Microbiology of winemaking
Eveline BartowskyLallemand Australia
PO Box 210
Edwardstown, SA 5039, Australia
Email: ebartowsky@lallemand.com
Microbiology Australia 38(2) 76-79 https://doi.org/10.1071/MA17033
Published: 7 April 2017
The production of alcoholic beverages, such as winemaking, has a long history, dating back well over 7000 years. The winemaking process is not vastly different to that used by the ancient Greeks and Egyptians. The main difference is that modern-day winemakers have much more control over the different steps; time and method of grape harvesting, use of selected yeast and bacteria, and maturation techniques. The various yeast and bacteria involved in winemaking originate in the vineyard, on grapes and winemaking equipment. Even though yeast and bacteria can impart desirable sensory characteristics to wine, this is not always the case – there are numerous microbes that are unwanted. This overview of wine microbiology will be limited to yeast and bacterial fermentations and microbiological spoilage by these microbes, and will not cover vineyard moulds.
Even though wine has been made for a very long time and the importance of fermentation was known, it was unclear until relatively recently (about 150 years ago) that yeast and bacteria were the vital drivers. Ancient winemakers knew of the significance of ‘the boiling’ during fermentation (from the Latin fevere, to boil). With the development of the microscope by Antonie van Leeuwenhoek, the first yeast were observed, albeit in beer wort. The connection with yeast was finally confirmed in studies by Pasteur in 1866. Bacteria also play an integral role in wine production. Early microbiologists, such as Pasteur and Müller-Thurgau observed the presence of bacteria in wine, and by early 1900’s their importance in winemaking was beginning to be understood.
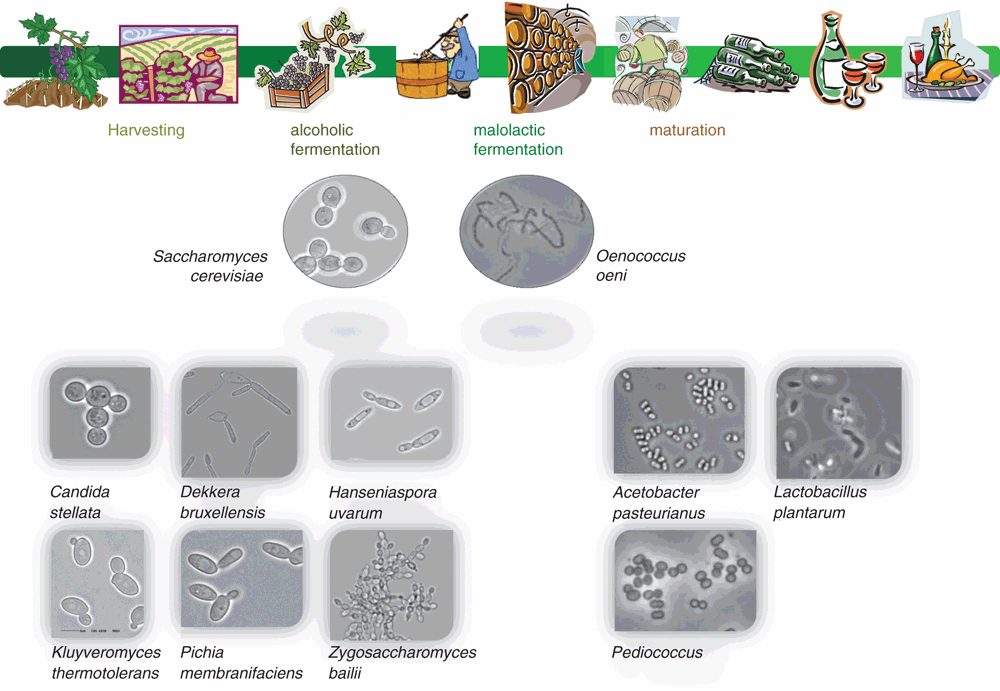
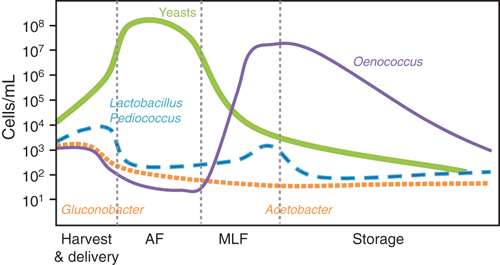
Low pH and high sugar content of grape must exert strong selective pressures on the microorganisms, such that only a few yeast and bacterial species can proliferate. Sulphur dioxide, used as both an anti-oxidant and anti-microbial preservative, imposes additional selection, particularly against undesirable oxidative microbes. Once anaerobic conditions are established in the fermenting grape must, further selectivity is established through the depletion of nutrients and the increasing concentration of ethanol. The growth of yeast and bacteria during grape vinification varies throughout the process; from harvest, through fermentation to wine maturation and storage (Figures 1, 2).
|
Wine yeast
Yeasts are the most important microorganisms, catalysing the conversion of grape sugars to ethanol – without yeast, there would not be wine. Although there are many yeast species associated with grapes and wine, it is Saccharomyces cerevisiae that is the work horse of winemaking.
There are numerous yeast genera and species that can be isolated from grapes and wine, including Brettanomyces, Candida, Kloeckera/Hanseniaspora, Kluyveromyces, Metschnikowia, Pichia, Saccharomyces, and Zygosaccharomyces (Figure 1). Many of these genera do not survive long in wine because of their low alcohol tolerance; most will die off over 5% ethanol v/v – S. cerevisiae strains will then dominate and complete the alcoholic fermentation. However, the metabolism of these non-Saccharomyces yeasts in the first few days of the fermentation can contribute to wine aroma and flavour.
With tremendous advances and greatly reduced costs of whole genome sequencing it is now possible to have a much better picture of the dynamics of yeast during fermentation. Metagenomic studies of grapes and wine fermentations have provided an insight to the number of yeast genera/species present3. These types of studies have also suggested links between the wine microbiota, fermentation characteristics and wine characteristics, and even possibly with wine regions4,5. Additionally, genome sequencing of S. cerevisiae strains has provided an insight into genetic relationships between wine and other beverage strains; wine strains are quite distinct from brewing and other fermentative strains6,7.
The ability to efficiently convert grape sugars (D-glucose and D-fructose) to ethanol and tolerate relatively higher concentrations of ethanol has ensured that S. cerevisiae is the main yeast in winemaking. In the past, winemakers relied solely on the indigenous yeast population to grow to sufficient numbers and metabolise grape sugar to alcohol. In order to have more control of both the fermentation and wine aroma and flavor, winemakers mostly inoculate grape juice with a selected S. cerevisiae strain.
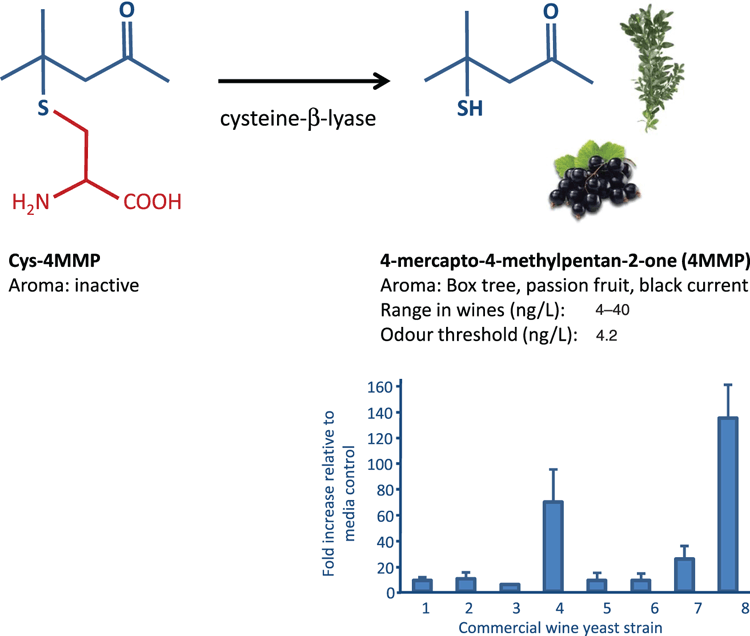
Yeast offer much more to wine than just the production of alcohol – through their metabolism of grape compounds they are able to greatly influence wine aroma and flavor, including higher alcohols, esters, monterpenes, norisoprenoids and sulphur-containing compounds8. For example, several compounds which provide characteristic aromas to Sauvignon blanc are due to yeast enzymatic activity to release the compounds from its precursor. One such aroma compound is 4-mercapto-4-methylpentan-2-one (4MMP), which is reminiscent of box tree and blackcurrant. There is variation in the ability of S. cerevisiae strains to release this volatile thiol compound from its non-volatile cysteinylated precursor (cys-4MMP)2 (Figure 3).
|
Wine bacteria
As a fermented beverage, the bacteria associated with grapes and wine are members of the lactic acid bacteria (LAB) family and specifically conduct malolactic fermentation (MLF); the decarboxylation of L- malic acid to L-lactic acid9. This benefits wine three-fold: it provides microbial stability through the removal of a fermentable carbon source from the wine, reduces acidity by increasing wine pH 0.2–0.5 units, and due to bacterial metabolism there are various sensory changes that can occur to the wine9. The major sensory impacting metabolite produced by LAB is diacetyl; it imparts butter characters to wine10. Other MLF sensory impacts include enhancing fruity-berry aromas and flavours and reduction in vegetative/green characters9.
Virtually all red wines undergo MLF and a range of white and sparkling-base wines will benefit from this bacterial driven fermentation. The preferred LAB species used in winemaking is Oenococcus oeni (formerly Leuconostoc oenos), specifically because of its ability to grow and perform in the low pH (>2.9) wine environment and capacity to survive in high ethanol (up to 16%), SO2, scarcity of nutrients and low temperature (>16°C)9,11. Species of Lactobacillus and Pediococcus (including Lb. plantarum, Lb. casei, Lb. brevis, Lb. hilgardii, Lb. kunkeei, P. damnosus and P. pentosaceus) have the ability to conduct MLF, however, they are usually more associated with wine spoilage12,13. Fortunately, members of these two genera are not very pH tolerant (>3.5), thus their numbers can usually be managed by ensuring pH 3.5 or lower12.
Oenococcus oeni has a small compact genome (1.84 Mb) that most probably has resulted from specialisation to the ecological niche of wine14. The genome sequences of over 200 O. oeni strains has shown an enormous genetic diversity in this species15,16.
MLF can be unpredictable when reliant on the natural bacterial microflora. As spontaneous MLF frequently follows the alcoholic fermentation, it often is not complete until late autumn or the following spring, when temperatures warm up enabling bacterial growth again. An efficient MLF is desirable so that the wine can be protected with SO2 to ensure no undesirable yeast or bacteria take hold and spoil the wine (examples are given below). As described above for alcoholic fermentation, it is common for winemakers to inoculate with a selected O. oeni strain to ensure an efficient MLF in order to stabilise wines as soon as possible. Even though Lactobacillus sp are not usually desired in winemaking, however, through careful selection there are examples of Lb. plantarum strains that are used as starter cultures for conducting MLF in red wine17.
The less pleasant yeast and bacteria
There are numerous yeast and bacteria that are naturally associated with grapes and wine, however, for some their presence is not desired8,12. Although these species are usually capable of alcoholic or malolactic fermentations, it is the production of various secondary metabolites with undesirable sensory qualities that places them into the ‘spoilage’ category. The more prevalent microbial spoilage examples will be described – for more information see8,12,18. Wine spoilage can usually be managed through good winemaking practices.
Brettanomyces bruxellensis readily proliferates and can survive in wine for long periods, particularly following alcoholic fermentation and during wine ageing in oak barrels19. This yeast can cause numerous wine faults, including turbidity in unfortified wines, excessive volatile acidity in sherry, mousy taint and rancid isovaleric acid. However, its greatest impact is through the production of 4-ethylphenol and 4-ethylguaiacol which impart bandaid®, phenolic, medicinal, barnyard and earthy aromas to the wine; often affectionately referred to as ‘Brett’ character20.
Zygosaccharomyces bailii has a unique combination of properties that contribute to its role as a spoilage microorganism; high tolerance to ethanol, growth at low pH and high sugar, and resistance to preservatives used in winemaking (including SO2 and sorbic acid)18.
Pediococcus species in wine is undesirable as these bacteria are able to produce excessive amounts of exopolysaccharides (such as β-D-glucans) which make wines viscous with a thick texture – referred to as ‘ropy wine’21. This type of spoilage tends to occur in low acid (high pH) wines.
Acetobacter and Gluconobacter species, members of the acetic acid bacteria family (AAB), are responsible for the conversion of ethanol through acetaldehyde to acetic acid (i.e. vinegar). Wines spoiled by AAB species will smell like vinegar, bruised apples and sherry-like (acetaldehyde) or nail polish remover (ethyl acetate). As these bacteria are aerobic, their numbers can be managed through ensuring barrels are topped up regularly, SO2 concentrations are maintained and minimising the presence of oxygen during the storage of wine22.
References
[1] Wibowo, D. et al. (1985) Occurrence and growth of lactic-acid bacteria in wine – a review. Am. J. Enol. Vitic. 36, 302–313.| 1:CAS:528:DyaL28Xmsleksg%3D%3D&md5=f6042e6182bf4256d444b055fd871793CAS |
[2] Howell, K.S. et al. (2004) Variation in 4-mercapto-4-methyl-pentan-2-one release by Saccharomyces cerevisiae commercial wine strains. FEMS Microbiol. Lett. 240, 125–129.
| Variation in 4-mercapto-4-methyl-pentan-2-one release by Saccharomyces cerevisiae commercial wine strains.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXptFOksLs%3D&md5=0765afc5001374f23b5566057027b909CAS |
[3] Bokulich, N.A. and Mills, D.A. (2012) Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 45, 377–389.
| Next-generation approaches to the microbial ecology of food fermentations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1agtLvM&md5=c6c410a2cd29e6ffbdf41b55642ff2abCAS |
[4] Bokulich, N.A. et al. (2014) Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 111, E139–E148.
| Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXps12ktg%3D%3D&md5=2e3bcde2877388d15eea2ebb16fe740aCAS |
[5] Bokulich, N.A. et al. (2016) Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 7, e00631-16.
| Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics.Crossref | GoogleScholarGoogle Scholar |
[6] Borneman, A.R. et al. (2011) Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae. PLoS Genet. 7, e1001287.
| Whole-genome comparison reveals novel genetic elements that characterize the genome of industrial strains of Saccharomyces cerevisiae.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXitV2rsLk%3D&md5=afd5a77b1bcfffd9a81017fca3eaeafdCAS |
[7] Borneman, A.R. et al. (2016) Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3 (Bethesda) 6, 957–971.
| Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae.Crossref | GoogleScholarGoogle Scholar |
[8] Swiegers, J.H. et al. (2005) Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 11, 139–173.
| Yeast and bacterial modulation of wine aroma and flavour.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXpsVarur4%3D&md5=b4d83069725c6eeb86f0d3cf588b4979CAS |
[9] Lonvaud-Funel, A. (1999) Lactic acid bacteria in the quality improvement and depreciation of wine. Antonie van Leeuwenhoek 76, 317–331.
| Lactic acid bacteria in the quality improvement and depreciation of wine.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXntVWgsrY%3D&md5=c14afd65ffe3a043fd1d1b276bb91324CAS |
[10] Bartowsky, E.J. and Henschke, P.A. (2004) The ‘buttery’ attribute of wine – diacetyl – desirability, spoilage and beyond. Int. J. Food Microbiol. 96, 235–252.
| The ‘buttery’ attribute of wine – diacetyl – desirability, spoilage and beyond.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXnvV2jt7Y%3D&md5=12ae3a3f3d1fbd38b5f3c110c723d10cCAS |
[11] Bartowsky, E.J. (2005) Oenococcus oeni and malolactic fermentation – moving into the molecular arena. Aust. J. Grape Wine Res. 11, 174–187.
| Oenococcus oeni and malolactic fermentation – moving into the molecular arena.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXpsVarur8%3D&md5=339646c68e6bc3773cabb5801aaa89feCAS |
[12] Bartowsky, E.J. (2009) Bacterial spoilage of wine and approaches to minimize it. Lett. Appl. Microbiol. 48, 149–156.
| Bacterial spoilage of wine and approaches to minimize it.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXjtlWgurs%3D&md5=1cbc32b134cb5e1ca6977cded6b9d39dCAS |
[13] Bartowsky, E.J. et al. (2015) Emerging trends in the application of malolactic fermentation. Aust. J. Grape Wine Res. 21, 663–669.
| Emerging trends in the application of malolactic fermentation.Crossref | GoogleScholarGoogle Scholar |
[14] Gibbons, J.G. and Rinker, D.C. (2015) The genomics of microbial domestication in the fermented food environment. Curr. Opin. Genet. Dev. 35, 1–8.
| The genomics of microbial domestication in the fermented food environment.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhtlSnt7rJ&md5=683ce5be5750a56c52e349afbc3eeb5dCAS |
[15] Sternes, P.R. and Borneman, A.R. (2016) Consensus pan-genome assembly of the specialised wine bacterium Oenococcus oeni. BMC Genomics 17, 1–15.
[16] Borneman, A. et al. (2015) Unravelling the capricious nature of Oenococcus oeni. Wine Vitic. J. 30, 34–37.
[17] du Toit, M. et al. (2011) Lactobacillus: the next generation of malolactic fermentation starter cultures—an overview. Food Bioprocess. Technol. 4, 876–906.
| Lactobacillus: the next generation of malolactic fermentation starter cultures—an overview.Crossref | GoogleScholarGoogle Scholar |
[18] Sponholz, W. (1993) Wine spoilage by microorganisms. In Wine Microbiology and Biotechnology (Fleet, G.H., ed). Taylor & Francis, pp. 395–420.
[19] Curtin, C.D. et al. (2015) Harnessing improved understanding of Brettanomyces bruxellensis biology to mitigate the risk of wine spoilage. Aust. J. Grape Wine Res. 21, 680–692.
| Harnessing improved understanding of Brettanomyces bruxellensis biology to mitigate the risk of wine spoilage.Crossref | GoogleScholarGoogle Scholar |
[20] Suárez, R. et al. (2007) The production of ethylphenols in wine by yeasts of the genera Brettanomyces and Dekkera: a review. Food Chem. 102, 10–21.
| The production of ethylphenols in wine by yeasts of the genera Brettanomyces and Dekkera: a review.Crossref | GoogleScholarGoogle Scholar |
[21] Walling, E. et al. (2005) Glucose fermentation kinetics and exopolysaccharide production by ropy Pediococcus damnosus IOEB8801. Food Microbiol. 22, 71–78.
| Glucose fermentation kinetics and exopolysaccharide production by ropy Pediococcus damnosus IOEB8801.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmvFCksbY%3D&md5=b4f0d0a5266491d4857ffe08012363a4CAS |
[22] Bartowsky, E.J. and Henschke, P.A. (2008) Acetic acid bacteria spoilage of bottled red wine—a review. Int. J. Food Microbiol. 125, 60–70.
| Acetic acid bacteria spoilage of bottled red wine—a review.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXnt1Oksb0%3D&md5=ace3d24f6f8c659e866c6c05b25ee9b9CAS |
Biography
Eveline Bartowsky is an Applied Microbiologist at Lallemand Australia. She provides the Australian and New Zealand wine industry with technical microbiological and fermentation support and oversees winemaking trials. Previously, Eveline was a Senior Research Microbiologist at The Australian Wine Research Institute leading the bacterial research team and managing the Culture Collection. She has over 23 years’ experience in wine microbiology with research interests focused on wine bacteria and malolactic fermentation, wine aroma and flavour, yeast and polyphenolics interactions and minimising wine spoilage by lactic acid and acetic acid bacteria. Following a PhD in microbiology from The University of Adelaide, Eveline undertook postdoctoral studies in Sweden (Umeå University, Umeå), USA (Washington University Medical School, St Louis) and University of Adelaide. She has wide experience with a diverse group of bacteria: Vibrio cholerae, Citrobacter freundii, and wine-associated lactic acid bacteria and acetic acid bacteria.