The Australian Group on Antimicrobial Resistance (AGAR)
Geoffrey Coombs A B E , Denise Daley C , Jan Bell D and on behalf of the Australian Group on Antimicrobial ResistanceA Antimicrobial Resistance and Infectious Diseases Laboratory, School of Veterinary and Life Sciences, Murdoch University, Murdoch, WA 6150, Australia
B Department of Microbiology, PathWest Laboratory Medicine-WA, Fiona Stanley Hospital, Murdoch, WA 6150, Australia
C Australian Group on Antimicrobial Resistance, Fiona Stanley Hospital, Murdoch, WA 6150, Australia
D The University of Adelaide, Adelaide, SA 5005, Australia
E Email: G.Coombs@murdoch.edu.au
Microbiology Australia 40(2) 69-72 https://doi.org/10.1071/MA19020
Published: 18 April 2019
The Australian Group on Antimicrobial Resistance (AGAR) is a collaboration of clinicians and scientists working in diagnostic medical microbiology laboratories located across Australia. The group gathers information on the level of antimicrobial resistance (AMR) in bacteria causing important and life threatening infections and is a key component of Australia’s response to the problem of increasing AMR. It defines where Australia stands with regard to antimicrobial resistance in human health.
History and governance
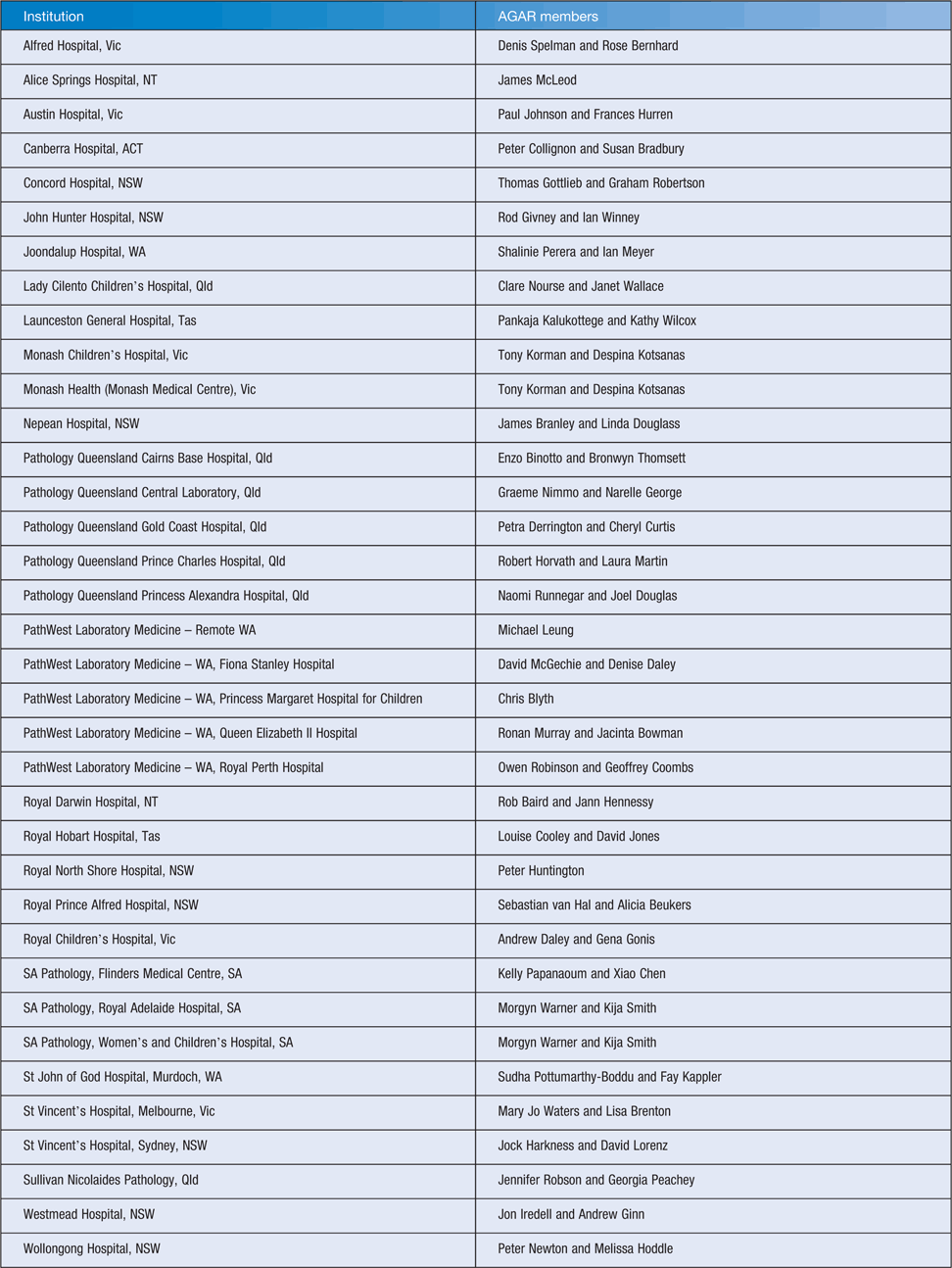
AGAR commenced in 1985 and at that time involved 13 teaching hospitals. It has subsequently grown to involve 40 institutions including five private laboratories (Table 1). This broadening of AGAR has meant that not only does the group have good information as to what is happening with major pathogens in the larger teaching hospitals in each State and Territory, but also has the ability to monitor what is happening with resistance rates in private hospitals and the community. Initially funded by Eli Lilly Pharmaceuticals, from 2003 AGAR has received funding from the Australian Government Department of Health to perform its AMR surveillance programs. Although AGAR is a working group of the Australian Society for Antimicrobials (ASA), the ASA Committee delegates the authority of running AGAR to the AGAR Executive.
|
AGAR is a key component of the Australian Commission on Safety and Quality in Health Care’s (ACSQHC) Antimicrobial Use and Resistance in Australia (AURA) Surveillance System. AURA provides essential information to develop and implement strategies to prevent and contain AMR in human health and improve antimicrobial use across hospital, residential care and community healthcare settings. AGAR complements two other AMR surveillance programs that contribute to AURA: the National Alert System for Critical Antimicrobial Resistances (CARAlert) and Australian Passive Antimicrobial Resistance Surveillance (APAS).
Surveillance programs
Historically, AGAR performed ‘snap shot’ targeted AMR surveillance programs that initially focused on antimicrobial resistance in Staphylococcus aureus and over time was broadened to include studies on Escherichia coli, Enterobacter species, Klebsiella species, Haemophilus influenzae, Streptococcus pneumoniae and Enterococcus species.
Since 2013 AGAR has primarily focused on performing ‘ongoing’ targeted AMR surveillance programs on bloodstream infections and conducts three annual programs:
-
Staphylococcus aureus (ASSOP – Australian Staphylococcal Sepsis Outcome Program)
-
Enterococcus species (AESOP – Australian Enterococcal Sepsis Outcome Program)
-
Enterobacterales, Pseudomonas aeruginosa and Acinetobacter species (GNSOP – Gram negative Sepsis Outcome Program)
The sepsis programs’ activities are overseen by three program committees which have the key advisory role of ensuring the highest quality of results is collected within an AGAR surveillance program. The program committees:
-
analyse the clinical data collected in the AGAR surveillance program with the aim of producing peer reviewed publications;
-
organise additional research activities related to the surveillance program; and
-
provide scientific advice to the AGAR Executive on matters that are related to the surveillance program.
Each year the AGAR laboratories collect all S. aureus and Enterococcus species isolates and up to 200 isolates of Enterobacterales, Acinetobacter species and P. aeruginosa from unique patient episodes of bacteraemia. Approval to conduct the prospective data collection, including de-identified demographic data, is given by the research ethics committees associated with each participating hospital.
All AGAR laboratories obtain basic laboratory information for each patient episode plus varying demographic information, depending on the level at which they are enrolled in the program. There are two levels of AGAR enrolment: Bronze and Silver. At Bronze level, participating laboratories provide date of collection, date of birth, sex, postcode and admission date. At Silver level, participating laboratories provide discharge date, device-related infection, principal clinical manifestation, outcome at seven and 30 days, and, if applicable, date of death.
Isolates are identified to species level using the routine method for each institution. This includes the Vitek® and Phoenix™ automated microbiology systems, and, if available, mass spectrometry (MALDI-TOF).
Susceptibility testing is performed using two commercial semi-automated methods: Vitek 2 (bioMérieux) and Phoenix (BD), which are calibrated to the ISO (International Organization for Standardization) reference standard method of broth microdilution. CLSI and EUCAST breakpoints are used in the analysis.
The AGAR data is submitted by AGAR laboratories using a web application portal. AGAR complies with all relevant privacy legislation and data security provisions.
Detailed annual reports on each program can be found on the AGAR website (www.agargroup.org). Reports are also published in the Australian Government Department of Health Communicable Diseases Intelligence (CDI) journal. An annual amalgamated report is produced by the ACSQHC and is available on their website (https://www.safetyandquality.gov.au/antimicrobial-use-and-resistance-in-australia/agar/).
In 2017, 11 562 episodes of bacteraemia across Australia were included in the AGAR programs.
ASSOP
The objectives of ASSOP are to determine the proportion of Staphylococcus aureus bacteraemia (SAB) isolates in Australia that are antimicrobial resistant, with particular emphasis on susceptibility to methicillin and to characterise the molecular epidemiology of the methicillin-resistant isolates.
Key findings from the 2017 ASSOP:
-
A total of 2515 SAB episodes were reported, of which 77% were community onset. One in five of all episodes were methicillin resistant (19%)
-
The 30-day all-cause mortality was 14.8% with a significant difference between methicillin-resistant (MRSA) (18.9%) and methicillin-sensitive S. aureus (MSSA) (13.9%) mortality, as well as community-onset (13.8%) and hospital-onset S. aureus bacteraemia (18.3%)
-
There is an increasing rate of community-associated MRSA (CA-MRSA) bacteraemia, and in Australia CA-MRSA dominate MRSA bacteraemia
-
With the exception of the β-lactams and erythromycin, antimicrobial-resistance in MSSA was rare. However, in addition to the β-lactams approximately 40% of MRSA were resistant to erythromycin and ciprofloxacin and approximately 15% resistant to co-trimoxazole, tetracycline and gentamicin. When applying the EUCAST breakpoints, teicoplanin resistance was detected in five S. aureus isolates. Resistance was not detected for vancomycin and linezolid
-
Three healthcare-associated MRSA (HA-MRSA) clones were identified of which EMRSA-15 (ST22-IV) was the major clone. The majority of EMRSA-15 episodes arose in the community, which is consistent with the prevalence of this clone in residential care facilities in Australia
-
Thirty-nine CA-MRSA clones were identified. The Queensland clone (ST93-IV) that harbours the Panton-Valentine leucocidin (PVL) associated genes has become the dominant CA-MRSA clone and is now seen throughout Australia; it is the most common CA-MRSA clone in Queensland, Western Australia and the Northern Territory
-
Overall, 49.7% of CA-MRSA isolates harboured the PVL associated genes
AESOP
The objectives of AESOP are to determine the proportion of E. faecalis and E. faecium bacteraemia isolates demonstrating antimicrobial resistance with particular emphasis on:
-
Assessing susceptibility to ampicillin
-
Assessing susceptibility to glycopeptides
-
Monitoring the molecular epidemiology of E. faecium
Key findings from the 2017 AESOP:
-
A total of 1,137 episodes of enterococcal bacteraemia were reported; the majority (95.3%) of episodes were caused by E. faecalis or E. faecium
-
The majority of E. faecalis bacteraemia were community-onset (71.3%), while in E. faecium bacteraemia only 30.1% were community onset
-
The combined 30-day all-cause mortality was 20.3%
-
There was significant difference in 30-day all-cause mortality between E. faecalis (14.3%) and E. faecium (27.7%)
-
Overall 50.9% of E. faecium harboured vanA or vanB genes or both, with 50% of vancomycin-resistant E. faecium bacteraemias due to vanA; this type of vancomycin resistance has emerged rapidly in the past six years, particularly in New South Wales, where it is now the dominant genotype
-
There were 64 E. faecium multilocus sequence types (STs), of which ST17, ST1421, ST796, ST1424, ST80, ST555, ST203, ST18, and ST78 were the nine most frequently identified
-
vanA genes were detected in nine STs, and vanB genes were detected in 12 STs. Two STs harboured vanA and vanB genes. The clonal diversity varied across Australia
-
The percentage of E. faecium bacteraemia isolates resistant to vancomycin is now much higher in Australia than in all European countries
GNSOP
The objectives of the 2017 surveillance program are to:
-
Monitor resistance in Enterobacterales, P. aeruginosa and Acinetobacter species isolated from blood cultures taken from patients presenting to the hospital or already in hospital
-
Study the extent of co-resistance and multidrug resistance in the major species
-
Detect emerging resistance to last-line agents such as carbapenems and colistin
-
Examine the molecular basis of resistance to third-generation cephalosporins, quinolones and carbapenems
-
Monitor the epidemiology of E. coli sequence type 131
Key findings from the 2017 GNSOP:
-
A total of 7910 episodes of gram-negative bacteraemia were reported, including Enterobacterales (89.8%), Pseudomonas aeruginosa (8.8%) and Acinetobacter species (1.4%). Three genera – Escherichia (61.6%), Klebsiella (19.9%) and Enterobacter (6.3%) – contributed 87.8% of all Enterobacterales bacteraemias
-
The all-cause 30-day mortality for gram-negative bacteraemia was 12.5% (10.1% in E. coli, 20.6% in P. aeruginosa)
-
Over 11% of E. coli isolates causing community-onset bacteraemia, which accounted for 84% of all E. coli bacteraemia cases, were ceftriaxone resistant
-
Extended-spectrum β-lactamase (ESBL) phenotypes were found in 12.6% of E. coli and 9.8% of Klebsiella pneumoniae and are more common in hospital onset episodes. The CTX-M type gene was present in 76.1% of E. coli with an ESBL phenotype
-
Increasing fluoroquinolone non-susceptibility in E. coli is a continuing concern and is most striking in hospital-onset bacteraemia, with a change from 16.1% to 21.1% between 2013 and 2017
-
Fluoroquinolone resistance is commonly linked to cephalosporin resistance caused by ESBLs of the CTX-M type. O25b-ST131 accounted for 57.3% of E. coli ESBL phenotypes that were ciprofloxacin resistant
-
Very low levels of carbapenemase-producing Enterobacterales (CPE) bacteraemia was observed (0.1% in E. coli and 0.7% in K. pneumoniae), although the Enterobacter cloacae complex hospital-onset figure is higher at 3.6%
-
The rate of colistin resistance – when tested for, but excluding species with intrinsic resistance – was 0.9% (7/752). No mobile colistin resistance genes were detected among all referred isolates
Individual 2017 program reports can found on the AGAR website. An amalgamated 2017 report will be available on the AGAR and the ACSQHC websites in the first half of 2019.
By using standardised methodology AGAR has been able to collect ongoing AMR data on what is happening in Australia over long periods of time. The group has also been very successful in being able to make this information available to the broader community both through publications in scientific journals and also numerous presentations at meetings and to groups around Australia and Internationally. This has led to important benefits within Australia. Among these benefits has been the ability to allow more rational use of antibiotics based on known Australia wide resistance patterns.
For further information on AGAR and its activities please contact Denise Daley, info@agargroup.org.au.
Reference laboratories
AGAR gratefully acknowledges the Australian Centre for Antimicrobial Resistance Ecology, The University of Adelaide, South Australia for the molecular characterisation of gram-negative isolates; and the Antimicrobial Resistance Reference Laboratory, Centre for Infectious Diseases and Microbiology Laboratory Services, Westmead Hospital, for performing whole genome sequencing on carbapenemase-producing isolates.
AGAR also gratefully acknowledges Dr Stanley Pang and Ms Yung Thin Lee at the Antimicrobial Resistance and Infectious Disease Laboratory, School of Veterinary Life Science, Murdoch University, Western Australia for performing the whole genome sequencing on E. faecium and MRSA isolates.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The AGAR programs are funded by the Department of Health via the Australian Commission on Safety and Quality in Health Care and the Australian Society for Antimicrobials.
Biographies
Professor Geoffrey Coombs, PhD, is Chair of Public Health and Murdoch University and Senior Clinical Scientist at PathWest Laboratory Medicine, WA. In addition he is the AGAR Chairman. His major research interest is on antimicrobial resistance and molecular epidemiology of S. aureus and Enterococcus faecium.
Denise Daley is the AGAR Scientific Officer for the Australian Staphylococcus and Enterococcus Sepsis Outcome Programs (ASSOP and AESOP).
Jan Bell is the AGAR Scientific Officer for the Gram Negative Sepsis Outcome Program.


