Antibiotic susceptibility testing methods and emerging bacterial resistance in hospitals
John MerlinoASM Antimicrobial SIG Chair/Convenor
Department of Infectious Diseases and Immunology
Faculty of Medicine
University of Sydney
Department of Microbiology and Infectious Diseases
Concord Hospital, NSW Pathology
Email: JMerlino@med.usyd.edu.au
Microbiology Australia 35(1) 9-12 https://doi.org/10.1071/MA14004
Published: 10 February 2014
Clinical microbiology laboratories, whether in hospitals or private institutions, have the important task of performing antimicrobial susceptibility testing on significant bacterial pathogens isolated from a variety of specimens. The aim of all this testing is to detect possible emerging antimicrobial drug resistance in unusual and common pathogens so that infections are treated with the appropriate antibiotics. Microbiologists and clinicians in hospitals are today more dependent on results from in vitro susceptibility testing. This signifies the importance of the diagnostic laboratory in clinical medicine. Hospital laboratories have the responsibility of reporting the antimicrobial agent(s) that are most appropriate for the organism(s) isolated, for the site of infection and the hospital pharmacy formulary.
Definitions used to describe antimicrobial resistance in hospitals
Antimicrobial resistance is the capacity of bacteria to survive exposure to a defined concentration of an antimicrobial substance1,2. However, in hospitals antimicrobial resistance may have multiple definitions according to the scientific discipline and the goals involved3,4:
-
Clinical definition: the bacteria survive an adequate treatment with an antibiotic.
-
Pharmacological definition: the bacteria survive a range of concentrations expressing the various amounts of an antibiotic present in the different compartments of the body when the antibiotic is administered at the recommended dose.
-
Microbiological and molecular definition: the bacteria have a mechanism or gene that governs a higher minimum inhibitory concentration (MIC) than the original or wild bacteria. In hospitals this may become an infection control issue with plasmid mediated resistant organisms and requires molecular testing, for example, detection of various genes such as MecA in Staphylococcus aureus, VanA or VanB genes in Enterococcus species, with carbapenemase resistance in Enterobacteriaceae family or Pseudomonas species with the detection of beta-lactamase resistant genes such as blaIMP/VIM, blaNDM, blaKPC etc.
-
Epidemiological definition: any group of bacterial strains that can be distinguished from the normal (Gauss) distribution of MIC(s) to an antibiotic.
In the past, for many clinical cases, hospital infections could be treated empirically based on the medical microbiologist’s past experiences. However, this has become more difficult recently with the emergence of new unpredictable resistances both in hospital and community patients. Empirical treatment continues to be effective for some bacterial pathogens since resistance mechanisms have not yet been seen, for example, penicillin susceptibility of Streptococcus pyogenes. Resistance to antimicrobial agents has been detected in many other organisms1,2. Antibiotic standardised susceptibility testing of individual isolates is important with species that may possess both intrinsic and acquired resistance including members of the Enterobacteriaceae family, Pseudomonas species, Staphylococcus species, Enterococcus species and Streptococcus pneumoniae2. New resistance phenotypes and genotypes are rapidly emerging with various intrinsic and acquired mechanisms among pathogens and even in some commensal organisms (Figure 1)5. In health-care settings, the spread of resistant clones can be rapid, becoming an infection control issue. Many studies also indicate that in a hospital environment for survival purposes, for a bacterium, it is better to be resistant than virulent6.

|
Antibiotic susceptibility testing methods used in Australia
Data from the Royal College of Pathologists of Australasia Quality Assurance Program Microbiology (RCPAQAP Microbiology) indicate that many laboratories in Australia use a number of different antibiotic susceptibility testing methods for various isolated organisms that are potential pathogen to detect resistance7. Methods range from rapid automated instruments that use commercial materials and devices (Vitek Systems – bioMerieux or Phoenix – BD Diagnostics) to manual methods that provide flexibility and cost savings, such as disc diffusion and gradient diffusion methods, for example, Etest (bioMerieux or AB BIODISK) or MICE (ThermoFisher, Scientific)1. Results from the RCPAQAP Microbiology data also show that in many laboratories the test method used also varies with the type of organism being tested and the site of isolation7.
Interpretation of antibiotic susceptibility test results in Australia
Susceptibility tests results are usually interpreted by the laboratory prior to releasing a report to the clinician or attending physician. In Australia current data from the RCPAQAP Microbiology indicates that three standardised susceptibility test methods are commonly used when interpreting susceptibility tests based on a quantifiable result either by disc zone sizes or MIC8. These three standardised methods are:
-
The American-based Clinical Laboratory Standard Institute (CLSI, formerly NCCLS) method;
-
The Australian-based Antibiotic Susceptibility Testing using the CDS method (first published in 1975, the Seventh Edition released in 2013)9; and
-
The European Committee on Antimicrobial Susceptibility Testing (EUCAST).
Most medical laboratories interpret susceptibility test using the CLSI method (approximately 80%) followed by the CDS method (approximately 16%) and a smaller number use EUCAST (approximately 3–4%); however, an update of this European EUCAST method is increasing in Australia. The RCPAQAP Microbiology accepts results from all three standardised methods when enrolled in their external proficiency program. Enrolment in such a program is a mandatory requirement for those laboratories assessed by the National Association of Testing Authorities (NATA) in Australia. It is important that guidelines and tables used for susceptibility test interpretations represent the most current criteria for accurate reporting.
Performance of antibiotic susceptibility test methods in laboratories
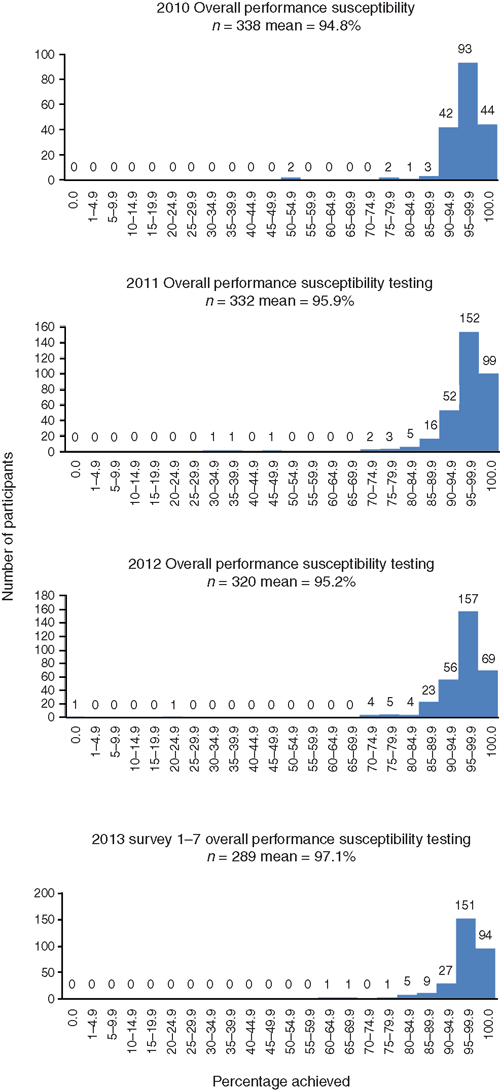
In most laboratories in Australia (and some overseas), as shown in Figure 2, the overall performance and accuracy of susceptibility testing reporting is high for those laboratories enrolled the RCPAQAP Microbiology. In 2010 from 338 laboratories the mean performance was 94.8%, in 2011 from 332 laboratories the mean was 95.8%, in 2012 from 320 laboratories the mean was 95.2% and in 2013 from 289 laboratories (not final as yet) the mean is approximately 97.1%7. When assessing the accuracy of various susceptibility testing methods this is done by comparing results of a wild isolate to a standardised isolate (e.g. ATCC strain) using guidelines and recommendations of reporting of the specific standardised method used (CLSI, CDS or EUCAST).
|
The emergence of new antimicrobial resistance mechanisms in some organisms in hospitals and the community may be difficult to detect with certain antibiotic susceptibility testing methods. In hospitals the best examples are vancomycin heterogeneous or intermediate susceptibility in S. aureus and low carbapenemase resistance in some Gram-negative organisms. This means that the performance of susceptibility devices needs to be constantly reassessed and updated. Many laboratories supplement susceptibility test results when required with additional testing methods such as placing discs on agar plates in certain orientation to detect extended-spectrum betalactamase (ESBL) or MBL (metallo-beta-lactamase) mechanisms in Gram-negative organisms with specific chemical inhibitory substrates (phenyl-boronic acid for AmpC, clavulanic acid for ESBL, EDTA for MBL resistance detection) or the inclusion of a screening agar as an indication of mechanisms of resistance for infection control purposes.
In most hospitals due to infection control priorities, phenotypic resistance is later confirmed by further using molecular testing in certain organisms showing unusual resistance. It is important to remember when using molecular methods to detect resistance that phenotypic test methods are based on antimicrobial activity (i.e. the MIC) and breakpoints that predict a quantifiable susceptibility and resistance, while genotypic test methods are based on the detection of a resistance gene (or its potential product) and are not quantifiable and predict potential resistance not susceptibility6.
Future directions and current methods of antibiotic susceptibility testing in hospitals in Australia
Standardised antimicrobial susceptibility testing methods such as CLSI, CDS or EUCAST provide reliable in vitro results when used according to the procedures defined, guidelines and recommendations. There is still a need for improvement in the area of rapid and accurate detection of bacterial resistance to antibiotics. Many hospitals with rapid identification of the bacterial isolate using matrix-assisted laser desorption/ionisation-time of flight mass spectrometry (MALDI-TOF MS) are tempted to perform a susceptibility test directly on the clinical specimens from a patient; this has the potential to save 24 hours in the provision of a result. However, there is no guarantee that the correct inoculum is used and this may result in a wrong susceptibility result. While many laboratories perform direct sensitivity from blood cultures, the result should be used as a presumptive guide for critical value in patient management and repeated using the correct inoculum after organism isolation. Some reference methods (such as the CDS method) allow acceptable direct susceptibility testing in urine only using a standardised procedure as described in their manual9. Some hospital laboratories use molecular methods to detect acquired plasmid mediated resistance or other resistance mechanisms on isolated organisms for infection control purposes; however, when attempted on direct clinical specimen there are hundreds of beta-lactamases, numerous mutations, acquisitions and expression mechanisms that result in many resistances and targeting an individual resistance mechanism is a problem1. Genome sequencing of isolates is fruitful in some instances and appears to work well for some laboratories that have access to such technology, but to add to my previous discussion, the detection of a gene or its product is not a quantifiable MIC and may (or may not) predict resistance not certain susceptibility or expression10.
Conclusion
In hospitals, a variety of laboratory methods can be used to measure the in vitro susceptibility of bacteria to antimicrobial agents. Standardised antibiotic susceptibility testing methods such as CLSI, CDS and EUCAST as phenotypic references indicate susceptibility and resistance of bacterial isolates. This will continue to be clinically important for years to come with the emergence of new unpredictable antimicrobial resistances. RCPAQAP Microbiology offers many laboratories an external susceptibility performance program were results can be compared with other participants and accuracy determined by using different standardised methods. The aim of the program is to assess the reliability of susceptibility methods used by participating laboratories and thereby promote a high standard of laboratory practice. The program is available to all laboratories in Australia, New Zealand and other countries7.
Acknowledgement
I thank Ms Debbie Walker and Dr Arthur Morris for providing valuable data from the RCPAQAP Microbiology surveys for this article.
References
[1] Jorgensen, J.H. and Ferraro, M.J. (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 49, 1749–1755.| Antimicrobial susceptibility testing: a review of general principles and contemporary practices.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhs1Sjt7fI&md5=19fe10fb1e75476b576146db575f60e6CAS | 19857164PubMed |
[2] Steward, C.D. et al. (2000) Ability of laboratories to detect emerging antimicrobial resistance in nosocomial pathogens: a survey of project ICARE laboratories. Diagn. Microbiol. Infect. Dis. 38, 59–67.
| Ability of laboratories to detect emerging antimicrobial resistance in nosocomial pathogens: a survey of project ICARE laboratories.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXntF2itb8%3D&md5=c39b76bda5323a7e3617106cdb95e06cCAS | 11025185PubMed |
[3] Acar, J. and Rostel, B. (2001) Antimicrobial resistance: an overview. Rev. Sci. Tech. 20, 797–810.
| 1:STN:280:DC%2BD3Mnos1OhsA%3D%3D&md5=9d04c5714ee382186e7eaf4a44939a6dCAS | 11732423PubMed |
[4] Acar, J.F. and Moulin, G. (2012) Antimicrobial resistance: a complex issue. Rev. Sci. Tech. 31, 23–31.
| 1:STN:280:DC%2BC38fksVSrtw%3D%3D&md5=2d1afee7e5c66f589ccfbc49a34b5d9aCAS | 22849265PubMed |
[5] Blondeau, J.M. (2008) STAT Steps to Antimicrobial Therapy (Second edn), North American Compendiums Inc.
[6] Varaldo, P.E. (2002) Antimicrobial resistance and susceptibility testing: an evergreen topic. J. Antimicrob. Chemother. 50, 1–4.
| Antimicrobial resistance and susceptibility testing: an evergreen topic.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XlsFGju70%3D&md5=92a609ce68a9849a768008aa83ac02bfCAS | 12095999PubMed |
[7] RCPA Quality Assurance Programs Pty Limited. RCPAQAP microbiology surveys (2010–2013). http://dataentry.rcpaqap.com.au/micro/
[8] Silley, P. (2012) Susceptibility testing methods, resistance and breakpoints: what do these terms really mean? Rev. Sci. Tech. 31, 33–41.
| 1:STN:280:DC%2BC38fksVSrtA%3D%3D&md5=459d7db2ee205b7ed2a99c033df43964CAS | 22849266PubMed |
[9] Bell, S.M. et al. (2013) Antibiotic Susceptibility Testing by the CDS Method: A Manual for Medical and Veterinary laboratories 2013 (Seventh edn), South Eastern Area Laboratory Services.
[10] Laxminarayan, R. et al. (2013) Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098.
| Antibiotic resistance-the need for global solutions.Crossref | GoogleScholarGoogle Scholar | 24252483PubMed |
Biography
Dr John Merlino is a senior scientist in the Department of Microbiology and Infectious Diseases at Concord Hospital, NSW Pathology and lectures at the University of Sydney, Faculty of Medicine, Department of Infectious Diseases and Immunology. He completed a Master of Science with honours in Microbiology and Biotechnology focusing on detection, identification and antimicrobial resistance in Enterococcus spp. at Macquarie University in the School of Biological Sciences. He later completed a PhD in Medicine in Bacteriology and Infectious Diseases focusing on the detection and expression of methicillin and oxacillin resistance in S. aureus in the Department of Infectious Diseases and Immunology, Faculty of Medicine at the University of Sydney. He is a Fellow of the Australian Society for Microbiology and a Founding Fellow of the Faculty of Science, RCPA. He is the current Convenor and Chairperson of the Antimicrobial Special Interest Group of the Australian Society for Microbiology.


