Life without water: how do bacteria generate biomass in desert ecosystems?
Sean Bay A , Belinda Ferrari B and Chris Greening CA School of Biological Sciences, Centre for Geometric Biology, Monash University, 25 Rainforest Walk, Clayton, Vic. 3800, Australia. Tel: +61 4 1355 5721, Email: sean.bay@monash.edu
B School of Biotechnology and Biomolecular Sciences, UNSW Sydney, Biosciences South E26, Kensington, NSW 2052, Australia. Tel: +61 2 9385 2032, Email: b.ferrari@unsw.edu.au
C School of Biological Sciences, Centre for Geometric Biology, Monash University, 25 Rainforest Walk, Clayton, Vic. 3800, Australia. Tel: +61 3 9905 1692, Email: chris.greening@monash.edu
Microbiology Australia 39(1) 28-32 https://doi.org/10.1071/MA18008
Published: 21 February 2018
Many of the world’s most arid deserts harbour surprisingly diverse communities of heterotrophic bacteria. These organisms persist in surface soils under extreme climatic conditions, despite lacking obvious energy inputs from phototrophic primary producers. A longstanding conundrum has been how these communities sustain enough energy to maintain their diversity and biomass. We recently helped to resolve this conundrum by demonstrating that some desert communities are structured by a minimalistic mode of chemosynthetic primary production, where atmospheric trace gases, not sunlight, serve as the main energy sources. These findings are supported by pure culture studies that suggest atmospheric trace gases are dependable energy sources for the long-term survival of dormant soil bacteria. We predict that atmospheric trace gases may be a major energy source for desert ecosystems worldwide.
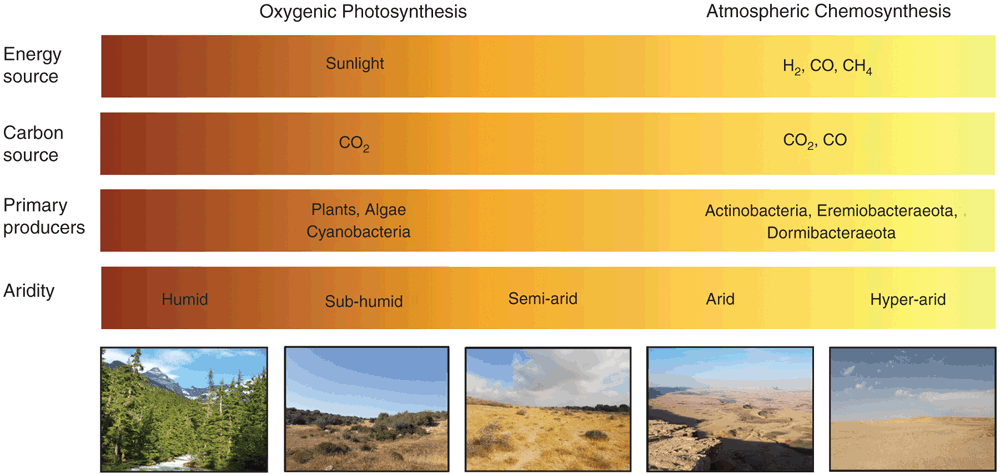
Deserts are one of the largest biomes. They cover one-fifth of the planet’s terrestrial land surface (33.7 × 106 km2) and occupy latitudinal ranges along the tropics, Arctic, and Antarctic. Deserts are defined as having a precipitation to evapotranspiration ratio (P/ET) of less than 1 and can be classified as sub-humid (0.5–0.65), semi-arid (0.2–0.5), arid (0.05–0.2) and hyper-arid (<0.05)1. With exception of hyper-arid deserts, these regions are collectively recognised as drylands and are critical for human development. However, the productivity and biodiversity of these regions is being increasingly threatened by anthropogenic land degradation and climate change.
Microbial community structure in desert soils
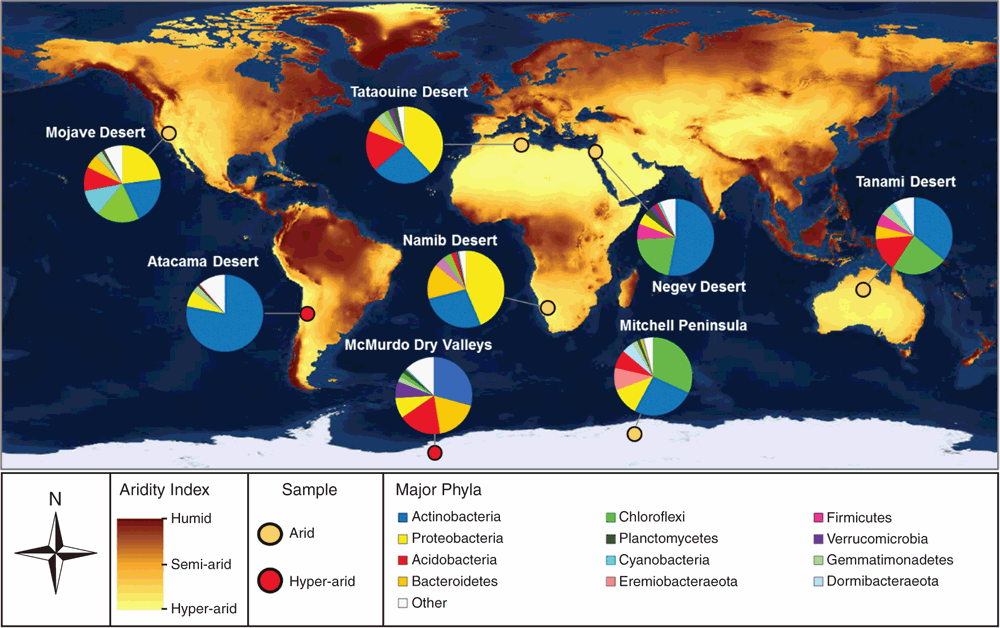
Organisms inhabiting arid and hyper-arid desert ecosystems face multiple physicochemical pressures, including water and organic carbon deficit, UV radiation damage, and often extreme temperature variations. Despite these stressors, these ecosystems host a surprising abundance and diversity of microorganisms2–4. Culture-independent surveys show microbial communities inhabiting both hot and cold deserts are similar on a phylum level to those inhabiting mesic soils, but are highly specialised at the species level and strongly shaped by physicochemical factors3,5,6. Aerobic heterotrophs from the Terrabacteria superphylum (including Actinobacteria and Chloroflexi) are particularly dominant in desert soils, with Proteobacteria, Acidobacteria, and Bacteroidetes phylotypes also common (Figure 1)7–13. It is thought that these communities are integral for supporting ecosystem services in desert regions, including nutrient turnover and fixation of carbon and nitrogen14.
|
The relative abundance and diversity of microbial taxa in desert ecosystems shows considerable variation across multiple spatial scales. This reflects both the influence of climatic factors and the inherent heterogeneity of surface soils in terms of physical structure, chemical composition, and nutrient bioavailability3,14. In desert communities, soil moisture and organic carbon content are thought to be particularly important factors driving niche processes. This reflects that organic carbon derived from photosynthetic primary production is a major energy source for the heterotrophic microorganisms that generally dominate these communities2–4. However, the combined effects of water deficit and damaging UV radiation inhibit photosynthetic processes and in turn limit primary production in arid and hyper-arid desert ecosystems. On a global scale, plant biomass per unit area is two to threefold less in drylands (6 kg km–2) compared to temperate ecosystems (10–18 kg km–2)15.
To withstand the physiochemical pressures of desert ecosystems, some photosynthetic bacteria (e.g. Cyanobacteria) and algae (e.g. Chlorophyta) have evolved cellular mechanisms to withstand the physicochemical pressures of desert ecosystems. Notably, many phototrophs can efficiently colonise cracks and fissures of translucent rocks and biological soil crusts2,16,17. These environmental refugia provide desiccation buffers and protection from UV radiation, allowing these specialised producers to fix carbon and nitrogen at sufficient rates to support associated heterotrophic communities. As a result, phototrophs are dominant primary producers in dryland ecosystems worldwide3,16. However, culture-independent studies indicate both arid and hyper-arid deserts, such as those in Atacama, Negev, and Antarctica, often harbour diverse communities of putative aerobic heterotrophic bacteria, despite very low abundances of Cyanobacteria and other phototrophs2,6,7,18. A longstanding conundrum has been how these heterotrophic bacteria sustain energy and biomass in the absence of obvious primary producers.
A minimalistic mode of primary production
Lacking obvious organic carbon inputs, most microorganisms within hyper-arid desert communities seemingly persist in various dormant states, where energy is directed towards persistence rather than growth19. While dormancy offers microorganisms a bet-hedging strategy to survive chemically and physically challenging conditions, it is not a cost-free state, as some maintenance energy is required for basic cellular functions such as macromolecular repair19,20. Through recent studies, we have provided evidence that some desert surface soil communities are structured by a minimalistic mode of primary production, where atmospheric gases, not sunlight, serve as the main energy source21.
We analysed the surface soil microbial communities of two coastal ice-free desert sites in Eastern Antarctica, Robinson Ridge and Adams Flat. Both sites had limited capacity for photosynthesis and were extremely low in organic carbon content. Despite this, they harboured diverse communities of bacteria belonging to the superphylum Terrabacteria, including Actinobacteria, Chloroflexi, and two candidate phyla, WPS-2 (Candidatus Eremiobacteraeota – desert bacterial phylum) and AD3 (Candidatus Dormibacteraeota – dormant bacterial phylum). To understand the metabolic potential of this community, shotgun metagenomics and differential coverage binning were used to construct 23 draft microbial genomes. Genes supporting energy conservation were widespread, with the majority of the bacteria encoding high-affinity lineages of the enzymes [NiFe]-hydrogenase and a carbon monoxide dehydrogenase21. Pure culture studies on multiple organisms have shown that these enzymes facilitate trace gas scavenging to support persistence of heterotrophic bacteria under organic carbon starvation22–25. Gas chromatography measurements confirmed that aerobic soil microcosms aerobically scavenged H2 and CO at rapid rates21. For dormant bacteria, atmospheric trace gases are favourable energy sources, given their ubiquity throughout the troposphere, low redox potential, and high diffusivity26.
In addition, we found that bacteria from the Actinobacteria, Eremiobacteraeota, and Dormibacteraeota clades encoded the genes for autotrophic CO2 fixation via the Calvin Benson-Bassham (CBB) cycle. We validated that the soil communities encoded and expressed type IE RuBisCO enzyme21, a recently discovered clade of the CO2-fixing enzyme that supports hydrogenotrophic growth in some Actinobacteria27 but is absent from known phototrophs. The co-occurrence of these genes with high-affinity hydrogenases and carbon monoxide dehydrogenases suggested that these communities were able to fix CO2 into biomass using atmospheric trace gases, rather than solely relying on exogenous inputs from photosynthetic organisms. To test this, we traced assimilation of 14C-labelled CO2 by these samples in microcosm experiments. We were able to demonstrate that, under H2-enriched conditions, chemosynthetic CO2 fixation increased up to tenfold. In contrast, no significant stimulation was observed following light illumination21. Based on these findings, we propose that, in desert ecosystems where photosynthetic organisms are excluded due to aridity, dormant bacterial communities are sustained by atmospheric chemosynthesis: members maintain energy and carbon needs by aerobically respiring atmospheric H2 and CO and, in some cases, using these gases to fix CO2 into biomass (Figure 2).
Aerobic gas scavengers
Pure culture studies have provided insights into the physiological role and biochemical basis of trace gas scavenging26,28. For example, our research has recently helped to resolve the biochemical basis and physiological significance of atmospheric H2 oxidation22,24,29. In the lower troposphere, H2 occurs at trace amounts (~530 ppbv) and is rapidly cycled between sources (e.g. methane photolysis, fossil fuel combustion) and sinks (i.e. bacterial scavenging, hydroxyl radical oxidation). Atmospheric H2 scavenging, in addition to being ecologically important, is of major biogeochemical significance given it is the primary sink in the global H2 cycle26,30.
To harness the energy of H2, bacteria employ specialised metalloenzymes called hydrogenases to catalyse the reversible reaction H2⇌ 2H+ + 2e–31. Historically, hydrogen metabolism was thought to primarily occur in low O2, high H2 environments such as oceanic sediments, gastrointestinal tracts, and hydrothermal systems. Reflecting this, the first isolated hydrogenotrophs had low affinities for H2, and the first structurally characterised hydrogenase enzymes were highly O2-sensitive31. However, recent studies have demonstrated that diverse soil bacteria can aerobically respire H2 even at atmospheric concentrations22,24,32. We now know of four [NiFe]-hydrogenase lineages (group 1h, 1d, 1f, 2a) that support aerobic respiration and have biochemical adaptations to function in the presence of O233,34. Of these, the group 1h [NiFe]-hydrogenase is a high-affinity enzyme that primarily mediates atmospheric H2 scavenging and is widely distributed in aerobic soil bacteria32,35,36, including those in Antarctica21.
Our recent pure culture studies have shown that the survival of bacteria belonging to dominant soil phyla such as Actinobacteria and Acidobacteria is enhanced by aerobic respiration of H2. For example, the model soil organism Mycobacterium smegmatis upregulates the expression of two high-affinity hydrogenases under carbon starvation and persists by oxidising H2 below atmospheric levels. Mutant strains, lacking the genes encoding hydrogenase structural subunits, have a 40% reduction in survival in carbon-limited batch and continuous cultures22,23,37. The physiological role of atmospheric H2 scavenging was further tested with a thermophilic isolate from an oligotrophic volcanic soil, namely Pyrinomonas methylaliphatogenes K22. Following the transition from exponential to stationary phase, this acidobacterium upregulated the expression of an eight-gene operon of the high affinity group 1h [NiFe]-hydrogenase. Depletion of its carbon sources triggered the transition to a non-replicative persistent state supported by atmospheric H2 scavenging24. Furthermore, pioneering work led by the Constant group has demonstrated that exospores of Streptomyces species express homologous enzymes and use them to support long-term survival25,32,35.
Conclusions and future directions
It is indisputable that microbial persistence requires energy. However, atmospheric substrates such as H2 and CO have long been overlooked as potential energy sources. We now have evidence that aerobic respiration of these gases is widespread and have a rationale for the adaptive advantage this offers to dormant bacteria living in conditions where persistence is favoured over growth. While trace gases serve as energy sources for bacteria in aerated soil ecosystems worldwide, they are particularly important for microbial communities in soils with low water and carbon content, where phototrophs are excluded. We have confirmed that trace gases serve as the primary energy sources supporting two Antarctic desert sites21. Moreover, there is evidence that the enzymes mediating atmospheric chemosynthesis are also encoded in other oligotrophic ecosystems, including the hyper-arid deserts of the Atacama38 and volcanic deposits of Hawaii39.
Our recent findings in the Antarctic, as well as ongoing research into trace gas scavenging, will form the basis of future investigations. We are particularly interested in answering how significant this process is in explaining microbial biodiversity and primary production in other desert ecosystems such as the Negev Desert, Israel and the Atacama Desert, Chile. A key question is how does the balance between photosynthetic primary production and chemosynthetic primary production change along aridity gradients. These ecological studies are being supported by ongoing work focused on understanding the physiology and biochemistry of trace gas scavenging using pure bacterial cultures and purified enzymes.
Acknowledgements
This work was supported by a Monash University PhD Scholarship (awarded to SB), ARC Future Fellowship (FT170100341; awarded to BF), ARC DECRA Fellowship (DE170100310; awarded to CG), and Australian Antarctic Science project grant (4406; awarded to BF and CG). We thank Steven Chown, Melodie McGeoch, Osnat Gillor, Mukan Ji, Don Cowan, Philip Hugenholtz, Paul Sunnucks, Dustin Marshall, Linda Blackall, and members of the Greening and Ferrari laboratories for helpful discussions.
References
[1] Zomer, R.J. et al. (2008) Climate change mitigation: a spatial analysis of global land suitability for clean development mechanism aforestation and reforestation. Agric. Ecosyst. Environ. 126, 67–80.| Climate change mitigation: a spatial analysis of global land suitability for clean development mechanism aforestation and reforestation.Crossref | GoogleScholarGoogle Scholar |
[2] Cary, S.C. et al. (2010) On the rocks: the microbiology of Antarctic Dry Valley soils. Nat. Rev. Microbiol. 8, 129–138.
| On the rocks: the microbiology of Antarctic Dry Valley soils.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXlvFGnug%3D%3D&md5=f492394c7ec3f777a3f863558d774908CAS |
[3] Caruso, T. et al. (2011) Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 5, 1406–1413.
| Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale.Crossref | GoogleScholarGoogle Scholar |
[4] Makhalanyane, T.P. et al. (2015) Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 39, 203–221.
| Microbial ecology of hot desert edaphic systems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhs1ans7vE&md5=c7679beeaf794ddcf381ecf326b95667CAS |
[5] Lee, C.K. et al. (2012) The Inter-Valley Soil Comparative Survey: the ecology of Dry Valley edaphic microbial communities. ISME J. 6, 1046–1057.
| The Inter-Valley Soil Comparative Survey: the ecology of Dry Valley edaphic microbial communities.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XlvVGksbk%3D&md5=874a014cfd3305e7f916650837e358adCAS |
[6] Angel, R. et al. (2010) Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 4, 553–563.
| Biogeography of soil archaea and bacteria along a steep precipitation gradient.Crossref | GoogleScholarGoogle Scholar |
[7] Ji, M. et al. (2015) Microbial diversity at Mitchell Peninsula, Eastern Antarctica: a potential biodiversity ‘hotspot’. Polar Biol. 33, 237–249.
[8] Fierer, N. et al. (2012) Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA 109, 21390–21395.
| Cross-biome metagenomic analyses of soil microbial communities and their functional attributes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXotVWgtA%3D%3D&md5=7029db20f9b1d4b538027d32195121cfCAS |
[9] Neilson, J.W. et al. (2017) Significant impacts of increasing aridity on the arid soil microbiome. mSystems 2, e00195-16.
| Significant impacts of increasing aridity on the arid soil microbiome.Crossref | GoogleScholarGoogle Scholar |
[10] Ronca, S. et al. (2015) Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities. Front. Microbiol. 6, 845.
| Namib Desert dune/interdune transects exhibit habitat-specific edaphic bacterial communities.Crossref | GoogleScholarGoogle Scholar |
[11] Chanal, A. et al. (2006) The desert of Tataouine: an extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria. Environ. Microbiol. 8, 514–525.
| The desert of Tataouine: an extreme environment that hosts a wide diversity of microorganisms and radiotolerant bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XjsFOqsbk%3D&md5=377fc1293588a600be0721c6fdefb536CAS |
[12] Mogul, R. et al. (2017) Microbial community and biochemical dynamics of biological soil crusts across a gradient of surface coverage in the central Mojave desert. Front. Microbiol. 8, 1974.
| Microbial community and biochemical dynamics of biological soil crusts across a gradient of surface coverage in the central Mojave desert.Crossref | GoogleScholarGoogle Scholar |
[13] Belbin, L. and Williams, K.J. (2016) Towards a national bio-environmental data facility: experiences from the atlas of living Australia. Int. J. Geogr. Inf. Sci. 30, 108–125.
| Towards a national bio-environmental data facility: experiences from the atlas of living Australia.Crossref | GoogleScholarGoogle Scholar |
[14] Fierer, N. (2017) Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579.
| Embracing the unknown: disentangling the complexities of the soil microbiome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhtlGktbfN&md5=b79aeb90c64d09f110fd78ab6a3d9586CAS |
[15] Ebeling, J. and Yasué, M. (2008) Generating carbon finance through avoided deforestation and its potential to create climatic, conservation and human development benefits. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 1917–1924.
| Generating carbon finance through avoided deforestation and its potential to create climatic, conservation and human development benefits.Crossref | GoogleScholarGoogle Scholar |
[16] Bahl, J. et al. (2011) Ancient origins determine global biogeography of hot and cold desert cyanobacteria. Nat. Commun. 2, 163.
| Ancient origins determine global biogeography of hot and cold desert cyanobacteria.Crossref | GoogleScholarGoogle Scholar |
[17] Christian, K. et al. (2017) Spatial patterns of hypolithic cyanobacterial diversity in Northern Australia. Ecol. Evol. 7, 7023–7033.
| Spatial patterns of hypolithic cyanobacterial diversity in Northern Australia.Crossref | GoogleScholarGoogle Scholar |
[18] Crits-Christoph, A. et al. (2013) Colonization patterns of soil microbial communities in the Atacama Desert. Microbiome 1, 28.
| Colonization patterns of soil microbial communities in the Atacama Desert.Crossref | GoogleScholarGoogle Scholar |
[19] Lennon, J.T. and Jones, S.E. (2011) Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9, 119–130.
| Microbial seed banks: the ecological and evolutionary implications of dormancy.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXlsFeguw%3D%3D&md5=9a9a83cda86710bd50d791c3c16edc6aCAS |
[20] Price, P.B. and Sowers, T. (2004) Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc. Natl. Acad. Sci. USA 101, 4631–4636.
| Temperature dependence of metabolic rates for microbial growth, maintenance, and survival.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXjtFKisLs%3D&md5=821189090e3bdd0dca2c4ea5f30cff5dCAS |
[21] Ji, M. et al. (2017) Atmospheric trace gases support primary production in Antarctic desert surface soil. Nature 552, 400–403.
| Atmospheric trace gases support primary production in Antarctic desert surface soil.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXhvFGlsbrK&md5=5d29fdab1187f4112907c4c42e16e2b8CAS |
[22] Greening, C. et al. (2014) A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases. Proc. Natl. Acad. Sci. USA 111, 4257–4261.
| A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXjtlynsLw%3D&md5=98909848907bda0a9ae56623cace3ea5CAS |
[23] Greening, C. et al. (2014) The growth and survival of Mycobacterium smegmatis is enhanced by co-metabolism of atmospheric H2. PLoS One 9, e103034.
| The growth and survival of Mycobacterium smegmatis is enhanced by co-metabolism of atmospheric H2.Crossref | GoogleScholarGoogle Scholar |
[24] Greening, C. et al. (2015) Persistence of the dominant soil phylum Acidobacteria by trace gas scavenging. Proc. Natl. Acad. Sci. USA 112, 10497–10502.
| Persistence of the dominant soil phylum Acidobacteria by trace gas scavenging.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXht1ygt73E&md5=ebbde2186c2714137dd3555e4ca4e64cCAS |
[25] Liot, Q. and Constant, P. (2016) Breathing air to save energy – new insights into the ecophysiological role of high-affinity [NiFe]-hydrogenase in Streptomyces avermitilis. MicrobiologyOpen 5, 47–59.
| Breathing air to save energy – new insights into the ecophysiological role of high-affinity [NiFe]-hydrogenase in Streptomyces avermitilis.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XjsFeit74%3D&md5=c9f28fde14e8522564e42ad66b44e3b7CAS |
[26] Greening, C. et al. (2015) Atmospheric hydrogen scavenging: from enzymes to ecosystems. Appl. Environ. Microbiol. 81, 1190–1199.
| Atmospheric hydrogen scavenging: from enzymes to ecosystems.Crossref | GoogleScholarGoogle Scholar |
[27] Grostern, A. and Alvarez-Cohen, L. (2013) RubisCO-based CO2 fixation and C1 metabolism in the actinobacterium Pseudonocardia dioxanivorans CB1190. Environ. Microbiol. 15, 3040–3053.
| 1:CAS:528:DC%2BC3sXhs12ls7vF&md5=a1d2d82ff4d1eacead71e3c66910f33fCAS |
[28] King, G.M. and Weber, C.F. (2007) Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5, 107–118.
| Distribution, diversity and ecology of aerobic CO-oxidizing bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXkslCgtw%3D%3D&md5=b364b9201bed6dc78e74ae63300233e8CAS |
[29] Greening, C. and Cook, G.M. (2014) Integration of hydrogenase expression and hydrogen sensing in bacterial cell physiology. Curr. Opin. Microbiol. 18, 30–38.
| Integration of hydrogenase expression and hydrogen sensing in bacterial cell physiology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXmslaksbk%3D&md5=26751ebf3e19d692d2b9b222e8c75441CAS |
[30] Constant, P. et al. (2009) Tropospheric H2 budget and the response of its soil uptake under the changing environment. Sci. Total Environ. 407, 1809–1823.
| Tropospheric H2 budget and the response of its soil uptake under the changing environment.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXit1SjsrY%3D&md5=3d8d68b2aa43aa6ac2f1946c8c35db1fCAS |
[31] Schwartz, E. et al. (2013) H2-metabolizing prokaryotes, Springer Berlin Heidelberg.
[32] Constant, P. et al. (2010) Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase. Environ. Microbiol. 12, 821–829.
| Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXkslSqtLg%3D&md5=6217d8086854e1ecbb0998d44dec8d2fCAS |
[33] Søndergaard, D. et al. (2016) HydDB: a web tool for hydrogenase classification and analysis. Sci. Rep. 6, 34212.
| HydDB: a web tool for hydrogenase classification and analysis.Crossref | GoogleScholarGoogle Scholar |
[34] Myers, M.R. and King, G.M. (2016) Isolation and characterization of Acidobacterium ailaaui sp. nov., a novel member of Acidobacteria subdivision 1, from a geothermally heated Hawaiian microbial mat. Int. J. Syst. Evol. Microbiol. 66, 5328–5335.
| Isolation and characterization of Acidobacterium ailaaui sp. nov., a novel member of Acidobacteria subdivision 1, from a geothermally heated Hawaiian microbial mat.Crossref | GoogleScholarGoogle Scholar |
[35] Constant, P. et al. (2011) Genome data mining and soil survey for the novel Group 5 [NiFe]-hydrogenase to explore the diversity and ecological importance of presumptive high-affinity H2-oxidizing bacteria. Appl. Environ. Microbiol. 77, 6027–6035.
| Genome data mining and soil survey for the novel Group 5 [NiFe]-hydrogenase to explore the diversity and ecological importance of presumptive high-affinity H2-oxidizing bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1OjurnM&md5=5beea14d00147b1a4f0229bbe4eefe8cCAS |
[36] Schäfer, C. et al. (2016) Structure of an actinobacterial-type [NiFe]-hydrogenase reveals insight into O2-tolerant H2 oxidation. Structure 24, 285–292.
| Structure of an actinobacterial-type [NiFe]-hydrogenase reveals insight into O2-tolerant H2 oxidation.Crossref | GoogleScholarGoogle Scholar |
[37] Berney, M. and Cook, G.M. (2010) Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia. PLoS One 5, e8614.
| Unique flexibility in energy metabolism allows mycobacteria to combat starvation and hypoxia.Crossref | GoogleScholarGoogle Scholar |
[38] Lynch, R.C. et al. (2014) Metagenomic evidence for metabolism of trace atmospheric gases by high-elevation desert Actinobacteria. Front. Microbiol. 5, 698.
| Metagenomic evidence for metabolism of trace atmospheric gases by high-elevation desert Actinobacteria.Crossref | GoogleScholarGoogle Scholar |
[39] Nanba, K. et al. (2004) Analysis of facultative lithotroph distribution and diversity on volcanic deposits by use of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase. Appl. Environ. Microbiol. 70, 2245–2253.
| Analysis of facultative lithotroph distribution and diversity on volcanic deposits by use of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXjtlOiu7o%3D&md5=d7b40dd8acd38c34a3eec53b51b97afeCAS |
Biographies
Sean Bay is a second year PhD student under the primary supervision of Dr Chris Greening at Monash University. He is investigating how microbial biodiversity and primary production strategies are influenced by aridity.
Dr Ferrari is an Associate Professor and ARC Future Fellow at UNSW Sydney. Her major interest is understanding the microbial diversity of soil bacteria, particularly in underexplored regions of Antarctica.
Dr Chris Greening is a Lecturer and ARC DECRA Fellow at the School of Biological Sciences and Centre for Geometric Biology, Monash University. He is a biochemist-turned-microbiologist investigating the metabolic basis of bacterial persistence.