Clostridium difficile infection in Australia
Niki F Foster A C and Thomas V Riley A B DA School of Pathology and Laboratory Medicine
The University of Western Australia
35 Stirling Highway
Nedlands, WA 6009, Australia
B PathWest Laboratory Medicine WA
Nedlands, WA 6009, Australia
C Tel: +61 8 6383 4361
Email: niki.foster@uwa.edu.au
D Tel: +61 8 6383 4355
Email: thomas.riley@uwa.edu.au
Microbiology Australia 35(1) 26-28 https://doi.org/10.1071/MA14008
Published: 5 February 2014
Clostridium difficile is the most common cause worldwide of infectious diarrhoea in hospitalised patients. It is also thought to be the number one healthcare-related infection in the USA costing >US$3 billion annually. In a recent report from the Centers for Disease Control and Prevention, C. difficile was described as ‘an immediate public health threat that requires urgent and aggressive action’1. Infection occurs following ingestion of C. difficile, probably as a spore, and usually after exposure to antibiotics. Since 2002 there has been a worldwide escalation in rates of C. difficile infection (CDI) with an epidemic strain of C. difficile (PCR ribotype [RT] 027) responsible for outbreaks of severe infection in North America and Europe. This strain is characterised by the production of greater amounts of toxins A and B, the putative major virulence factors and an additional toxin (binary toxin or CDT), as well as resistance to fluoroquinolone antimicrobials. In Quebec Province in Canada (population 7.5 million in 2003) the Health Ministry reported a total of 7004 cases of CDI between 1 April 2003 and 31 March 2004, with 1270 deaths (a crude mortality rate of 18%) and an attributable mortality of greater than 10% in those aged over 60 years – a remarkably high figure. RT 027 then spread quickly to the UK where it caused several highly publicised outbreaks with significant mortality, increasing in prevalence to 26% of all C. difficile in 2005–06 and rising to 42% in 2007–082.
While there have been three separate known introductions of RT 027 into Australia in 2008 and two in 2010, the strain has not established here, possibly because of Australia’s conservative policies regarding fluoroquinolones. Nevertheless, there has been a significant increase in the rate of CDI in Australia over the past 3–4 years. Surveillance of hospital-identified (HI) CDI was mandated for public healthcare facilities in Western Australia in January 2010. The quarterly aggregate rate climbed from 1.47 per 10,000 bed days in the first quarter of 2010 to 2.64 per 10,000 bed days in the fourth quarter of 2010 and 4.59 per 10,000 bed days a year later, falling slightly to 4.27 per 10,000 bed days in the fourth quarter of 2012. Rates were approximately 1–3 times higher for tertiary hospitals alone3,4. In a laboratory-based, retrospective review of all cases of CDI identified in four acute care public hospitals in Tasmania between July 2006 and June 2010 inclusive, the annual rate increased from 2.5 per 10,000 patient-care days in 2006–07 to 4.2 per 10,000 patient-care days in 2009–105. The first report from Victoria following the commencement of targeted surveillance for CDI in 2010 was for the period October 2010 to March 2011 inclusive6. They reported a monthly increase in the number of HI-CDI cases, falling only in December 2010, and an overall rate of 2.2 per 10,000 occupied bed days.
Over this period, laboratory testing has increased and diagnostic methods have changed, potentially influencing the rates of infection being reported. Historically, C. difficile laboratory diagnosis involved cell culture cytotoxicity testing (the application of stool filtrates to a cell monolayer and the observation for cytopathic effects caused by toxin B that could be neutralised with anti-toxin) or toxigenic culture (growth of C. difficile from stool and confirmation that the isolate could produce toxin, usually by tissue culture). While these methods remain the ‘gold standards’, they were slow and so were replaced in many clinical laboratories by rapid and commercially available toxin enzyme immunoassays (EIA) targeting first toxin A, then toxins A and B7. Toxin EIAs began to lose favour in the late 2000s with reports of reduced sensitivity and the advent of highly sensitive and rapid nucleic acid amplification tests (NAATs). It is now common for laboratories to use a two-step algorithm incorporating an inexpensive, rapid and sensitive, but not necessarily highly specific, screening test (e.g. an assay for the glutamate dehydrogenase common antigen) followed by a more specific NAAT that detects C. difficile toxin genes8. A recent report demonstrated that toxin detection correlated with the severity of CDI and that this had prognostic value9. The detection of toxin rather than the genes that encode them is now mandatory for the diagnosis of CDI in the UK10. Unfortunately, the sensitivity of toxin EIAs remains suboptimal; however, their positive predictive value is increased substantially as part of a two-step algorithm. This does not mean the end of NAATs. NAATs still offer a rapid and sensitive screen for C. difficile and may be particularly useful for identifying carriers who could be of significance for infection control. It is this sensitivity and the high prevalence of C. difficile carriers in the hospital population11 that may mean false positive diagnoses of disease leading to increased rates of CDI. Since the introduction of NAATs in Australia, CDI rates have continued to rise; however, these changes in testing procedures are unlikely to explain the full magnitude of the increases seen recently.
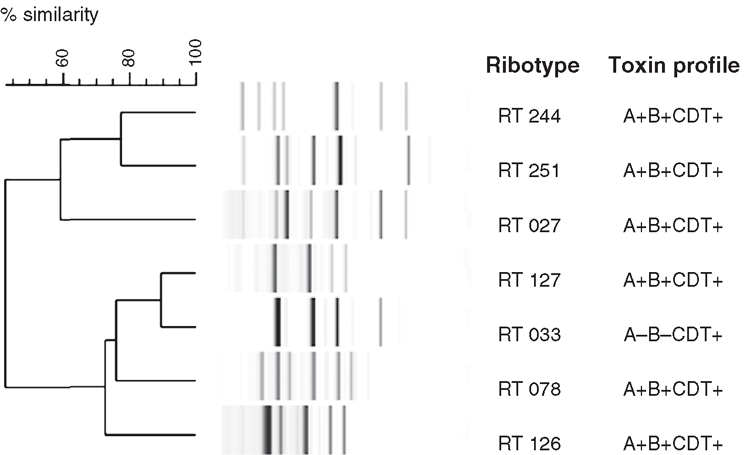
Another potential explanation for the increase in CDI rates is the emergence of new strains. RT 244 was first identified in Australia in 2011 and preliminary data suggested it was associated with more severe disease. This was recently supported by De Almeida et al.12 who, after identifying a new strain in New Zealand as RT 244, performed a case-control study of cases with CDI due to this RT. Cases had more severe disease (OR 9.33; P = 0.015) and were more likely to have community-associated infections (prevalence ratio 3.33; P = 0.078) when compared with controls who were infected with other C. difficile strains. Like RT 027, RT 244 produces more toxins A and B as a result of a single base pair deletion in tcdC (a gene involved in regulation of toxin A and B production), as well as binary toxin. RT 244 is therefore identified as a putative RT 027 with the Cepheid Xpert® C. difficile/Epi system. Other emerging RTs in Australia include RT 251 (A+B+CDT+), which also appears to be closely related to RT 027, and RT 033 (A–B–CDT+), RT 126 (A+B+CDT+) and RT 127 (A+B+CDT+), which are closely related to RT 078 (A+B+CDT+), a RT originally only isolated from animals, particularly livestock, in the Northern hemisphere and also associated with more severe disease (Figure 1)13. Interestingly, RT 078 is not found in Australian livestock14. The clinical significance and extent of infection with RT 033 remains to be determined. The problem for diagnostic laboratories is that tests designed to detect toxin A or toxin B, or the genes that encode them, will not detect RT 033.
CDI surveillance is important in identifying increases in disease occurrence that might suggest: (1) a more susceptible population; (2) circulation of particularly harmful strains; or (3) an increase in exposure to the organism. There has been mandatory surveillance in public hospitals in Australia of ‘hospital-identified’ CDI since 2010. It is unlikely that increased testing and updated laboratory diagnostic methods are entirely responsible for the increase in CDI rates recently seen in Australia. The population is ageing and patients in hospitals are sicker as they replace those who can be treated at home. However, those patients being treated outside the hospital are also at risk and there has been a significant increase in community-acquired CDI. We are seeing new C. difficile strains emerging in Australia, either strains seen previously in foreign countries or apparently novel strains. How they got to Australia and how the population is being exposed is still not clear.
References
[1] Centers for Disease Control and Prevention (2013) Antibiotic Resistance Threats in the United States, 2013, U.S. Department of Health and Human Services. http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf[2] Riley, T.V. (2006) Epidemic Clostridium difficile. Med. J. Aust. 185, 133–134.
| 16893351PubMed |
[3] Healthcare Infection Surveillance Western Australia (2013) Quarterly Aggregate Report, Quarter 3 – 2013, Number 33, Department of Health, Government of Western Australia. http://www.public.health.wa.gov.au/cproot/5574/2/hiswa-agg-report-q3-2013.pdf
[4] Healthcare Infection Surveillance Western Australia (2013) Annual Report 2011–2012, Department of Health, Government of Western Australia. http://www.public.health.wa.gov.au/cproot/5173/2/hiswa-2011-12-ar-final.pdf
[5] Mitchell, B.G. et al. (2012) An increase in community onset Clostridium difficile infection: a population-based study, Tasmania, Australia. Healthc. Infect. 17, 127–132.
| An increase in community onset Clostridium difficile infection: a population-based study, Tasmania, Australia.Crossref | GoogleScholarGoogle Scholar |
[6] Bull, A.L. et al. (2012) Implementation of standardised surveillance for Clostridium difficile infections in Australia: initial report from the Victorian Healthcare Associated Infection Surveillance System. Intern. Med. J. 42, 715–718.
| Implementation of standardised surveillance for Clostridium difficile infections in Australia: initial report from the Victorian Healthcare Associated Infection Surveillance System.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC38jhs1ylsQ%3D%3D&md5=734fec56e364af01065df583d4b83fb2CAS | 22697155PubMed |
[7] Carroll, K.C. (2011) Tests for the diagnosis of Clostridium difficile infection: the next generation. Anaerobe 17, 170–174.
| Tests for the diagnosis of Clostridium difficile infection: the next generation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtVyrsb7E&md5=a5974c0ec91258dcf09a7fe4d971aa16CAS | 21376826PubMed |
[8] Cheng, A.C. et al. (2011) Australasian Society for Infectious Diseases guidelines for the diagnosis and treatment of Clostridium difficile infection. Med. J. Aust. 194, 353–358.
| 21470086PubMed |
[9] Planche, T.D. et al. (2013) Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C. difficile infection. Lancet Infect. Dis. 13, 936–945.
| Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C. difficile infection.Crossref | GoogleScholarGoogle Scholar | 24007915PubMed |
[10] Wilcox, M.H. (2012) Overcoming barriers to effective recognition and diagnosis of Clostridium difficile infection. Clin. Microbiol. Infect. 18, 13–20.
| Overcoming barriers to effective recognition and diagnosis of Clostridium difficile infection.Crossref | GoogleScholarGoogle Scholar | 23121550PubMed |
[11] Eyre, D.W. et al. (2013) Asymptomatic Clostridium difficile colonisation and onward transmission. PLoS ONE 8, e78445.
| Asymptomatic Clostridium difficile colonisation and onward transmission.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhslylsLzI&md5=c8ae4d5e93a6c15aa147071397280889CAS | 24265690PubMed |
[12] De Almeida, M. et al. (2013) Severe Clostridium difficile infection in New Zealand associated with an emerging strain, PCR-ribotype 244. N. Z. Med. J. 126, 9–14.
| 24126745PubMed |
[13] Freeman, J. et al. (2010) The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23, 529–549.
| The changing epidemiology of Clostridium difficile infections.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtFWgtLjI&md5=fcb4a8ad4b4d9d203385f265a5be8040CAS | 20610822PubMed |
[14] Knight, D.R. et al. (2013) Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter. Appl. Environ. Microbiol. 79, 2630–2635.
| Cross-sectional study reveals high prevalence of Clostridium difficile non-PCR ribotype 078 strains in Australian veal calves at slaughter.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXlvVKlsb8%3D&md5=90848f88f3a882017e0210eb451a5de0CAS | 23396338PubMed |
Biographies
Niki Foster is a Post-doctoral Research Associate at The University of Western Australia. Her interests are in infectious disease epidemiology, diagnostics and infection control. She is currently employed on NHMRC Project Grant No. 1006243.
Thomas Riley is a Professor at The University of Western Australia and a Senior Clinical Scientist in the Division of Microbiology and Infectious Diseases at PathWest Laboratory Medicine. His interests are the diagnosis, pathogenesis and epidemiology of CDI.