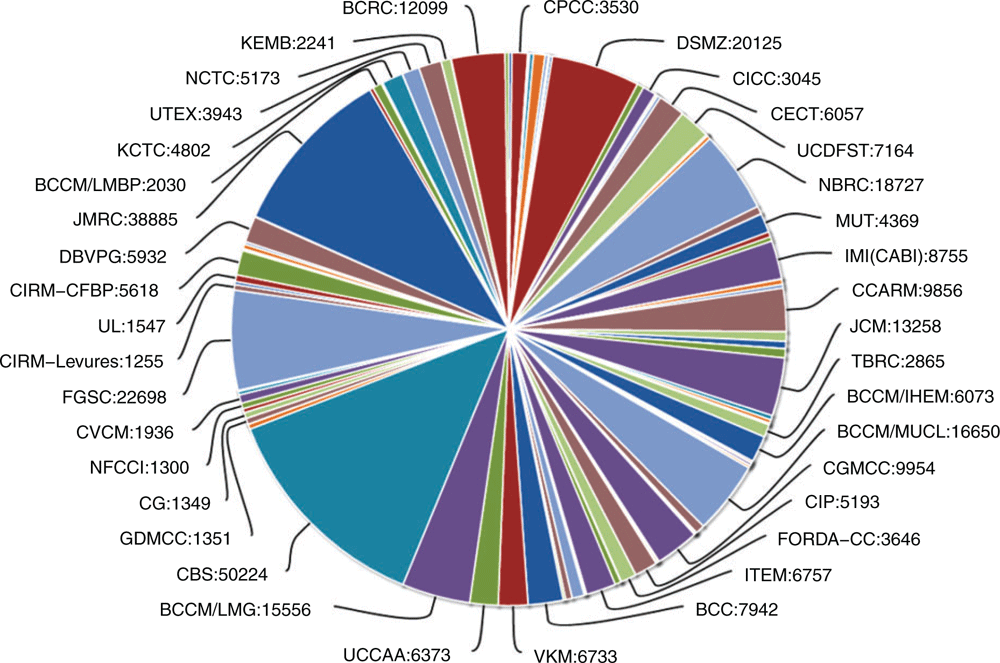
Culture Collections Information Worldwide (CCINFO) is a registration system and metadata archive for culture collections around the world. WFCC recommends that every culture collection register in the CCINFO database before providing public services. CCINFO serves as a metadata recorder. It provides a unique identifier for each culture collection and lists the species names of collections’ holdings. Currently, 783 culture collections from 76 countries have registered with CCINFO and 131 of these collections have registered with WFCC (http://www.wfcc.info/) as affiliate members representing 49 countries. The foundational structure of the WDCM ensuring information transfer from key organisations is highlighted in Figure 1. Using the unique strain numbers and species names, WDCM developed ABC, a data mining tool to extract information from public resources such as Pubmed, WIPO, Genome Online database and NCBI nucleotide sequence database. After catalogue information is submitted from individual culture collection, WDCM automatically links this catalogue information with the available knowledge on each strain extracted by ABC, which is subsequently accessible to the public through the Global Catalogue of Microorganisms (GCM). Currently, 127 collections from 48 countries are part of the GCM and information on the 447 512 strains and 54 736 species is available. The original isolation places of the isolates are also listed: Asia, 44; Africa, 46; Europe, 42; North America, 17; Oceania, 9; and South America, 13. Such information is of importance for accessing, tracking, monitoring and benefit sharing, and compliance with the Nagoya Protocol (https://www.cbd.int/abs/). The GCM can aid culture collections in monitoring the utilisation of their microorganisms. GCM can also facilitate access to strains that might otherwise have been unavailable prior to becoming a part of GCM such as recent accessibility into the 1000 strains at the Vietnam Type Culture Collection. GCM has also provided support to 40 different culture collections to create their online catalogues (Figure 2). Moreover, regional organisations such as the Asian Network of Research Resource Centres (ANRRC) and the Asian Consortium for the conservation and Sustainable Use of Microbial Resources (ACM) are using the data stored in the GCM to create their online catalogues.
|
In the advance stage of the GCM 2 development the following points are targeted: (1) From strain data to ‘omics’ data and to improved database platform, (2) from database to knowledge base and to improved personalised services to microbiologists, (3) from data search to analysis to cloud-based analysis pipeline integrated.
In addition, the GCM has initiated an international project titled ‘The global catalogue of microorganisms 10K type strain sequencing project’ to close the genomic gaps for the validly published prokaryotic and fungi species2. This project has two core subprojects: (1) to sequence 10 000 bacterial and archaeal type strains; and (2) to sequence selected fungal type strains. The outcomes of the project will close currently existing large gaps in the available genomic sequence information published for bacterial and archaeal species. This gap is even larger for fungal type strains. The GCM led and internationally coordinated effort will facilitate the generation of a more comprehensive genomic information platform to be used for research via in-depth genome mining. Information to be generated on the taxonomic, phylogenetic, and functional genes of microorganisms will be of immense value for the advancement of biological sciences and biotechnology.
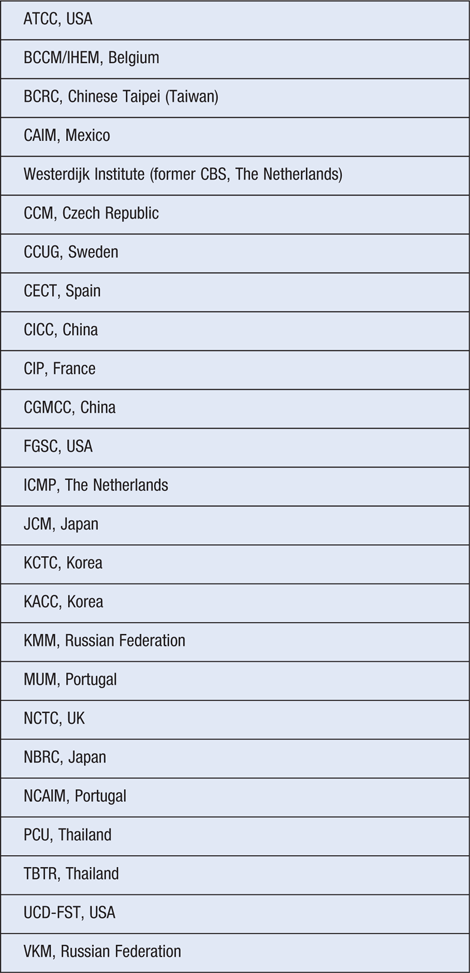
In this project, the genomes of 10 000 type strains will be sequenced (http://gcm.wdcm.org/typestrain/) with the WDCM covering the costs for sequencing services, database system and data analysis. Upon completion, raw data and analysed results will also be published online and made freely available. So far, 25 collections from 16 different countries have agreed to take part in the project (Table 1).
|
The project has established standard operational procedures for DNA extraction, sample submission, sequencing, and data processing to ensure that all genetic resources, data, and metadata associated with type strains are appropriately obtained, recorded, and stored. A project proposed by the WDCM, ‘CD 20170: Specification on Data Integration and Publication in Microbial Resource Centers’, is currently under development and will meet the standards of the International Organization for Standardization (ISO).
International Working Groups have been established to provide expert advice during the selection of type strains for genome sequencing. In addition, SOPs, database establishment and intellectual property rights and legal issues have been clarified. WDCM will only use strains and DNA samples provided by formal collaborators, for sequencing, data mining and integration into the data platform for microbial resources. Collaborative research agreements were also put into place.
All sequencing is currently being conducted at the BGI and IMCAS, which has the largest sequencing capacity in the world at >30 Tb/day with Sequencers (295+): BGISeq-500, Illumina/HiSeq, Illumina/MiSeq, AB/3730xi, Roche/454, PacBio RS, Sequel, Bionano Irys System, Life Tech/Ion Torrent. Out of the 719 type strains received to date, 465 of them have been genome sequenced.
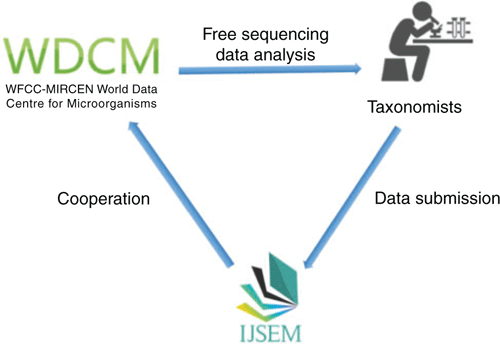
The GCM type strain sequencing project encourages all culture collections to participate in this international collaborative project. Interested parties should be willing to provide DNA for type strains held in their collections. All microbiologists and institutions from related fields are welcome to submit subprojects for genomic data-related research questions. In addition, the WDCM has established a MOU with the International Journal of Systematic and Evolutionary Microbiology and the Bergey’s Manual Trust in March 2019 and will provide free services for genome sequencing and annotation required for description of new species and publication3 (Figure 3).
|
In the light of the above presented information, we now would like to bring the focus to Australia and ask the Council of Heads of Australian Collections of Microorganisms (CHACM), who were part of the original Australian Microbial Resources Information Network (AMRIN)4, to work closely with the GCM to establish their online catalogues. We look forward to the fulfilment of Skerman’s vision of the GCM and to revitalise it in Australia by following in the footsteps of Professor Lindsay Sly who created the AMRIN and CHACM as a vision for the future interlinked-development of microbial collections in Australia (also see article by Sly5).
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA19050301), the Bureau of International Cooperation of the Chinese Academy of Sciences (grants 153211KYSB20160029 and 153211KYSB20150010), the National Key Research Program of China (grants 2017YFC1201202, 2016YFC1201303, and 2016YFC0901702), the 13th Five-year Informatization Plan of the Chinese Academy of Sciences (grant XXH13506), and the National Science Foundation for Young Scientists of China (grant 31701157).
References
[1] Kurtböke, D.İ. (Ed.) (2017) Microbial Resources: From Functional Existence in Nature to Applications. Elsevier, Academic Press.[2] Wu, L. et al. (2018) The global catalogue of microorganisms 10K type strain sequencing project: closing the genomic gaps for the validly published prokaryotic and fungi species. Gigascience 7, giy026.
| The global catalogue of microorganisms 10K type strain sequencing project: closing the genomic gaps for the validly published prokaryotic and fungi species.Crossref | GoogleScholarGoogle Scholar | 30239706PubMed |
[3] Wu, L. and Ma, J. (2019) The Global Catalogue of Microorganisms (GCM) 10K type strain sequencing project: providing services to taxonomists for standard genome sequencing and annotation. Int. J. Syst. Evol. Microbiol. 69, 895–898.
| The Global Catalogue of Microorganisms (GCM) 10K type strain sequencing project: providing services to taxonomists for standard genome sequencing and annotation.Crossref | GoogleScholarGoogle Scholar | 30832757PubMed |
[4] Sly, L.I. (2010) Biodiversity and the role of microbial resource centres. Paper prepared for presentation at the ‘Biodiversity and World Food Security: Nourishing the planet and its people’ conference conducted by the Crawford Fund for International Agricultural Research, Parliament House, Canberra, Australia, 30 August–1 September 2010 (No. 656-2016-44603).
[5] Sly, L.I. (2019) Historical perspectives and new opportunities for Australian collections of microorganisms in the microbiome era. Microbiol. Aust. 40, 140–143.
| Historical perspectives and new opportunities for Australian collections of microorganisms in the microbiome era.Crossref | GoogleScholarGoogle Scholar |
Biographies
Dr Juncai Ma is director of the Center for Microbial Resource and Big Data, Institute of Microbiology, Chinese Academy of Sciences (CAS), director of World Data Center for Microorganisms (WDCM), executive member at the World Federation for Culture Collections (WFCC), chair of the Mirrors Working Group of International Barcode of Life Project (iBOL), and Convener of the Information Technology Committee, Asian Network of Research Resource Centers (ANRRC). Dr Ma has initiated the international cooperation project of the Global Catalogue of Microorganisms (GCM), which assists culture collections across the world in the managing, disseminating and sharing of information, and which greatly increases the visibility as well as the accessibility of microbial strains.
Dr Linhuan Wu is a young data scientist working at the Institute of Microbiology, Chinese Academy of Sciences and WDCM (WDS Regular Member). To improve the accuracy and efficiency of data sharing among the microbial community, Dr Wu has designed and established an international data standard system for microbial resources information management and data sharing. As the team leader of the Global Catalogue of Microorganisms (GCM), Dr Wu has implemented the first uniform database management system for culture collections worldwide. She also works as a principal scientist of WDCM and the secretary of the former CODATA Advancing Informatics for Microbiology Task Group.
Dr İpek Kurtböke has been working in the field of biodiscovery and has been an active member of the international actinomycete research community since 1982. She currently conducts research and teaches in the field of applied microbiology and biotechnology and is senior lecturer at the University of the Sunshine Coast (USC), Queensland. She has also been an active member of the World Federation for Culture Collections (WFCC) including serving as the Vice-President of the Federation (2010–2013) and currently is the President of the Federation (2017–2020).



