Emerging microbiome technologies for sustainable increase in farm productivity and environmental security
Brajesh K Singh A B E , Pankaj Trivedi C , Saurabh Singh D , Catriona A Macdonald A and Jay Prakash Verma A DA Hawkesbury Institute for the Environment, Western Sydney University, Penrith, NSW, Australia
B Global Centre for Land-Based Innovation, Western Sydney University, Penrith, NSW, Australia
C Bioagricultural Sciences and Pest Management, Colorado State University, Fort Collins, CO, USA
D Institute of Environment and Sustainable Development, Banaras Hindu University, Varanasi-221005, U.P. India
E Tel: +61 2 4570 1329, Email: b.singh@westernsydney.edu.au
Microbiology Australia 39(1) 17-23 https://doi.org/10.1071/MA18006
Published: 21 February 2018
Farming systems are under pressure to sustainably increase productivity to meet demand for food and fibre for a growing global population under shrinking arable lands and changing climatic conditions. Furthermore, conventional farming has led to declines in soil fertility and, in some cases, inappropriate and excessive use of chemical fertilisers and pesticides has caused soil degradation, negatively impacting human and environmental health. The soil and plant microbiomes are significant determinants of plant fitness and productivity. Microbes are also the main drivers of global biogeochemical cycles and thus key to sustainable agriculture. There is increasing evidence that with development of appropriate technologies, the plant microbiome can be harnessed to potentially decrease the frequency of plant diseases, increase resource use efficiencies and ultimately enhance agricultural productivity, while simultaneously decreasing the input of chemical fertilisers and pesticides, resulting in reduced greenhouse gas emissions and promoting environmental sustainability. However, to successfully translate potential to practical outcomes, both fundamental and applied research are needed to overcome current constraints. Research efforts need to be embedded in industrial requirements and policy and social frameworks to expedite the process of innovation, commercialisation and adoption. We propose that learning from the advancement in the human microbiome can significantly expedite the discovery and innovation of effective microbial products for sustainable and productive farming. This article summarises the emergence of microbiome technologies for the agriculture industry and how to facilitate the development and adoption of environmentally friendly microbiome technologies for sustainable increase in farm productivity.
The global population is expected to reach 9 billion by 2050 and an increase of 70–100% in farm productivity is needed to meet the demand for food and fibre. This increase in agriculture productivity needs to be met from a shrinking arable land area due to multiple demands (e.g. food, fuel, fibre and climate mitigation) and land-degradation. Current farming practices that use chemical fertilisers and pesticides have contributed significantly to increase farm productivity but have also contributed, in some cases, to chemical contamination, soil degradation, loss of biodiversity and compromised soil and water quality, which together impact overall environmental sustainability and can impact human health. Agriculture productivity faces additional major challenges including structural decline in soil fertility (i.e. increase in inputs does not result in proportional yield gain) and negative impact of climate change including extreme weather events1,2. Emerging microbiome approaches have potential to address most of these challenges as a complimentary approach to conventional farming.
The plant microbiome, which consists of microbiota associated with all plant compartments (e.g. root, stem, leaves, flowers, seed), many of which have a wide and beneficial impact on plant fitness and productivity, if exploited appropriately, can boost agricultural productivity and environmental outcomes. The plant microbiome affects host physiology and productivity by improving resistance to biotic (e.g. disease and pest attack) and abiotic (e.g. nutrient and water limitation, heavy metal contamination) stresses3. The plant microbiome is immensely diverse and comprises mainly mutualistic partners where microbes receive carbon and habitats in return for supply nutrients and defense provision against plant pests and pathogens. Manipulating the plant microbiome has great potential to increase farm productivity by enhancing resource (e.g. water and nutrients) use efficiency and reducing the impact of disease and pest incidences. Because plant and microbial associations have evolved together for millions of years, they have well-developed mutual recognition, association and communication mechanisms4. Identifying drivers of microbial assembly and communication molecules can therefore significantly advance our ability to manipulate microbiomes for better outcomes.
Concept of core and hub microbiome
There is growing evidence that different plant species harbor distinct microbiomes that are significant determinants of their survival and fitness3. However, not all plant microbiota are beneficial. In fact, a significant proportion are opportunistic microbes which are there only to exploit available nutrients and a small number are plant pathogens3,5, which may dominate under certain environmental conditions and limit crop productivity. Therefore, identifying beneficial microbes is a critical first step to harness them to sustainably increase farm productivity. In this regard, applying the core microbiome (persistent members of microbiota that appear in all communities associated with a particular crop or plant species under different environments and management practices) approach is gaining scientific attention6,7. The core microbiome is considered a critical component for essential functions for holobionts (i.e. plant plus microbiota) as they are enriched, selected and inherited by evolutionary steps6. The core microbiome of a number of crops including maize, barley, rice, soybean, lettuce, and sugarcane have been reported7–9 with some taxa present in most of the studied crop hosts. However, few biogeography studies have questioned the universal distribution of taxonomic core microbiome under various environmental conditions. It has been suggested that the microbiota recruited by a given plant genotype in different environments seems to share greater functional than taxonomic similarity6. According to this view, elucidating a functional core microbiome either by directly looking for functional attributes (using metagenomics such as shotgun sequencing) or indirectly through taxonomic information (using methods such as PICRUSt and Tax4Fun) will provide better understanding of the role of the microbiome in plant performance and health that can be harnessed for improving farm productivity for multiple crops.
The identification of a core microbiome of various crop species will help to identify plant-associated microbes that should be prioritised for further research, inclusion in culture collections, and manipulative experiments to improve crop productivity further. Fast moving omics technologies (genomics, metagenomics, metatranscriptomics and metaproteomics) can provide the information on key functional roles of uncultivable microbes within the plant microbiome10 and identify those that are adaptive to environmental pressures. Comparison of the core microbiomes between plant species and genotypes within a species reveals host-driven differences in microbiome assembly. The mechanisms by which hosts assemble microbial community are not fully understood, although plant biochemical traits such as hormones, secondary metabolites, cuticle composition, root length and exudates, and plant defences (immunity) have been identified as important determinants. Because the crop microbiome, plant phenotype, and environment interact to affect yield, comparing the microbiomes of plants grown in contrasting environments can potentially provide key insights of the microbial role in plant fitness. Microbes that are especially common in challenging environments are more likely to protect yield under biotic and abiotic stresses. The core microbiome still contains hundreds of microbial ‘species’ and therefore, it is logistically difficult to manipulate the systems. To address this challenge, a ‘hub microbiota approach’ has been used11. This approach is based on the concept that microbiomes are a complex and inter-connected network where different populations have different roles and some ‘keystone or hub species’ are crucial for maintenance of the functioning network7,11,12. The finding that the effect of host and abiotic factors can cascade through communities via ‘hub’ microbes is important to understand the fluctuations in community structure and functions that can be linked to plant performance. Theoretically, these hub species are highly interconnected and centre of the microbial network and therefore, changing any of the hub microbiome can have a significant impact on the core and overall microbiome of plant species. Thus hub microbiota are prime targets for in situ manipulation of the crop microbiome for better productivity and environmental outcomes.
Current status and challenges
Use of microbes for agriculture has been practiced for several decades, mainly in the form of bio-fertilisers and bio-pesticides. These are mainly one-species products that either provide nutrients, particularly nitrogen (e.g. use of symbiotic rhizobia or free-living Azotobacter), mobilise phosphorus (e.g. Penicillium species) or protect against pests; insect (e.g. Bacillus thurengenesis) or fungus (e.g. Trichoderma viride). In recent years, a number of start-up companies (e.g. Indigo Ag, Chr-Hansen, NewLeaf Symbiotics, Growcentia) and large multi-national companies (e.g. Bayer Ltd, Nufarm, Monsanto BioAg) have commercialised microbial products for enhancing farm productivity. In fact, microbial products are one of the fastest growing start-ups and are expected to have global market of $6.4 billion by 2022. It is estimated by 2020, there will be more bio-pesticides in the European market than chemical pesticides and within the next few years, microbial products will have complementary markets of chemical pesticides ($55 billion1). This projection is based on the fact that current microbial products are based on a small proportion of cultivable species (~5% of the total microbiome) and biochemical characterisation of whole microbiomes for agriculture products is in its infancy. Further, the majority of cultivable microbes have yet to be explored for their plant beneficial activities. For example, more than 50% of human medicines come from natural resource13 but only 11% of pesticides have biological origin, suggesting that most pesticidal properties from microbes are yet to be discovered. These possibilities have attracted significant investments from both government agencies and private companies. However, to realise the full potential, a number of technical, regulatory and social challenges need to be addressed.
The technical challenges are significant. First, we are unable to cultivate most environmental microbes (>95%) and that means most microbial metabolisms involved in plant health are not yet characterised. This heavily constrains our ability to harness them for agriculture productivity. Second, the performance of microbial products in field conditions has been mixed and in some cases effective performance in greenhouse studies was not replicated in field conditions. In several cases microbial products were not able to colonise plants or were outcompeted by indigenous microflora. Sustaining the efficiency of microbial products for the duration of the crop cycle is another major challenge. Activities of several microbes are influenced by abiotic (e.g. soil types, pH), climatic (e.g. drought) and biotic (e.g. competition with indigenous microflora, recognition of host-microbial interactions) conditions. Microbes not only need to survive but colonise crop plants and maintain activities for at least the duration of the crop cycle. In other words, microbial products, in several cases, perform short of the gold standard for industry, i.e. works in all environmental conditions and at all crop stages. These are serious challenges that need to be overcome if microbial products can be used alongside or as a substitute to agrochemicals. In addition to these technical challenges, different collection and analysis techniques, reagents, and parameters may introduce variations in microbiome results further compounding the biologically relevant role of the microbiome in practical settings14. Equally important, with the present ‘microbiome potential’, it should be emphasised that the structure and function of the microbiome are only one component in the multi-trophic cascades that determines host response. Thus, only an integrative multivariable approach, which integrates the physiology and genetics of both host and microbiome, as well as other environmental variables (including stress such as drought), may ensure that microbiome-based approaches are implemented to their fullest potential to influence plant production and health.
Two key approaches for harnessing the plant microbiomes
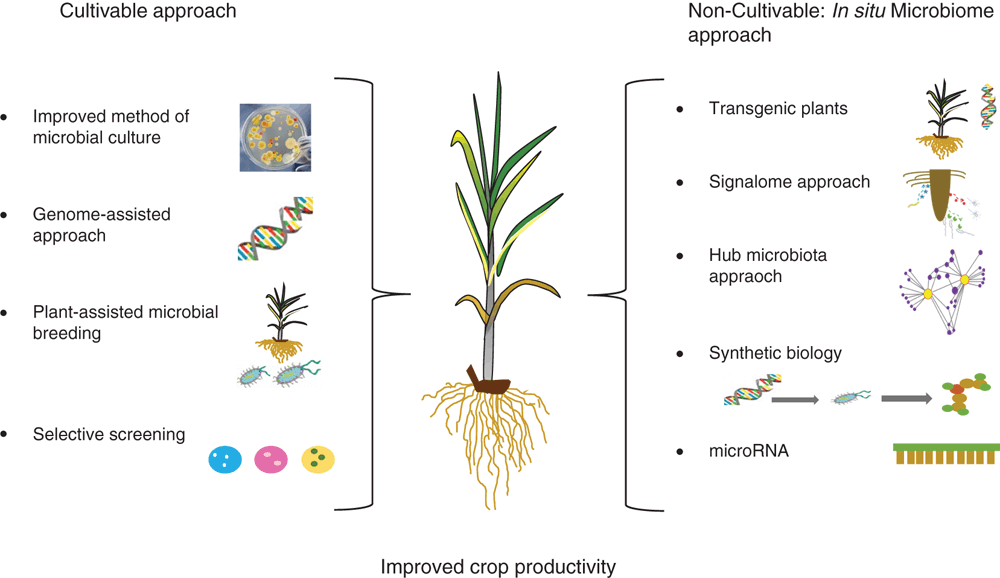
A simplified approach for harnessing plant microbiomes (we used this term both for isolated consortium and in situ microbiome) is outlined in Figure 1. First, characterisation of plant beneficial microbes can be achieved by the isolation from the rhizosphere, phyllosphere or endosphere. Isolates can be screened for their plant growth-promoting properties, and interspecific interaction assessed. Through selection of those microbes that demonstrate synergistic interactions between each other and with plants (as opposed to those that are antagonistic), a core microbiome that leads to plant and environmental benefits can be identified and harnessed directly.
|
Improvised traditional approaches
Traditional methods of microbial screening and isolation and their use in agriculture have provided some success both in nutrient supply and pest management. However, improvement in isolation and screening methods has been frustratingly slow and we are still not able to culture the vast majority of microbes. Some progresses have been made, for example in formulating optimised media (e.g. shell vial procedure)15, automated sorting and imaging techniques16, and use of helper strains17 to cultivate novel microbes from complex environmental settings. Emerging technologies such as genomics have allowed the cultivation of previously uncultivable microbes by identifying special nutritional or co-factor requirements18. Bai et al.19 have shown that through selective screening protocols a culture collection of microbes can be generated that represents the majority of bacterial species that are reproducibly detectable by culture independent community sequencing. As the number of genomes obtained from binning of metagenomics sequence data is rapidly growing, this genome-assisted cultivation approach has potential to significantly improve microbial cultivation fields. Success of microbial products in field conditions can be enhanced either by the improvement of strains or using local microflora which are adapted to a particular region. Furthermore, plant-assisted microbial breeding can improve the mutual recognition of host and microbial products that can help the colonisation in field conditions.
Use of endophytes provides another avenue for better efficacy, particularly if the endophyte can colonise in the early stage of crop development. In such scenarios, the competition with indigenous microflora is minimised, which improves their ability to maintain activities. For example, Mitter et al.20 discovered that the introduction of beneficial endophytes to the flower of parent plants can drive its inclusion in the progeny seed microbiome, thereby inducing vertical inheritance to the offspring generations. There is growing evidence that the use of consortia provides better performance than single species products and future products should target this approach where multiple species can be harnessed1,21. However, it is important to examine the synergy of survival, lifestyle and activities of individual members for successful outcomes under field conditions. The success of these and probiotic approaches (see below) depends on addressing key fundamental questions, i.e. identification of requirements of recognition, colonisation, persistence, and continuous activities of introduced microbiota. This is a critical knowledge gap that needs to be addressed in order to provide consistent efficacy of microbial products. Framing ‘invasion ecological theory’ in creating unique niches for the introduced microbes can be useful to address this challenge.
Emerging approaches to manipulate plant microbiome in situ
A number of approaches are being currently developed and trialled to harness the whole plant microbiome without a need to culture, including use of transgenic or more conventional approaches. For example, using engineered plants with root traits that stimulate beneficial microbes such as mycorrhiza; nitrogen fixers, phosphorous, potassium and zinc solubilisers; siderophore and phyto-hormone producing microbes can have direct positive impacts on farm productivities22. However, given the public perception of transgenic plants, use of this approach for food crops remains limited. Other non-transgenic approaches which can manipulate the microbiome in situ are gaining more attention. These include:
-
exploiting plant-microbial communication molecules: plants and microbes both produce a number of communication molecules to communicate their requirements to their partners. For example, when a plant is starved of phosphorus, it produces signal molecules which rhizosphere microbes respond to by upregulating their phosphorus-mobilising genes23. Similar signaling mechanisms are also evident for attack of pathogens and pests4. Plants also communicate with each other through volatile organic compounds (VOCs) to induce responses that facilitate colonisation with beneficial microbes24. However, given the extremely low quantity of signal molecules produced, only a few such molecules have been characterised. If the detection and characterisation of signal molecules can be improved, it will provide an important tool to introduce a directional change in microbial activities which is beneficial for plant performance.
-
use of microbial cocktails, which does not have direct beneficial impacts on plants, can increase the activity of indigenous plant-beneficial microflora. These cocktails mainly contain microbes with high amounts of signal molecules.
-
identification of hub microbiota of crop species, and their role in microbiome assembly and activities, provides an important tool to manipulate the whole microbiome in situ.
-
synthetic biology provides another important tool to engineer novel and predictable functions in crop probiotics, which upon release on a plant can enhance the activities of beneficial microbes in a predictable fashion.
-
in situ genome engineering tools25 can be used to directly engineer the genome of the in situ microbiome. Here mobile genetic materials (e.g. plasmid) can be delivered to indigenous microflora where they promote desired and directional functions.
-
plants have micro-RNA (miRNA) which is responsible for regulating the structure and function of plant-microbe interactions and the rhizosphere microbiome. This has been utilised to restore healthy digestive systems26 and could be a significant tool to target beneficiary microbes for enhanced crop production.
We envisage that the use of microfluidics-based technologies will be instrumental in providing unique insights into the microscale plant-microbiome interactions in complex root microenvironments by allowing dynamic imaging of these interactions. This technology is likely to enhance the current rate of discoveries in the field of microbiome research27 with tremendous applications towards harnessing beneficial interactions in large field settings. All above emerging technologies in combination with ecological engineering (use of management tools such as crop rotation, non-tillage) and plant breeding (e.g. the integration of the plant breeding with a particular microbiome) has a significant potential to manipulate host microbiomes to enhance the efficiency of controlling plant diseases and increasing farm productivity2. However, these technologies are still in their infancy and need to be further tested for their efficacy under field conditions as well as for any non-intended impact, for example, negative impact on overall environmental outcomes.
Learning from the human microbiome and future perspectives
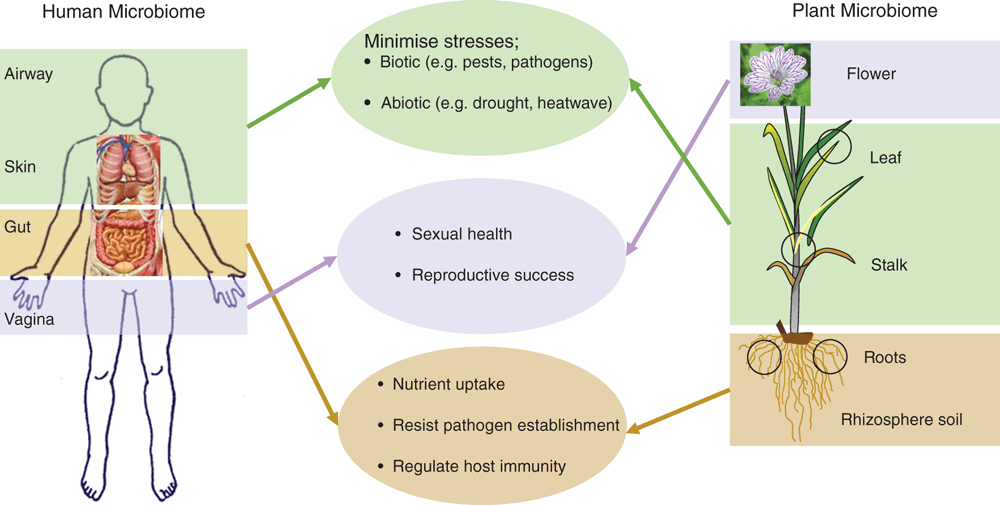
The fundamental principle of microbial assembly in humans and plants is identical and is based on selection enrichment and evolutionary processes and there are important similarities between plant and human microbiomes in their functional roles. There is growing evidence that human microbiomes play an essential role in physiological, emotional and evolutionary aspects and therefore overall health and fitness of humans. Plant microbiomes have a similar role for overall plant fitness and health (Figure 2). Learning from the advancement in human microbiome research (which is at a significantly more advanced stage) can significantly expedite discovery and innovations in agricultural microbiology because there are striking similarities in the functional role between human and plant microbiomes (Figure 2). For example, there is increasing evidence that dysregulation of human and microbiota is associated with multiple human diseases including diabetes, colorectal cancer, liver cirrhosis28. The essential role of the gut microbiome in effectiveness of cancer chemo- and immunotherapy has been found and recent studies suggest that humans can be grouped to responsive and non-responsive groups of therapy based on their intestinal microbiomes. More importantly, transfer of microbiomes from responsive to non-responsive groups can improve the efficacy of the cancer medicines29, indicating the direct role of microbiota. A similar framework can be developed to identify the mechanisms of pesticide resistance in weeds, insects and pathogens and intervention can be developed (e.g. pesticide + responsive microbiota) for effective pest control with minimal use of chemical or biological pesticides. Similarly, the role of gut and genital microbiomes in enteric and HIV infections is well documented30, and if the key (core and hub) microbiota of a crop species that protects or promotes immunity against pathogens can be identified, an appropriate intervention (e.g. microbial cocktails, probiotics, microbial transplant) can be developed to minimise the rate of infections and hence improve farm productivity. In human microbiome research, the next avenue is the utilisation of the microbiome information to assist personalised diagnostic assessment, risk stratification, disease prevention, and treatment-decision-making14. Once this concept is developed and successfully implemented, it can also be used for tailor-made microbiome interventions in context based situations for increased plant performance and health.
Conclusion
The microbiome approaches provide significant opportunities to increase farm productivity in an environmentally sustainable way. However, plant microbiome research is still in its infancy and further research is needed to advance both theoretical and experimental frameworks in order to convert potential into reality. Systematic and concerted efforts are required to identify core and hub microbiota of important crop species and how they respond to biotic and abiotic stresses. Although use of microbial products has been growing rapidly, transformational changes in the industry will come from our ability to manipulate the whole microbiome in situ. There are a number of technologies being developed but a major challenge will be efficacy of these technologies in field conditions. Integrating effective microbiome approaches with emerging precision agriculture, synthetic biology, satellite, big data and genomic approaches can provide a strong framework to realise the true potential of plant microbiome technologies in agriculture and environmental sectors. With these challenges met, incorporating microbiome-related interventions for increasing plant productivity in an environmentally sustainable way, by promoting resilience/resistance to abiotic and biotic stresses may emerge as an integral part of modern agriculture.
Acknowledgements
BKS acknowledge funding from Australian Research Council (DP170104634). JPV is grateful to DBT for providing the fellowship of Indo-Australia Carrier Boosting Gold Fellowship for working on microbiome research at Hawkesbury Institute for the Environment, Western, Sydney University, Penrith, NSW, Australia. PT acknowledges funding from Cutrale Citrus Industry and National Institute for Food and Agriculture, USA.
References
[1] Singh, B.K. and Trivedi, P. (2017) Microbiome and the future for food and nutrient security. Microb. Biotechnol. 10, 50–53.| Microbiome and the future for food and nutrient security.Crossref | GoogleScholarGoogle Scholar |
[2] Trivedi, P. et al. (2017) Tiny microbes, big yields: enhancing food crop production with biological solutions. Microb. Biotechnol. 10, 999–1003.
| Tiny microbes, big yields: enhancing food crop production with biological solutions.Crossref | GoogleScholarGoogle Scholar |
[3] Mendes, R. et al. (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663.
| The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsVSqtLrP&md5=6526bb0d00524382d13846cc526cbab2CAS |
[4] Leach, J.E. et al. (2017) Communication in the phytobiome. Cell 169, 587–596.
| Communication in the phytobiome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXnt1Cmsb8%3D&md5=e1afc132f5dfe69ee7c9fcffbcfd71aaCAS |
[5] Berg, G. et al. (2016) The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002.
| The plant microbiome explored: implications for experimental botany.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28Xhs1Oqtb3I&md5=aed1a25870693cdcfe93a29abf954aeeCAS |
[6] Lemanceau, P. et al. (2017) Let the core microbiota be functional. Trends Plant Sci. 22, 583–595.
| Let the core microbiota be functional.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXnsl2gu7k%3D&md5=01deb7553fdd1e80a859776784348407CAS |
[7] Hamonts, K. et al. (2018) Field study reveals core plant microbiota and relative importance of their drivers. Environ. Microbiol. , .
| Field study reveals core plant microbiota and relative importance of their drivers.Crossref | GoogleScholarGoogle Scholar |
[8] Peiffer, J.A. et al. (2013) Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 110, 6548–6553.
| Diversity and heritability of the maize rhizosphere microbiome under field conditions.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXnvVOlu7o%3D&md5=41704d798641490428fbfbcd21f09cb9CAS |
[9] Edwards, J. et al. (2015) Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 112, E911–E920.
| Structure, variation, and assembly of the root-associated microbiomes of rice.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtFKhsbw%3D&md5=2a397048da354667302daeabde30c240CAS |
[10] Vandenkoornhuyse, P. et al. (2015) The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206.
| The importance of the microbiome of the plant holobiont.Crossref | GoogleScholarGoogle Scholar |
[11] Agler, M.T. et al. (2016) Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14, e1002352.
| Microbial hub taxa link host and abiotic factors to plant microbiome variation.Crossref | GoogleScholarGoogle Scholar |
[12] Trivedi, P. et al. (2017) Keystone microbial taxa regulate the invasion of a fungal pathogen in agro-ecosystems. Soil Biol. Biochem. 111, 10–14.
| Keystone microbial taxa regulate the invasion of a fungal pathogen in agro-ecosystems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXlsVCitbY%3D&md5=4a87a80e0e3c3aea273cdc1d8c3a46ecCAS |
[13] Singh, B.K. and Macdonald, C.A. (2010) Drug discovery from uncultivable microbes. Drug Discov. Today 15, 792–799.
| Drug discovery from uncultivable microbes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtFCrurfN&md5=49709402aecb99e4708a58b2cac3463dCAS |
[14] Zmora, N. et al. (2016) Taking it personally: personalised utilization of the human microbiome in health and disease. Cell Host Microbe 19, 12–20.
| Taking it personally: personalised utilization of the human microbiome in health and disease.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhvFCltA%3D%3D&md5=a5603261adb6141f7bd6b5c28d2f5fd5CAS |
[15] Lagier, J.-C. et al. (2015) The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 28, 237–264.
| The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota.Crossref | GoogleScholarGoogle Scholar |
[16] Dong, B. et al. (2016) Superresolution intrinsic fluorescence imaging of chromatin utilizing native, unmodified nucleic acids for contrast. Proc. Natl. Acad. Sci. USA 113, 9716–9721.
| Superresolution intrinsic fluorescence imaging of chromatin utilizing native, unmodified nucleic acids for contrast.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhtlGmtrfF&md5=44a0c0a4407d8c9e3e59711c2eec0e8eCAS |
[17] Nichols, D. et al. (2008) Short peptide induces an ‘uncultivable’ microorganism to grow in vitro. Appl. Environ. Microbiol. 74, 4889–4897.
| Short peptide induces an ‘uncultivable’ microorganism to grow in vitro.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXpslKktLc%3D&md5=b33b4d18e04c9eefd9462cfb47ca0145CAS |
[18] Garza, D.R. and Dutilh, B.E. (2015) From cultured to uncultured genome sequences: metagenomics and modelling microbial ecosystems. Cell. Mol. Life Sci. 72, 4287–4308.
| From cultured to uncultured genome sequences: metagenomics and modelling microbial ecosystems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtlSntbzN&md5=556e9f308d1b1fdade401795b03478e1CAS |
[19] Bai, Y. et al. (2015) Functional overlap of the arabidopsis leaf and root microbiota. Nature 528, 364–369.
| Functional overlap of the arabidopsis leaf and root microbiota.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhvFemsbjL&md5=004aa48fa95e00231358a2b44f3cdc37CAS |
[20] Mitter, B. et al. (2017) A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 8, 11.
| A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds.Crossref | GoogleScholarGoogle Scholar |
[21] Wallenstein, M.D. (2017) Managing and manipulating the rhizosphere microbiome for plant health. A systems approach. Rhizosphere 3, 230–232.
| Managing and manipulating the rhizosphere microbiome for plant health. A systems approach.Crossref | GoogleScholarGoogle Scholar |
[22] Macdonald, C. and Singh, B. (2014) Harnessing plant-microbe interactions for enhancing farm productivity. Bioengineered 5, 5–9.
| Harnessing plant-microbe interactions for enhancing farm productivity.Crossref | GoogleScholarGoogle Scholar |
[23] Zysko, A. et al. (2012) Transcriptional response of Pseudomonas aeruginosa to a phosphate-deficient Lolium perenne rhizosphere. Plant Soil 359, 25–44.
| Transcriptional response of Pseudomonas aeruginosa to a phosphate-deficient Lolium perenne rhizosphere.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtlGnu7fL&md5=6cc46ecb75b2af5c40f328ab68442408CAS |
[24] Sweeney, C. et al. (2017) Interplant aboveground signalling prompts upregulation of auxin promoter and malate transporter as part of defensive response in the neighbouring plants. Front. Plant Sci. 8, 595.
| Interplant aboveground signalling prompts upregulation of auxin promoter and malate transporter as part of defensive response in the neighbouring plants.Crossref | GoogleScholarGoogle Scholar |
[25] Sheth, R.U. et al. (2016) Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32, 189–200.
| Manipulating bacterial communities by in situ microbiome engineering.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XitlSgtbw%3D&md5=5c627b40e940984b0ae0fd748c736b4dCAS |
[26] Liu, S. et al. (2016) The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 19, 32–43.
| The host shapes the gut microbiota via fecal microRNA.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC28XhvFClug%3D%3D&md5=89e910a45f9742ca108b029fba1abd0fCAS |
[27] Massalha, H. et al. (2017) Live imaging of root-bacteria interactions in microfluidics setup. Proc. Natl. Acad. Sci. USA 114, 4549–4554.
| Live imaging of root-bacteria interactions in microfluidics setup.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2sXkvF2itbg%3D&md5=51f92bda5b068957d1ee7453b7efc562CAS |
[28] Jobin, C. (2018) Precision medicine using microbiota Science 359, 32–34.
| Precision medicine using microbiotaCrossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC1cXhtFCntL0%3D&md5=af2d34d655b4026b6ecbc78bdbba1209CAS |
[29] Routy, B. et al. (2018) Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97.
| Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC1cXjslOrsw%3D%3D&md5=50306a80435249932a286040fe3bf0cbCAS |
[30] Harris, V.C. et al. (2017) The intestinal microbiome in infectious diseases: the clinical relevance of a rapidly emerging field. Open Forum Infect. Dis. 4, ofx144.
| The intestinal microbiome in infectious diseases: the clinical relevance of a rapidly emerging field.Crossref | GoogleScholarGoogle Scholar |
Biographies
Professor Brajesh Singh’s research interests encompass soil biology and ecosystem ecology. His research identifies the quantitative relationships between biodiversity and ecosystem functions and how natural/anthropogenic pressures such as land-use and climate change affect these. His applied research harnesses the knowledge gained in fundamental research to achieve sustainable development, environmental protection and food security.
Dr Pankaj Trivedi is an Assistant Professor in the Bioagricultural Sciences and Pest Management at Colorado State University, USA. He is a microbial ecologist and his research program addresses the assembly, fitness and roles of plant, insect, and soil-associated microbiomes; how these are influenced by various biotic and abiotic factors; and what are their consequences on plant productivity, agro-ecosystems sustainability, and local and global level ecological processes.
Dr Catriona Macdonald is a research lecturer at the Hawkesbury Institute for the Environment at the University of Western Sydney. She has >10 years of experience in the field of soil microbiology and nutrient cycling. Her research interests are focussed on understanding microbial-mediated function under environmental change and interaction with plant communities, and how we can utilise the microbial community to overcome associated constraints on productivity and ecosystem function.
Mr Saurabh Singh is a PhD student at Banaras Hindu University India and his research interests include environmental microbiology, and plant-microbe interaction, phosphate solubilisation.
Dr Jay Prakash is an Assistant Professor at Banaras Hindu University India, and his research interests include use of microbes for climate resilient sustainable agriculture, biofertiliser, soil fertility management, remediation of agrochemical, and cellulose degradation for bioethanol production.