Future prospects of structural studies to advance our understanding of phage biology
Pavol Bárdy A * , Dominik Hrebík B * , Roman Pantůček A C and Pavel Plevka B DA Department of Experimental Biology, Faculty of Science, Masaryk University, Kamenice 5, 625 00, Brno, Czech Republic
B Central European Institute of Technology, Masaryk University, Kamenice 5, 625 00, Brno, Czech Republic
C Tel: +420 54949 6379, Email: pantucek@sci.muni.cz
D Tel: +420 54949 7756, Email: pavel.plevka@ceitec.muni.cz
Microbiology Australia 40(1) 42-46 https://doi.org/10.1071/MA19009
Published: 19 February 2019
Bacteriophages, being the most abundant biological entities on the Earth, play a major role in regulating populations of bacteria and thus influence the evolution and stability of ecosystems. Phage infections of pathogenic bacteria can both exacerbate and alleviate the severity of the disease. The structural characterisations of phage particles and individual proteins have enabled the understanding of many aspects of phage biology. Due to methodological limitations, most of the structures were determined from purified samples in vitro. However, studies performed outside the cellular context cannot capture the complex and dynamic interactions of the macromolecules that are required for their biological functions. Current developments in structural biology, in particular cryo-electron microscopy, allow in situ high-resolution studies of phage-infected cells. Here we discuss open questions in phage biology that could be addressed by structural biology techniques and their potential to enable the use of tailed phages in industrial applications and human therapy.
State-of-the-art structural biology methods in studies of phage replication
Bacteriophages are a diverse group of viruses that infect bacteria. Bacteriophages are research models for molecular biology and have the potential to be used in modern biotechnology and phage therapy. After ejecting their genome into a cell, bacteriophages can establish two types of infection. The lytic cycle leads to the production of virion progeny and cell death, whereas in lysogenic infection the phage genome integrates into the bacterial one and replicates when the bacterium divides. Knowledge of the high-resolution structures of phage particles and their assembly intermediates has played an important role in our understanding of phage attachment to receptors, genome ejection, virion assembly and genome packaging. Furthermore, the structures of non-structural proteins and their complexes have explained the mechanisms of the lytic-lysogeny switch, genome transcription and replication, and the degradation of the cell envelope. However, nearly all of the structural studies performed to date were limited to analyses of purified macromolecular samples in vitro. In contrast, phage macromolecules perform their functions in vivo by interacting with other phage or cellular components. Current technological developments in the cryo-preservation of cells and cryo-electron tomography (cryo-ET) have enabled structural studies of replicating phages in bacteria. These reports described the ultrastructure of bacteriophages penetrating the cell wall of Gram-negative and Gram-positive bacteria, the formation of a nucleus-like structure during phage replication, and changes in the structure of the cell wall before lysis1–3.
Structural virology beyond purified proteins and phage particles
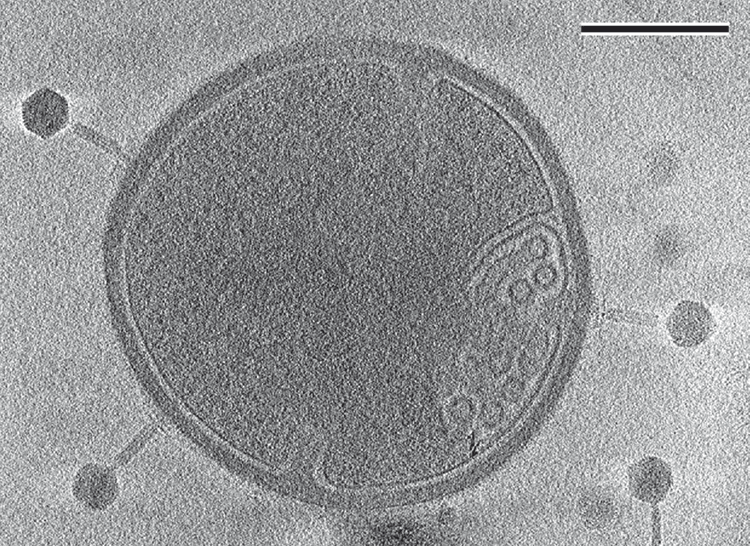
In the single-particle cryo-electron microscopy (cryo-EM) approach, a sample is deposited on a grid in a thin layer of aqueous solution and rapidly plunged into liquid ethane4. This results in the formation of vitreous ice with a structure similar to that of liquid water. Rapid cooling is required to prevent the formation of crystalline ice, which may damage cellular structures. This limits the thickness of samples vitrified under ambient pressure to a few micrometres. Samples up to a few hundred micrometres thick can be cryo-preserved by high-pressure freezing. Individual molecules or macromolecular complexes embedded in vitreous ice are photographed using a transmission electron microscope. Information from many thousands of the projection images of the macromolecules is used to reconstruct their three-dimensional structures. The images are aligned and averaged to improve the signal-to-noise ratio of the reconstructed structure. Pleomorphic objects, such as cells or irregular virus particles can be studied by cryo-ET5. Cells have to be thinned to about 200 nm by cryo-sectioning or focused ion beam (FIB)-milling before imaging in a transmission electron microscope because of the limited penetration of electrons through biological samples (Figure 1). In cryo-ET, samples are imaged from different directions by tilting the stage of the microscope. The resulting tilt series of images is used to calculate the three-dimensional reconstruction of the object. The sensitivity of biological objects to an electron beam limits the overall dose that can be used to image one sample, resulting in a low signal to noise ratio in the reconstructed tomograms. However, sub-tomogram averaging can be used to resolve the structures of regular components of the tomograms with high contrast and resolution.
Currently, high-resolution structures can only be routinely determined for protein complexes or phage particles that can be prepared with high purity and at high concentration. These experimental constraints limit the knowledge that can be gained from the resulting structures, because the complexes may display different conformations in vivo. Determining the structures of macromolecular complexes in situ without the need to purify them from cells would avoid these experimental limitations. Structural analyses of macromolecular complexes in situ are becoming practical thanks to developments in: (1) sample preparation methods, including correlative light and electron microscopy (CLEM), focused ion beam milling, and localised mass spectrometry6; and (2) software for data processing including sub-tomogram classification and averaging7. CLEM is particularly useful in the combination with FIB-milling technique. Events of interest in the cell can be pre-selected by cryo-fluoresce microscopy and subsequently milled with high-precision to open a ‘window’ into the cell for transmission electron microscopy. The localised-mass spectroscopy utilises imaging of single particles from a cell extract. First, the cell lysate is chromatographically separated into fractions. Subsequently, the fractions are characterised by mass spectrometry and macromolecules present in the sample are structurally analysed by transmission electron microscopy. The new advancements in software for classification of particles allow classification of particles in a sample based on their structures. This can be imagined as an in silico ‘purification’ of the macromolecular complexes.
Open questions in phage biology that may be addressed by structural studies
Mechanism of phage genome delivery
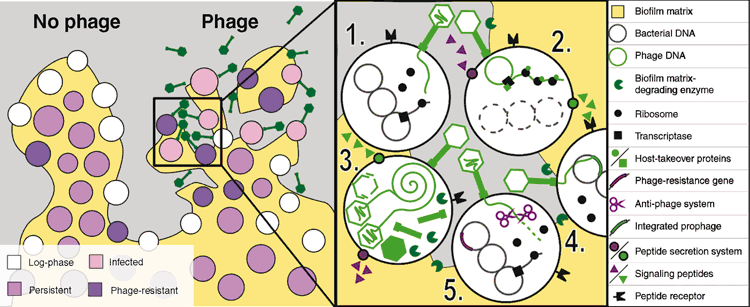
Tailed phages eject their genomes into bacterial cells, however, several aspects of this process are not well understood (Figure 2), including: (1) How is the phage genome ejection triggered? (2) How is the genome transported across bacterial membrane(s)? (3) What is the mechanism for the completion of phage genome ejection after the pressures inside the phage head and cell equalise? (4) How is the transcription machinery recruited to the phage genome? These questions may be addressed by cryo-EM observations of interactions of phages with liposomes8, nevertheless, more biologically relevant answers will be obtained by time-resolved cryo-ET studies of phage genome ejection into mini-cells or focused ion beam milling-prepared sections of bacteria9,10. Changes in the phage particle prior to genome ejection, formation of channels in the membrane, possible genome-uptake machinery or phage genome localisation in the cell may be characterised by such studies. Single particle cryo-EM and X-ray crystallography can be used to determine the structures of the complexes of phage receptor-binding proteins with the receptors. Such knowledge may make it possible to design a group of genetically modified phages with a receptor range so wide that bacteria would be incapable of becoming resistant to the phage infection. Phage receptor-binding proteins themselves may be used as tools for the rapid detection and identification of pathogenic bacteria in environmental samples.
Bacterial resistance to phage infection
Mutations enabling bacteria to avoid phage attachment or block genome ejection can have secondary effects on the cellular phenotype. Phage receptors are often bacterial virulence factors or play roles in substances intake11, and thus phages targeted to bind to specific cellular receptors could be used to shift bacterial population towards lesser virulence12.
Bacterial anti-phage defense systems, such as restriction-modification, CRISPR/Cas, bacteriophage exclusion, or the defense island system associated with the restriction-modification system, degrade phage DNA during delivery13–15 (Figure 2). However, many phages have acquired anti-defense proteins16,17. Structural understanding of the anti-phage defense complexes will enable the preparation of phages capable of protecting their DNA during delivery, which may be important for the development of phage therapy. Similarly, the systems by which bacteria abort late stages of phage infection are assumed to be widespread, but many of them have not been characterised in detail18. The CLEM and cellular cryo-ET could be used to visualise these processes in vivo and explain the functions of these complexes. These findings will enable the engineering or phages with exceptionally broad host-ranges.
Subverting cell resources for phage replication
Some myoviruses, such as T2 and T4 infecting Gram-negative bacteria and K and SPO1 of Gram-positive bacteria, degrade the host genome and block cell division in minutes after the initiation of genome ejection19,20. This mechanism enables them to complete their lytic cycle quickly and thus gain a reproductive advantage over slower replicating phages. The proteins encoded by the host takeover region of the phage genome, which is the first part of its DNA that enters the cell, enable the rapid shutdown of the host’s transcription21. When expressed on their own, these proteins are toxic for the natural host of the phage but also for other bacterial species20. In contrast, some podoviruses such as φ29 do not inhibit cell growth22. Genomes of viruses from the family Podoviridae contain fewer than 20 early genes and during phage infection only affect the expression of a minor number of host genes22. Therefore, podoviruses are a suitable model system for studying the minimal set of phage products that are required to hijack the host resources for phage replication. The interactions of phage proteins with host complexes could be studied by the time-lapse cryo-EM of macromolecular complexes pre-sorted by mass spectrometry23. Identification of the host takeover protein-machinery may enable design of antibiotics inspired by phage proteins24.
Phage spread through biofilm
Biofilm inactivation is a major healthcare and food-hygiene challenge. It has been shown that some phages can eliminate a biofilm thanks to their ability to (1) bind to and accumulate within the biofilm matrix; (2) infect dormant cells; (3) express phage biofilm de-polymerases or induce bacteria-encoded biofilm depolymerases25. Nevertheless, biofilm infection by some lytic phages can lead to accelerated biofilm growth with an increased concentration of extracellular DNA in the biofilm matrix26.
The metabolic heterogeneity of cells within a biofilm presents a challenge for analysing the impact of the phage infection on the biofilm (Figure 2). Identifying the genetic markers of fast proliferating cells, anaerobically growing cells, starving cells, and persister cells will enable the differentiation of bacteria by fluorescence microscopy to study their unique interactions with phages by cryo-ET. However, some phages can form particles with different propensities to infect starved cells27. CLEM and cryo-ET studies of biofilm infection by bacteriophages will determine whether and how phage particles distinguish between metabolically distinct host cells. Mechanisms that allow phages to diffuse through the biofilm matrix are of interest because they may be used to enhance penetration of antibiotics into biofilms. Furthermore, phage-derived nano-vehicles may be used for the targeted delivery of drugs into biofilms.
Phage-phage communication in the regulation of lysis-lysogeny decisions
It is beneficial for phages to establish lysogeny in an environment with a shortage of non-infected host cells28. For the application of phage therapy, however, lysogeny is not desired because of the possible associated acquisition of bacterial virulence factors29. The communication among phage-infected cells and phages was proven in several cases, either in the most simple form of super-infection exclusion regulated by changes in the bacterial membrane potential30, complex communication by the production of Arbitrium-like peptides31, or through a host-produced quorum-sensing system32. Even lytic phages were shown to modify the speed of their reproduction cycle based on available nutrients and enter the ‘hibernation’ phase in starved cells33. X-ray crystallography and nuclear magnetic resonance spectroscopy can be used to determine the structural interactions between the complexes responsible for such communication. Understanding this communication may enable the use of temperate phages for phage therapy in bacterial pathogens for which there are no available strictly virulent phages. In lytic phages it can lead to design of small-molecule additives, ensuring the lytic cycle will be rapid and robust.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The bacteriophage research by the authors is supported by grants from the Ministry of Health of the Czech Republic grant 16-29916A to RP, Grant Agency of the Czech Republic grants 18-13064S to RP, 15-21631Y and 18-17810S to PP and from EMBO installation grant 3041 to PP.
References
[1] Farley, M.M. et al. (2017) Ultrastructural analysis of bacteriophage phi29 during infection of Bacillus subtilis. J. Struct. Biol. 197, 163–171.| Ultrastructural analysis of bacteriophage phi29 during infection of Bacillus subtilis.Crossref | GoogleScholarGoogle Scholar | 27480510PubMed |
[2] Chaikeeratisak, V. et al. (2017) Assembly of a nucleus-like structure during viral replication in bacteria. Science 355, 194–197.
| Assembly of a nucleus-like structure during viral replication in bacteria.Crossref | GoogleScholarGoogle Scholar | 28082593PubMed |
[3] Fu, X. et al. (2014) Controlled bacterial lysis for electron tomography of native cell membranes. Structure 22, 1875–1882.
| Controlled bacterial lysis for electron tomography of native cell membranes.Crossref | GoogleScholarGoogle Scholar | 25456413PubMed |
[4] Thompson, R.F. et al. (2016) An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology. Methods 100, 3–15.
| An introduction to sample preparation and imaging by cryo-electron microscopy for structural biology.Crossref | GoogleScholarGoogle Scholar | 26931652PubMed |
[5] Wan, W. and Briggs, J.A. (2016) Cryo-electron tomography and subtomogram averaging. Methods Enzymol. 579, 329–367.
| Cryo-electron tomography and subtomogram averaging.Crossref | GoogleScholarGoogle Scholar | 27572733PubMed |
[6] Oikonomou, C.M. et al. (2016) A new view into prokaryotic cell biology from electron cryotomography. Nat. Rev. Microbiol. 14, 205–220.
| A new view into prokaryotic cell biology from electron cryotomography.Crossref | GoogleScholarGoogle Scholar | 26923112PubMed |
[7] Zivanov, J. et al. (2018) New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166.
| New tools for automated high-resolution cryo-EM structure determination in RELION-3.Crossref | GoogleScholarGoogle Scholar | 30412051PubMed |
[8] Xu, J. et al. (2016) The bacteriophage phi29 tail possesses a pore-forming loop for cell membrane penetration. Nature 534, 544–547.
| The bacteriophage phi29 tail possesses a pore-forming loop for cell membrane penetration.Crossref | GoogleScholarGoogle Scholar | 27309813PubMed |
[9] Farley, M.M. et al. (2016) Minicells, back in fashion. J. Bacteriol. 198, 1186–1195.
| Minicells, back in fashion.Crossref | GoogleScholarGoogle Scholar | 26833418PubMed |
[10] Rigort, A. et al. (2010) Micromachining tools and correlative approaches for cellular cryo-electron tomography. J. Struct. Biol. 172, 169–179.
| Micromachining tools and correlative approaches for cellular cryo-electron tomography.Crossref | GoogleScholarGoogle Scholar | 20178848PubMed |
[11] Nobrega, F.L. et al. (2018) Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 16, 760–773.
| Targeting mechanisms of tailed bacteriophages.Crossref | GoogleScholarGoogle Scholar | 30104690PubMed |
[12] León, M. and Bastias, R. (2015) Virulence reduction in bacteriophage resistant bacteria. Front. Microbiol. 6, 343.
| Virulence reduction in bacteriophage resistant bacteria.Crossref | GoogleScholarGoogle Scholar | 25954266PubMed |
[13] Dupuis, M.È. et al. (2013) CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance. Nat. Commun. 4, 2087.
| CRISPR-Cas and restriction-modification systems are compatible and increase phage resistance.Crossref | GoogleScholarGoogle Scholar | 23820428PubMed |
[14] Goldfarb, T. et al. (2015) BREX is a novel phage resistance system widespread in microbial genomes. EMBO J. 34, 169–183.
| BREX is a novel phage resistance system widespread in microbial genomes.Crossref | GoogleScholarGoogle Scholar | 25452498PubMed |
[15] Ofir, G. et al. (2018) DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat. Microbiol. 3, 90–98.
| DISARM is a widespread bacterial defence system with broad anti-phage activities.Crossref | GoogleScholarGoogle Scholar | 29085076PubMed |
[16] Rifat, D. et al. (2008) Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target. J. Mol. Biol. 375, 720–734.
| Restriction endonuclease inhibitor IPI* of bacteriophage T4: a novel structure for a dedicated target.Crossref | GoogleScholarGoogle Scholar | 18037438PubMed |
[17] Borges, A.L. et al. (2017) The discovery, mechanisms, and evolutionary impact of anti-CRISPRs. Annu. Rev. Virol. 4, 37–59.
| The discovery, mechanisms, and evolutionary impact of anti-CRISPRs.Crossref | GoogleScholarGoogle Scholar | 28749735PubMed |
[18] Dy, R.L. et al. (2014) A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism. Nucleic Acids Res. 42, 4590–4605.
| A widespread bacteriophage abortive infection system functions through a Type IV toxin-antitoxin mechanism.Crossref | GoogleScholarGoogle Scholar | 24465005PubMed |
[19] Kutter, E. et al. (2018) From host to phage metabolism: hot tales of phage T4’s takeover of E. coli. Viruses 10, 387.
| From host to phage metabolism: hot tales of phage T4’s takeover of E. coli.Crossref | GoogleScholarGoogle Scholar |
[20] Tevdoradze, E. et al. (2014) Bactericidal genes of Staphylococcal bacteriophage Sb-1. Curr. Microbiol. 68, 204–210.
| Bactericidal genes of Staphylococcal bacteriophage Sb-1.Crossref | GoogleScholarGoogle Scholar | 24077954PubMed |
[21] Stewart, C.R. et al. (2009) Roles of genes 38, 39, and 40 in shutoff of host biosyntheses during infection of Bacillus subtilis by bacteriophage SPO1. Virology 392, 271–274.
| Roles of genes 38, 39, and 40 in shutoff of host biosyntheses during infection of Bacillus subtilis by bacteriophage SPO1.Crossref | GoogleScholarGoogle Scholar | 19665746PubMed |
[22] Mojardín, L. and Salas, M. (2016) Global transcriptional analysis of virus-host interactions between phage φ29 and Bacillus subtilis. J. Virol. 90, 9293–9304.
| Global transcriptional analysis of virus-host interactions between phage φ29 and Bacillus subtilis.Crossref | GoogleScholarGoogle Scholar | 27489274PubMed |
[23] Verbeke, E.J. et al. (2018) Classification of single particles from human cell extract reveals distinct structures. Cell Reports 24, 259–268.e3.
| Classification of single particles from human cell extract reveals distinct structures.Crossref | GoogleScholarGoogle Scholar | 29972786PubMed |
[24] Wagemans, J. et al. (2014) Functional elucidation of antibacterial phage ORFans targeting Pseudomonas aeruginosa. Cell. Microbiol. 16, 1822–1835.
| Functional elucidation of antibacterial phage ORFans targeting Pseudomonas aeruginosa.Crossref | GoogleScholarGoogle Scholar | 25059764PubMed |
[25] Pires, D.P. et al. (2016) Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 100, 2141–2151.
| Bacteriophage-encoded depolymerases: their diversity and biotechnological applications.Crossref | GoogleScholarGoogle Scholar | 26767986PubMed |
[26] Fernández, L. et al. (2017) Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus. Sci. Rep. 7, 40965.
| Low-level predation by lytic phage phiIPLA-RODI promotes biofilm formation and triggers the stringent response in Staphylococcus aureus.Crossref | GoogleScholarGoogle Scholar | 28102347PubMed |
[27] Golec, P. et al. (2011) Persistence of bacteriophage T4 in a starved Escherichia coli culture: evidence for the presence of phage subpopulations. J. Gen. Virol. 92, 997–1003.
| Persistence of bacteriophage T4 in a starved Escherichia coli culture: evidence for the presence of phage subpopulations.Crossref | GoogleScholarGoogle Scholar | 21177930PubMed |
[28] Sinha, V. et al. (2017) In silico evolution of lysis-lysogeny strategies reproduces observed lysogeny propensities in temperate bacteriophages. Front. Microbiol. 8, 1386.
| In silico evolution of lysis-lysogeny strategies reproduces observed lysogeny propensities in temperate bacteriophages.Crossref | GoogleScholarGoogle Scholar | 28798729PubMed |
[29] Howard-Varona, C. et al. (2017) Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 11, 1511–1520.
| Lysogeny in nature: mechanisms, impact and ecology of temperate phages.Crossref | GoogleScholarGoogle Scholar | 28291233PubMed |
[30] Abedon, S.T. (2015) Bacteriophage secondary infection. Virol. Sin. 30, 3–10.
| Bacteriophage secondary infection.Crossref | GoogleScholarGoogle Scholar | 25595214PubMed |
[31] Erez, Z. et al. (2017) Communication between viruses guides lysis-lysogeny decisions. Nature 541, 488–493.
| Communication between viruses guides lysis-lysogeny decisions.Crossref | GoogleScholarGoogle Scholar | 28099413PubMed |
[32] Silpe, J.E. and Bassler, B.L. (2019) A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision. Cell 176, 268–280.e13.
| A host-produced quorum-sensing autoinducer controls a phage lysis-lysogeny decision.Crossref | GoogleScholarGoogle Scholar | 30554875PubMed |
[33] Bryan, D. et al. (2016) Bacteriophage T4 infection of stationary phase E. coli: life after log from a phage perspective. Front. Microbiol. 7, 1391.
| Bacteriophage T4 infection of stationary phase E. coli: life after log from a phage perspective.Crossref | GoogleScholarGoogle Scholar | 27660625PubMed |
* These authors contributed equally.
Biographies
Pavol Bardy is a PhD student in the field of Molecular and Cell Biology at the Masaryk University, Brno. He is utilising structural biology techniques to understand assembly process of bacteriophages and phage-like particles.
Dominik Hrebik is a PhD student in the group of Structural virology at Central European Institute of Technology, Masaryk University, Brno. He is exploiting structural biology techniques to understand cell entry mechanisms of bacteriophages and eukaryotic viruses. His current research focuses on the study of mechanisms of membrane penetration by different viruses. He is also interested in novel methodological developments in the field of cryo-electron microscopy.
Roman Pantucek is an associate professor at the Department of Experimental Biology, Masaryk University, Brno. His research interests are focused on molecular biology of staphylococcal bacteriophages, horizontal gene transfer mediated by phages, phage therapy, and genomics and evolutionary biology of staphylococci.
Pavel Plevka is a research group leader at Central European Institute of Technology, Masaryk University, Brno. His research interests are focused on structural characterisation of the replication of human viruses from the families Picornaviridae and Flaviviridae and of bacteriophages from the family Myoviridae in infected cells. He uses cryo-electron microscopy, high-pressure freezing, focused ion beam milling, cryo-electron tomography, correlative light-electron microscopy, and time-resolved fluorescence microscopy.