Molecular diversity of black corals from the Saudi Arabian Red Sea: a first assessment
Silvia Vicario A B * , Tullia Isotta Terraneo B , Giovanni Chimienti B C , Davide Maggioni
A B * , Tullia Isotta Terraneo B , Giovanni Chimienti B C , Davide Maggioni  D E , Fabio Marchese B , Sam J. Purkis F G , Ameer Abdulla Eweida H , Mattie Rodrigue I and Francesca Benzoni A B
D E , Fabio Marchese B , Sam J. Purkis F G , Ameer Abdulla Eweida H , Mattie Rodrigue I and Francesca Benzoni A B
A
B
C
D
E
F
G
H
I
Abstract
Black corals occur as part of benthic assemblages from shallow to deep waters in all oceans. Despite the importance in many benthic ecosystems, where these act as biodiversity aggregators, antipatharians remain poorly studied, with 75% of the known species occurring below recreational SCUBA diving depth limits. Currently, information regarding the diversity and evolutionary history is limited, with most studies focusing on Hawaii and the South Pacific Ocean. Other regions of the world have received less attention, such as the Red Sea, where only two black coral families and four genera have been recorded. We provide the first analysis of the molecular diversity of black corals in the eastern Gulf of Aqaba and the northern and central Saudi Arabian Red Sea, based on a dataset of 161 antipatharian colonies collected down to 627 m deep. Based on specimen morphology, we ascribed our material to 11 genera belonging to 4 of the 7 known Antipatharia families, i.e. Antipathidae, Aphanipathidae, Myriopathidae and Schizopathidae. The genus level phylogeny of three intergenic mitochondrial regions, the trnW-IGR-nad2 (IgrW), nad5-IGR-nad1 (IgrN) and cox3-IGR-cox1 was reconstructed including previously published material. Overall, we recovered six molecular clades that included exclusively Red Sea sequences, with the highest diversity occurring at mesophotic depths. This study highlights that diversity of black corals in the Red Sea is much higher than previously known, with seven new generic records, suggesting that this basin may be a hotspot for antipatharian diversity as is known for other taxa. Our results recovered unresolved relationships within the order at the familial and generic levels. This emphasises the urgent need for an integration of genomic-wide data with a re-examination of informative morphological features necessary to revise the systematics of the order at all taxonomic levels.
Keywords: Antipatharia, cox3-cox1, deep sea, IgrN, IgrW, mesophotic, phylogeny, Saudi Arabia.
Introduction
Antipatharia Milne-Edwards & Haime, 1857 is an order of anthozoans within the subclass Hexacorallia, commonly known as black corals. These are slow-growing, ahermatypic sessile organisms, potentially representing some of the longest-living animals on Earth, with colonies over 4000 years old (Roark et al. 2009; Wagner et al. 2012). Despite being important organisms in many benthic ecosystems, providing three-dimensional structure and hosting a rich associated fauna (Bo et al. 2019), antipatharians remain poorly studied, with 75% of the species occurring below recreational SCUBA diving depths limits (Cairns 2007). Antipatharia taxonomy and systematics have traditionally relied on morphological characters (Brugler et al. 2013). For example, family-level taxa are defined on the basis of the number of mesenteries inside the polyps, the size and shape of the polyps and tentacles, and the morphology of the axial spines, whereas genera are distinguished based on the morphology of the corallum and associated branching pattern (e.g. Opresko 2004; Bo et al. 2019). However, problems arise both for species level identification and phylogenetic reconstruction when relying exclusively on morphology. This has been well exemplified by the plethora of works addressing similar questions in other anthozoan clades such as Scleractinia (see for example Fukami et al. 2004, 2008; Stolarski et al. 2011) and Octocorallia (Fabricius and Alderslade 2001; McFadden et al. 2022) and to a lesser extent in black corals. For example, Warner (1977, 1981) revealed the presence of environmentally driven plasticity at the colony level in Antipathes atlantica Gray, 1857, where the colonies were hypothesised to increase the degree of branching to improve drag under strong currents. Moreover, a study from Molodtsova and Budaeva (2007) showed that symbiotic polychaetes living in the stems of a wide taxonomic range of black corals could warp and influence the morphology of the corallum and skeletal spines. In addition, as shown for Scleractinia, cases of convergent evolution and homoplasy of morphological characters may hinder the systematics of the order, since morphologically similar taxa might not be phylogenetically closely related (see for example, van Oppen et al. 2001; Fukami et al. 2004).
Several studies in the past 15 years have relied on an integrated morpho-molecular approach to clarify the phylogenetic relationships of the order Antipatharia using a combination of mitochondrial and nuclear genes, including 5.8S, 18S, 28S, ITS1, ITS2, cox3-IGR-cox1, nad5-IGR-nad1 (IgrN) and trnW-IGR-nad2 (IgrW), (Lapian et al. 2007; Wagner et al. 2010; Bo et al. 2012; Brugler et al. 2013; Macisaac et al. 2013; Bo et al. 2018; Opresko et al. 2020; Chimienti et al. 2022). Most of the studies focused on inferring relationships among and within the families Antipathidae, Aphanipathidae and Myriopathidae (Lapian et al. 2007; Lapian 2009; Wagner et al. 2010), whereas Macisaac et al. (2013), Lü et al. (2021) and Chimienti et al. (2022) investigated the relationships within the family Schizopathidae. The advent of high throughput sequencing technology allowed Barrett et al. (2020) to reconstruct family-level relationships within the order on the basis of mitochondrial genomes. Indeed, the authors sequenced 18 complete and 5 partial mitochondrial genomes from 14 genera and 6 families (Antipathidae, Aphanipathidae, Cladopathidae, Leiopathidae, Stylopathidae and Schizopathidae) from the Northeast Atlantic. This work confirmed that the families Antipathidae, Cladopathidae and Schizopathidae are not monophyletic and further confirmed the phylogenetic position of Leiopathidae as a sister clade to the other six Antipatharia families, as already indicated by Brugler et al. (2013). Recently, the use of UCEs and exon loci captured with a targeted-enrichment approach has proven useful to infer phylogenetic relationships at genus and species-level in black corals (Horowitz et al. 2020, 2022). Using targeted sequence capture of conserved loci, Horowitz et al. (2022) provided a phylogenetic reconstruction of 80 antipatharians, clarifying systematic relationships within the order. However, the need for integrating sampling from understudied localities and the detailed study of type material of antipatharian taxa remains a necessary step to resolve taxonomic issues related to this problematic order.
Most Antipatharia studies are focused on Hawaii and the South-east Pacific Ocean. In other regions of the world such as the Red Sea, the order has received little attention to date. Indeed, studies on the diversity and systematics of Red Sea corals remain mainly focused on shallow-water Scleractinia and Alcyonacea (Fabricius and Alderslade 2001; Haverkort-Yeh et al. 2013; Bouwmeester et al. 2015; Terraneo et al. 2014, 2016, 2019, 2021; Arrigoni et al. 2016, 2017, 2021; McFadden et al. 2017). Sporadic information regarding black corals in the Red Sea can be retrieved from a few studies regarding the reproduction and associated fauna (Totton 1923; Rees 1969; Okamura and Habe 1976; Richmond and Hunter 1990; Herler 2007). Antipatharians were recently mentioned in scientific works regarding the effect of sewage discharge tracking near the city of Jeddah in the central Red Sea (Risk et al. 2009) or assessing the potential role in cancer therapy (Ghandourah and Alorfi 2019). However, to date, the diversity, abundance and distribution of the order Antipatharia in the basin have not been assessed, and only the genera Antipathes Pallas, 1766 (Antipatharia: Antipathidae) (Brook 1889; Jones et al. 2000; Risk et al. 2009; Ghandourah and Alorfi 2019; Morgulis et al. 2022), Cirrhipathes de Blainville, 1830 (Antipatharia: Antipathidae) (Brook 1889; Cooper 1903; Herler 2007), Stichopathes Brook, 1889 (Antipatharia: Antipathidae) (Qurban et al. 2014; Morgulis et al. 2022) and Bathypathes Brook, 1889 (Antipatharia: Schizopathidae) (Qurban et al. 2014, but see Chimienti et al. 2022 for the identification) are known to live in the Red Sea. Recently, Chimienti et al. (2022) revealed the presence of a new species, Bathypathes thermophila Chimienti, 2022, found at bathyal depths in the northern Saudi Arabian Red Sea.
We provide (a) the first assessment of the molecular diversity of the order Antipatharia in the Saudi Arabian Red Sea, (b) a reconstruction of genus-level phylogenetic relationships among 161 samples covering a 627-m range in water depth and (c) genus-level morphological identifications of the collected specimens.
Materials and methods
Collection
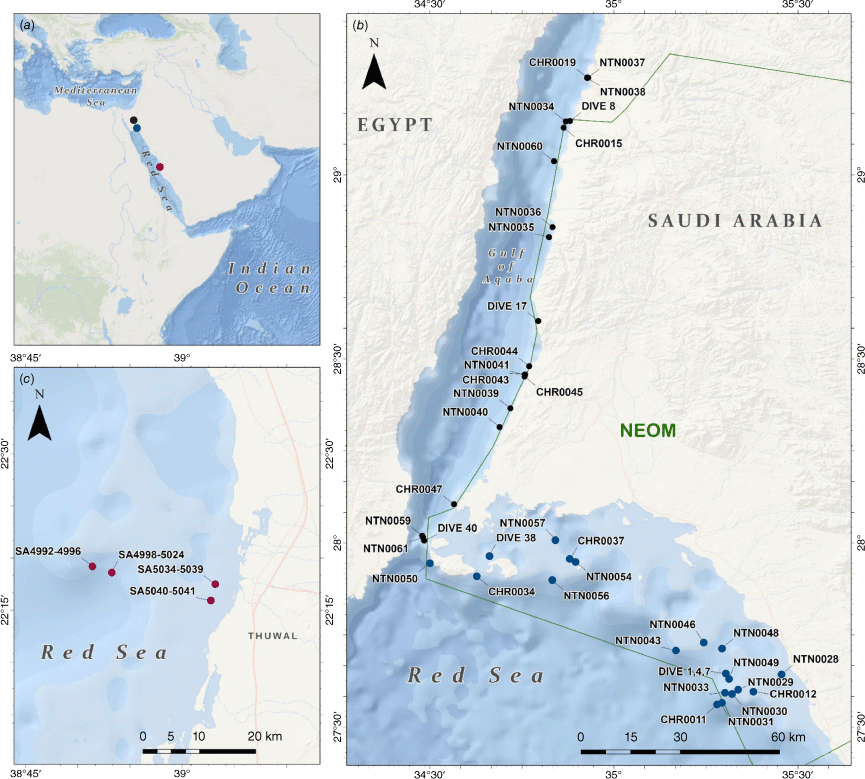
A total of 161 black coral colonies were collected at three main localities in the Saudi Arabian Red Sea, including the Gulf of Aqaba (85 specimens), the northern Red Sea (46 specimens) and the central Red Sea (25 specimens) (Fig. 1, Supplementary Table S1). During the OceanX-NEOM Deep Blue Expedition, aboard the R/V OceanXplorer in 2020, shallow (above 30 m), mesophotic (from 30 to 200 m) and deep sea (>200 m) explorations were conducted in the NEOM (one of the Saudi Arabian development giga-projects) area along the Saudi Arabian coast of the Gulf of Aqaba and northern Red Sea. Sampling occurred from 4- to 627-m depth using a combination of SCUBA diving, Argus Mariner XL108 remotely operated vehicle (ROV) and two Triton 3300/3 submersibles. Images of corals in situ were taken with a Canon PowerShot SX260 HS for colonies collected by SCUBA diving. Mesophotic and deep colony in situ images were taken with a complex light-cameras apparatus with one DSPL Super wide-angle CCD camera for landscape view and one HDTV 1080p F/Z Colour camera for detailed observations mounted on the ROV. Several light and camera systems including a Wide Angle Red DSMC2 Helium 8k Canon CN-E15.5–47-mm lens and macro Red DSMC2 Helium 8k Nikon ED 70–180-mm F4.5–5.6D mounted on each submersible were also used. Black corals were collected with pliers while SCUBA diving. Sampling from both ROV and submersible occurred with a Schilling T4 hydraulic manipulator. For each black coral specimen collected below 30 m, high-definition (HD) videos were recorded with the cameras mounted on ROVs and submersibles and a series of images from the HD video were extracted using the software Media Player Classic Home Cinema (MPC – HC, ver. 1.9.10, see https://mpc-hc.org). Moreover, between September and November 2021, black coral colonies were collected from the central Saudi Arabian Red Sea while SCUBA diving. Images of living corals were taken in situ with a Canon PowerShot SX260 HS.
Maps of the study area. (a) Sampling regions for our study. (b) Boundaries of NEOM (green polygon) including the Gulf of Aqaba and northern Red Sea coastline and major islands, and position of the sampling sites at which Antipatharia specimens were collected (black dots correspond to dives in the Gulf of Aqaba; blue dots correspond to dives in the northern Red Sea. (c) Sampling sites in the central Saudi Arabian Red Sea (Thuwal).
All the colonies collected were tagged soon after collection and the apical 10 cm of a branch or whip were preserved in 99% ethanol. The remainder of the corallum was air-dried in the shade for 24 h. The samples examined in this work are catalogued and currently deposited at the King Abdullah University of Science and Technology (KAUST), Thuwal, Saudi Arabia and the Zoological Museum of the University of Bari Aldo Moro, Bari, Italy (MUZAC). Images and metadata are available upon request.
Morphological analyses and identifications
When available, the whole dry colony (voucher) was photographed using an Olympus TG-6 camera and macromorphological characters were examined. Optical images of dry skeletons were taken using a Leica M205 A stereomicroscope equipped with a Leica DMC 5004 camera to observe skeletal features, including corallum morphology, presence and arrangement of pinnules and subpinnules, shape and transverse diameter of polyps, and interpolypar distance. Micromorphology of skeletal structures (shape, size and arrangements of the spines) was investigated using scanning electron microscopy (SEM) with a ThermoFisher Scientific Quattro S Environmental SEM at KAUST Imaging Core Lab on selected specimens per taxon. A subsample of the skeleton was hydrated in distilled water, cleaned in diluted sodium hypochlorite (NaClO), washed in distilled water and dehydrated in ethanol (Chimienti et al. 2020) for these. Once dry, samples were mounted on stubs and coated with a 5-nm-thick layer of iridium using a Quorum Q150T S turbomolecular pumped coater.
Specimens included in this study were identified to genus level following original descriptions and recent taxonomic revisions. We referred to Brook (1889), Opresko and Cairns (1994), Opresko (2001), Bo et al. (2009) and Wagner (2015a, 2015b) for the family Antipathidae. The taxonomic revisions of Opresko (2001, 2004) were respectively referenced for the families Aphanipathidae and Myriopathidae and for Schizopathidae we referred to Brook (1889), Opresko (2002), Molodtsova and Opresko (2017) and Chimienti et al. (2022).
DNA extraction, amplification and sequencing
Total genomic DNA of each sample stored in ethanol was extracted using the QIAGEN DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). The concentration and quality of DNA were assessed using a NanoDrop 2000 C spectrophotometer (ThermoFisher Scientific, Wilmington, DE, USA).
Polymerase chain reactions (PCRs) were used to amplify three mitochondrial genetic markers: trnW-IGR-nad2 (IgrW), nad5-IGR-nad1 (IgrN) and cox3-IGR-cox1. All amplifications were conducted in a 15-μL PCR volume composed of 7.5 μL of 1× Multiplex PCR Master Mix (Qiagen Inc., Hilden, Germany), 1.5 μL of each primer at 10-μM concentration, 3.3 μL of RNase free water and 1.2 μL of DNA. A summary of primer details and PCR cycle profiles is provided in Supplementary Table S2. All successfully amplified sequences were purified using Illustra ExoStar (GE Healthcare, Buckinghamshire, UK) and Sanger sequenced in both directions using an ABI 3730xl DNA Analyser (Applied Biosystems, Carlsbad, USA) at KAUST Bioscience Core Lab.
Sequences were checked manually, edited and assembled using Geneious (ver. 10.1.3, Biomatters Ltd, Auckland, New Zealand). A total of 89 sequences for IgrN, 102 for IgrW and 98 for the cox3-cox1 marker were downloaded from the GenBank Database and added to the dataset (Supplementary Table S1). Newly produced sequences from our study and sequences downloaded from GenBank were aligned using the software MAFFT (ver. 7.130b, Biopython, see http://mafft.cbrc.jp/alignment/software/; Katoh and Standley 2013) with the E-INS-i method. All sequences produced in our study were deposited in GenBank (see https://www.ncbi.nml.nih.gov/genbank/), with accession numbers listed in Supplementary Table S1.
Phylogenetic analyses
Phylogenetic analyses were conducted on the concatenated dataset including the IgrN and IgrW, and separately for each of the three molecular markers. Maximum likelihood (ML) and Bayesian inference (BI) analyses were run using the online CIPRES Science Gateway (ver. 3.3, San Diego Supercomputer Center, see https://www.phylo.org; Miller et al. 2011). The selection of models of nucleotide substitution were determined using JModeltest2 on XSEDE (ver. 2.1.6, see https://github.com/ddarriba/jmodeltest2; Guindon and Gascuel 2003; Darriba et al. 2012). Partitioning scheme and models were determined with PartitionFinder 2 (ver. 2.1.1, see https://www.phylo.org; Lanfear et al. 2017) using the Akaike information criterion (AIC) that selected the General Time Reversible (GTR) + gamma model for the three markers. Maximum Likelihood analyses were performed using RaxML-HPC2 on XSEDE (ver. 8.2.12, see https://www.phylo.org/; Stamatakis 2014) with 1000 bootstrap replicates. Bayesian Inference analyses were performed using MrBayes on XSEDE (ver. 3.2.7a, see https://github.com/NBISweden/MrBayes/; Huelsenbeck and Ronquist 2001) with 10,000,000 generations, saving one tree every 1000 generations to obtain 10,000 trees, of which the first 2500 were discarded as burn-in, following parameter estimations and convergence examined by Tracer (ver. 1.7, see https://github.com/beast-dev/tracer/releases/tag/v1.7.1; Rambaut et al. 2018)
Results
Morphological results
Based on macro and micromorphological examinations, the 161 Antipatharia colonies collected from the Saudi Arabian Red Sea were ascribed to 4 families (Antipathidae, Aphanipathidae, Myriopathidae and Schizopathidae) and 11 genera (Supplementary Table S1). A detailed description of the morphological characters used to identify the specimens at genus level can be found in the Supplementary Text S1, Supplementary Fig. S1–S4).
Family ANTIPATHIDAE Ehrenberg, 1834
Overall, 104 colonies belonged to the family Antipathidae. The genera Allopathes Opresko & Cairns, 1994, Antipathes, Cirrhipathes, Stichopathes, Pseudocirrhipathes Bo & Bavestrello, 2009, cf. Pseudocirrhipathes were identified within the family.
Family APHANIPATHIDAE Opresko, 2004
In our study, the 21 specimens identified as Aphanipathidae belong to the genera Acanthopathes Opresko, 2004, Aphanipathes Brook, 1889, Asteriopathes Opresko, 2004, Tetrapathes Opresko and cf. Tetrapathes.
Family MYRIOPATHIDAE Opresko, 2001
The 31 representatives identified as Myriopathidae belong to the genus Myriopathes Opresko, 2001 and cf. Myriopathes.
Family SCHIZOPATHIDAE Brook, 1889
The four representatives of the family Schizopathidae detected in this study belong to the genus Bathypathes and correspond to those examined by Chimienti et al. (2022).
Molecular results
The mitochondrial intergenic region IgrW dataset included 258 sequences, 102 of which were retrieved from GenBank (Supplementary Fig. S5) and 156 from this study. The IgrN dataset included 254 sequences of Antipatharia, 89 of which were retrieved from GenBank (Supplementary Fig. S6) and 165 sequences newly produced in this work. The cox3-cox1 dataset included 132 sequences, of which 98 were retrieved from GenBank (Supplementary Fig. S7) and 34 from this study. Finally, the IgrN-IgrW concatenated dataset included 228 sequences of Antipatharia, 161 of which were newly produced in this work and 67 sequences retrieved from GenBank. Information on final alignment and statistics for each molecular marker used in this study can be found in Table 1. Regarding single marker reconstructions, the topologies obtained from BI and ML analyses for IgrW and IgrN were mostly concordant (Fig. 2, Supplementary Fig. S5, S6), but this was not the case for the cox3-cox1 reconstructions (Supplementary Fig. S7) as detailed below. Only the Bayesian phylogram of the combined IgrN-IgrW dataset with branch support indicated by Bayesian posterior probability (PPBI) and ML bootstrap support (BTML) is shown in Fig. 2.
| Locus | Sequences from this study | Sequences from GenBank | FA (bp) | SC | SV | SPI | |
|---|---|---|---|---|---|---|---|
| IgrW | 156 | 102 | 616 | 406 | 210 | 205 | |
| IgrN | 165 | 89 | 705 | 371 | 334 | 315 | |
| cox3-cox1 | 34 | 98 | 838 | 575 | 252 | 234 | |
| IgrN-IgrW | 161 | 67 | 1321 | 707 | 535 | 493 |
Bayesian inference phylogenetic reconstruction of Antipatharia based on the concatenated dataset of the mitochondrial intergenic regions IgrW and IgrN. Numbers at nodes indicate Bayesian posterior probability (≥0.7) and maximum likelihood bootstrap (≥70). Specimens analysed in this study are in bold. Representatives of the Antipathidae that clusters in another family-level clade are marked with single asterisks. Representatives of the Stylopathidae that clusters in another family-level clade are marked with double asterisks. Representatives of the Cladopathidae that cluster in another family-level clade are marked with triple asterisks.
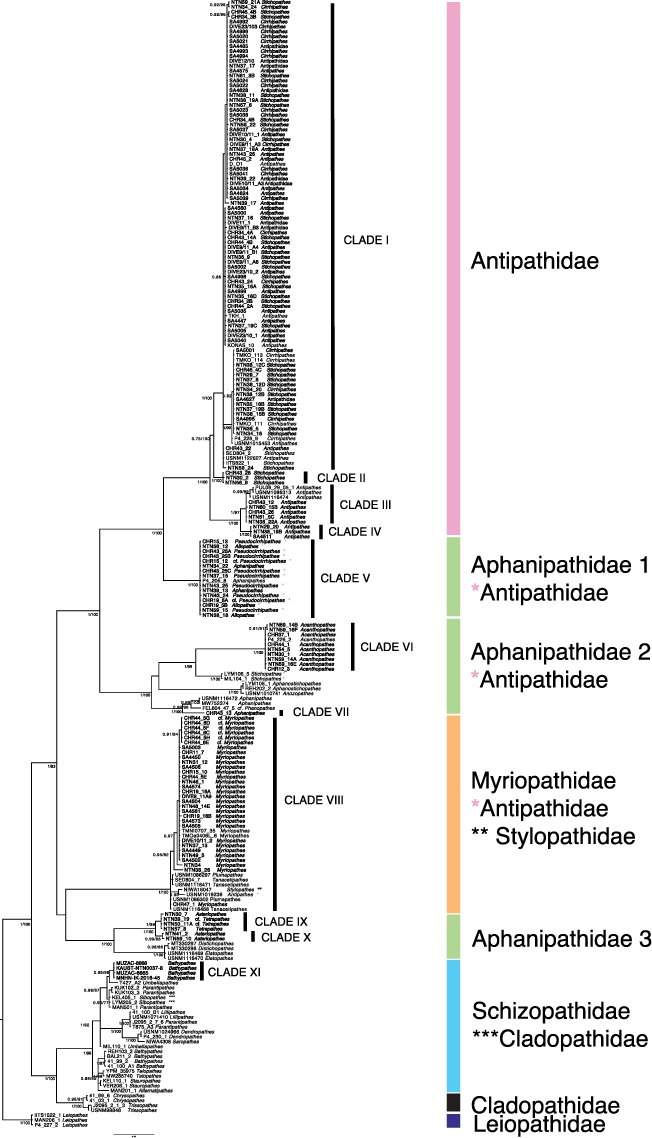
The IgrW and IgrN phylogeny reconstructions included specimens ascribed to the families Antipathidae, Cladopathidae, Leiopathidae, Myriopathidae and Schizopathidae, and the examined representatives of the family Aphanipathidae were split into three clades (Aphanipathidae 1, Aphanipathidae 2 and Aphanipathidae 3). Our newly produced sequences nested within the lineages corresponding to the families Antipathidae, Aphanipathidae 1, Aphanipathidae 2, Aphanipathidae 3, Myriopathidae and Schizopathidae. In particular, the concatenated phylogenetic reconstruction clustered the newly produced Red Sea sequences into 11 clades (clade I to clade XI), six of which included only samples from the Red Sea (clades II, IV, VII, IX, X, XI), whereas five included sequences from both our study and GenBank (clades I, III, V, VI, VIII) (Fig. 2).
The combined IgrN-IgrW phylogeny recovered specimens identified as Antipathidae in four distinct molecular clades (from clade I to clade IV) (Fig. 2). Clade I (PPBI = 1, BTML = 100) included sequences obtained from the genera Antipathes, Cirrhipathes and Stichopathes. Clade II included representatives of Stichopathes and was highly supported by BI posterior probability and ML bootstrap values (PPBI = 1 BTML = 100 respectively). Clades III and IV both comprised sequences of Antipathes. Specimens ascribed to the family Aphanipathidae based on morphology were split into three major clades (Aphanipathidae 1, Aphanipathidae 2 and Aphanipatidae 3). Aphanipathidae 1 (clade IV) included representatives of the genera Aphanipathes (also clustering in the Aphanipathidae 2), Allopathes, Pseudocirrhipathes and cf. Pseudocirrhipathes (the latter two genera are currently classified in the family Antipathidae). The Aphanipathidae 2 lineage consisted of seven molecular clades, yet only two included sequences from the Red Sea (clade VI and clade VII). Clade VI (PPBI = 1, BTML = 100) included sequences of Acanthopathes, whereas clade VII included the second group of representatives of the genus Aphanipathes. Sequences of the unbranched whip Stichopathes, Aphanostichopathes Bo and Opresko, 2021, two further groups of the genus Aphanipathes and Phanopathes Opresko, 2004 from GenBank also clustered within the Aphanipathidae 2. The Aphanipathidae 3 lineage included a total of four molecular clades, with Red Sea representatives grouped in clades IX and X. The highly supported clade IX (PPBI = 1, BTML = 99) was composed of four Red Sea sequences identified as Asteriopathes, Tetrapathes and cf. Tetrapathes, whereas two specimens identified as Asteriopathes clustered within clade X (PPBI = 0.99, BTML = 88).
In the Myriopathidae lineage, we recovered only one group, clade VIII (PPBI = 1, BTML = 100), that included Red Sea sequences of the genus Myriopathes and cf. Myriopathes, and GenBank sequences of the genera Plumapathes Opresko, 2001, Tenancipathes Opresko, 2001, Antipathes and Stylophates Opresko, 2006 (the latter two currently classified within the families Antipathidae and Stylopathidae respectively) (Fig. 2). These phylogenetic relationships were concordant in the IgrN and IgrW single-locus reconstructions but not in the cox3-cox1 one. In particular, in the latter tree (Supplementary Fig. S7), a clade clustering Stylopathes sequences downloaded from GenBank was sister to the Myriopathidae lineage, rather than clustering within this. Moreover, in the cox3-cox1 reconstruction, the Myriopathidae formed a sister clade to Aphanipathidae 2 rather than Aphanipathidae 3, as retrieved in the other phylogenetic reconstructions.
The family Schizopathidae included eight molecular clades, among which clade XI comprised Red Sea sequences (PPBI = 0.99, BTML = 99) recently identified as Bathypathes (Chimienti et al. 2022) (Fig. 2). The other sequences clustering in the Schizopathidae belonged to the genera Umbellapathes Opresko, 2005, Parantipathes Brook, 1889, Lillipathes Opresko, 2002, Dendropathes Opresko, 2005, Saropathes Opresko, 2002, Telopathes MacIsaac & Best, 2013, Stauropathes Opresko, 2002, Alternatipathes Molodtsova & Opresko, 2017 and Sibopathes van Pesch, 1914 (the latter classified within the family Cladopathidae). The family Cladopathidae included only GenBank sequences of the genera Crysopathes Opresko, 2003 and Trissopathes Opresko, 2003.
Geographical and bathymetric distributions
In this work, we integrated newly collected material from the shallow to the deep Red Sea with published sequence data from several localities in the Atlantic and Pacific Oceans. Specimens analysed for this study were sampled in two main areas of the Saudi Arabian Red Sea, the northern Red Sea and the Gulf of Aqaba (Fig. 1), with a minor sampling effort in the central Red Sea. The geographical distributions of the recovered clades is shown in Table 2. In all reconstructions, six clades recovered samples from the Red Sea exclusively. Specifically, clade VII – Aphanipathes and clade X – Asteriopathes, comprised specimens collected in the Gulf of Aqaba only, clade II – Stichopathes, clade IV – Antipathes, clade IX – Asteriopathes, Tetrapathes and cf. Tetrapathes and clade XI – Bathypathes, recovered samples from the northern Red Sea and the Gulf of Aqaba. In the combined IgrN-IgrW dataset, a total of five clades that comprised samples from the Red Sea clustered with published sequences from the Atlantic and Pacific Oceans. In detail, clade I – Antipathes, Cirrhipathes and Stichopathes presented a disjunct distribution including the Red Sea (Antipathes, Cirrhipathes and Stichopathes), Northeast Pacific (Hawaii – Antipathes and Cirrhipathes), Caribbean Sea (Antipathes), Northwest Pacific (South China Sea – Antipathes) and Northwest Atlantic (Florida – Stichopathes). Clade III – Antipathes clustered samples of Antipathes from the Red Sea and Gulf of Mexico. Notably, clade V – Allopathes, Pseudocirrhipathes, cf. Pseudocirrhipathes, Aphanipathes and clade VI – Acanthopathes, comprised samples from the Red Sea and Northeast Pacific (Hawaii – Aphanipathes and Acanthopathes). Finally, clade VIII – Myriopathes, cf. Myriopathes, Plumapathes, Tanacetipathes, Antipathes and Stylopathes included samples from the Red Sea (Myriopathes and cf. Myriopathes), Northwest Pacific (Hawaii – Myriopathes), Northwest Atlantic (Gulf of Mexico – Plumapathes, Tanacetipathes and Florida – Tanacetipathes), North–central Pacific (Stylopathes) and Northeast Pacific (Antipathes).
| Clade | Genus | RS | HW | GM | C | NWAO | NWPO | NCPO | NEPO | |
|---|---|---|---|---|---|---|---|---|---|---|
| I | Antipathes | × | × | × | × | |||||
| Cirrhipathes | × | × | ||||||||
| Stichopathes | × | × | ||||||||
| II | Stichopathes | × | ||||||||
| III | Antipathes | × | × | |||||||
| IV | Antipathes | × | ||||||||
| V | Allopathes | × | ||||||||
| Aphanipathes | × | × | ||||||||
| Pseudocirrhipathes | × | |||||||||
| cf. Pseudocirrhipathes | × | |||||||||
| VI | Acanthopathes | × | × | |||||||
| VII | Aphanipathes | × | ||||||||
| VIII | Myriopathes | × | × | |||||||
| cf. Myriopathes | × | |||||||||
| Plumapathes | × | |||||||||
| Tanacetipathes | × | × | ||||||||
| Antipathes | × | |||||||||
| Stylopathes | × | |||||||||
| IX | Asteriopathes | × | ||||||||
| Tetrapathes | × | |||||||||
| cf. Tetrapathes | × | |||||||||
| X | Asteriopathes | × | ||||||||
| XI | Bathypathes | × |
RS, Red Sea; HW, Hawaii; GM, Gulf of Mexico; C, Caribbean Sea; NWAO, Northwest Atlantic Ocean; NWPO, Northwest Pacific Ocean; NCPO, North–central Pacific Ocean; NEPO, Northeast Pacific Ocean.
Table 3 reports a comparison of the bathymetric distribution of the black coral genera from our study and data from GenBank. In the concatenated phylogenetic reconstruction, from a bathymetric perspective, no clades were composed exclusively of material collected in shallow water environments (0–30 m) (Supplementary Table S1). Four out of the six exclusive Red Sea lineages comprised depth specialists. Particularly, clade VII – Aphanipathes included a single sample found in mesophotic waters (143 m), clade IX – Asteriopathes, Tetrapathes and cf. Tetrapathes included samples collected from 94 to 163 m, whereas clade X – Asteriopathes was composed of two sequences from colonies collected between 171 and 193 m. Notably, clade XI – Bathypathes was a deep depth specialist, being found between 278 and 627 m. Clade IV – Antipathes included specimens from shallow and mesophotic waters distributed from 5 to 83 m, whereas clade II – Stichopathes was distributed between 164 and 217 m. As for the five clades that included sequences from GenBank, clade III – Antipathes comprised samples collected in the Red Sea from 74 to 155 m, whereas GenBank representatives ranged between 60 and 69 m. Clade V – Allopathes, Aphanipathes, Pseudocirrhipathes and cf. Pseudocirrhipathes comprised Red Sea specimens collected between 76 and 165 m, whereas the only GenBank sequence clustering (Aphanipathes) was obtained from a sample found at a depth range of 88–102 m. Clade VI – Acanthopathes comprised sequences from the mesophotic and deep, with samples from the Red Sea ranging from 95 to 328 m and data from GenBank at 263 m. Clade VIII – Myriopathes, cf. Myriopathes, Plumapathes, Tanacetipathes, Antipathes and Stylopathes had a distribution range that spanned the shallow, mesophotic and deep environments. Red Sea specimens ascribed to the genus Myriopathes were recorded from 5 to 412 m, and representatives of GenBank were ascribed to the same genus and the other genera clustering here, ranged from 25 to 1174 m. Ultimately clade I – Antipathes, Cirrhipathes and Stichopathes was also retrieved from the shallow, mesophotic and deep environments, with Red Sea sequences ranging between 6 and 492 m, whereas representatives of GenBank ranged between 6 and 332 m.
| Clade | Genus | Source | Depth (m) | ||||
|---|---|---|---|---|---|---|---|
| 1–30 | 30–150 | 150–300 | >300 | ||||
| I | Antipathes | Red Sea | × | × | |||
| GenBank | × | × | |||||
| Cirrhipathes | Red Sea | × | × | ||||
| GenBank | × | × | |||||
| Stichopathes | Red Sea | × | × | × | × | ||
| GenBank | × | ||||||
| II | Stichopathes | Red Sea | × | ||||
| III | Antipathes | Red Sea | × | ||||
| GenBank | × | ||||||
| IV | Antipathes | Red Sea | × | ||||
| V | Allopathes | Red Sea | × | ||||
| Aphanipathes | Red Sea | × | |||||
| GenBank | × | ||||||
| Pseudocirrhipathes | Red Sea | × | |||||
| cf. Pseudocirrhipathes | Red Sea | × | |||||
| VI | Acanthopathes | Red Sea | × | × | × | ||
| GenBank | × | ||||||
| VII | Aphanipathes | Red Sea | × | ||||
| VIII | Myriopathes | Red Sea | × | × | × | × | |
| GenBank | × | ||||||
| cf. Myriopathes | Red Sea | × | × | ||||
| Plumapathes | GenBank | × | |||||
| Tanacetipathes | GenBank | × | |||||
| Antipathes | GenBank | × | |||||
| Stylopathes | GenBank | × | |||||
| IX | Asteriopathes | Red Sea | × | ||||
| Tetrapathes | Red Sea | × | |||||
| cf. Tetrapathes | Red Sea | × | |||||
| X | Asteriopathes | Red Sea | × | ||||
| XI | Bathypathes | Red Sea | × | × | |||
Discussion
Our work represents the first molecular study of the order Antipatharia in the Red Sea on a dataset spanning a 627-m bathymetric range. Our results confirmed the presence of the families Antipathidae, Aphanipathidae, Myriopathidae and Schizopathidae in the northern and central Red Sea, and provided a large molecular database for genetic comparisons between the Red Sea and other biogeographical provinces in the Indo-Pacific and Atlantic Ocean.
Phylogenetic relationships within the order
In recent decades, the advent of molecular tools has revealed the incongruence of morphology based taxonomy and classification of Antipatharia, and the evolutionary relationships. Studies that involved the use of an integrated systematic approach revealed widespread polyphyly at family and genus levels (e.g. Brugler et al. 2013; Macisaac et al. 2013; Horowitz et al. 2022). For example, the families Cladopathidae and Schizopathidae are not resolved because the cladopathid genus Sibopathes clusters within Schizopathidae (Lü et al. 2021; Chimienti et al. 2022). Moreover, a more complex situation is retrieved for the families Antipathidae and Aphanipathidae, for which several genera are currently intermixed based on molecular data (Barrett et al. 2020; Horowitz et al. 2022). Overall, our analyses confirmed the polyphyly of several taxa within the order. Indeed, specimens ascribed to the genera Antipathes, Stichopathes and Cirrhipathes based on traditional morphological diagnostic features clustered together indistinctly in clade I and could not be differentiated based on molecular results (Fig. 2, Supplementary Fig. S5, S6). This result is consistent with Horowitz et al. (2022) who reconstructed the phylogeny of black corals from a target capture dataset of UCEs and exon loci. Barrett et al. (2020) also found, based on complete mitochondrial genomes, a lack of phylogenetic signal between Antipathes and Stichopathes. Interestingly, within the same family, further analyses might highlight that the species clustering within the Red Sea clades II – Stichopathes and IV – Antipathes, represent Red Sea endemics. This would be consistent with the Red Sea’s recognition as a marine biodiversity hotspot (DiBattista et al. 2016b). Moreover, genera Allopathes, Pseudocirrhipathes and cf. Pseudocirrhipathes (all of the family Antipathidae) clustered in clade V, together with the aphanipatid genus Aphanipathes. Accordingly, the morphological distinction among these genera is not supported by our molecular data. Different hypotheses can be put forth to explain this result, including low-resolution of molecular markers, incipient speciation, hybridisation or non-informative morphological characters (Brugler et al. 2013; Terraneo et al. 2016). Although the aphanipatid genus Aphanipathes falls within clade V, this clade notably exhibits a closer phylogenetic relationship to the Antipathidae lineage than the remaining two lineages of Aphanipathidae. Brugler et al. (2013) first evidenced this result by highlighting the sister relationship between Aphanipathes verticillata mauiensis Opresko, Wagner, Montgomery & Brugler, 2012 and the Antipathidae. Our phylogenetic reconstructions also confirmed that the unbranched morphology typical of the Antipathidae genera Cirrhipathes, Stichopathes and Pseudocirrhipathes, and the aphanipathid Aphanostichopathes, evolved several times over the evolutionary history of the order (Bo et al. 2009; Horowitz et al. 2022). Indeed, Pseudocirrhipathes diverged from the two other whip-like genera of the family Antipathidae (Cirrhipathes and Stichopathes). Furthermore, an additional representative of the genus Aphanipathes clustered in clade VII, within the lineage Aphanipathidae 2. To clarify the phylogenetic position of the family Antipathidae, we need to refer to sequences of the type species of Antipathes, Antipathes dichotoma Pallas, 1766. Even if excluded from our study, this species has been shown to be closely related to representatives of the family Aphanipathidae (Brugler et al. 2013; Bo et al. 2018; Horowitz et al. 2020, 2022; Opresko et al. 2020). This evidence suggests that the clade in which sequences of A. dichotoma are nested represents the real phylogenetic position of the family Antipathidae. What falls outside of this is unrepresentative of the genus and consequently the family needs to be revised. The same applies for the family Aphanipathidae in which only studies based on nuclear loci have included the type species, Aphanipathes sarothamnoides Brook, 1889. These studies revealed a close relationship of A. cf. sarothamnoides with the taxa Aphanipathes pedata and Phanopathes cf. rigida (Bo et al. 2009, 2018; Terrana et al. 2021). In our reconstructions these species were sister to clade VII, leading to the hypothesis that this is the real phylogenetic position of the Aphanipathidae and the other lineages are ripe for revision. As additional support, the third lineage of the family Aphanipathidae, Aphanipathidae 3, was not closely related to the lineages in which the other representatives of the Aphanipathidae clustered (Aphanipathidae 1 and 2). Instead, this had a sister relationship with the Myriopathidae (Horowitz et al. 2022). Considering the evolutionary distance among the three Aphanipathidae lineages, a revision of the morphological characters in delineating different taxonomic levels is crucial. As an example, the close phylogenetic relationship of the genera clustering in the lineage Aphanipathidae 3 (Asteriopathes, Tetrapathes, Distichopathes Opresko, 2004 and Elatopathes Opresko, 2004) corresponds with the fact that these share a simple pinnulation pattern (arranged in different rows among the genera) that is not found in any other genus of the family Aphanipathidae. Our concatenated mtDNA-based reconstruction clustered myriopathids from the Red Sea, ascribed to the genus Myriopathes and cf. Myriopathes, into clade VIII. In our reconstructions the family was not resolved because representatives of both the Antipathidae and Stylopathidae grouped within the lineage. All topologies agreed in placing four Red Sea sequences of Bathypathes in clade XI within the Schizopathidae lineage. The comprehensive phylogenetic reconstruction we provided confirmed the already known problematic position of this genus, as discussed by Chimienti et al. (2022).
Overall, this work has highlighted the absence of genetic differentiation among most of the genera analysed that might be explained either by the scarce resolution of the chosen markers or the fact that some morphological characters traditionally used to discriminate family and genera in black corals are not evolutionarily meaningful. Yet nuclear markers previously employed in the same contexts have not shown a higher degree of resolution. The mitochondrial markers amplified in our present study (IgrN, IgrW and cox3-cox1) are among the most widely used in the reconstruction of phylogenetic relationships in the order Antipatharia (Wagner et al. 2010; Brugler et al. 2013; MacIsaac et al. 2013; Chery et al. 2018; Horowitz et al. 2020; Opresko et al. 2020, 2021). These have proven useful in reconstructing higher taxonomic level relationships in the order, as for the families Myriopathidae, Leiopathidae and Schizopathidae but inferring relationships at lower taxonomic level remains problematic. The inclusion of type material in future analyses will be crucial for improving the systematics of the order, alongside updating identifications for certain molecular data deposited in GenBank. Moreover, high throughput sequencing techniques coupled with detailed observations of macro and micromorphological characters should be helpful in determining genera and species boundaries in the order (Cowman et al. 2020; Quattrini et al. 2017).
Unravelling Antipatharia taxonomy: the next steps
Several morphologies typical of different nominal genera of black coral were spread among separate clades and no clear genetic boundaries were observed among several different genera (Fig. 2). Albeit apparently disconcerting, this situation has been encountered in other groups of cnidarians (e.g. Kitahara et al. 2016) and invertebrates that have been undergoing a molecular re-examination prior to black corals. As an example, the taxonomy of the order Scleractinia traditionally relied on macroscopic characters and microstructure (e.g. Chevalier and Beauvais 1987). However, the advent of molecular tools revealed a pervasive polyphyly spread throughout all taxonomic levels (Kitahara et al. 2016), emphasising the need for a reclassification. However, this reclassification was prevented by the evolutionary convergence of morphological characters typically used to define taxa (e.g. Fukami et al. 2008; Kitahara et al. 2016). Since the beginning of the 2000s, the integration of molecular data with traditional lines of evidence such as macro and micromorphology resulted in the identification of problematic taxa that have subsequently been revised (Kitahara et al. 2010; Stolarski et al. 2011) leading to a more robust classification of the order (Budd et al. 2012; Huang et al. 2014). Antipatharian taxa remain defined based on traditional macro and micromorphological characters, generating taxonomic instability when compared to phylogeny. As shown for Scleractinia, cases of convergent evolution and homoplasy of morphological characters likely hide the real systematics of the order, as taxa with similar morphological characters might be phylogenetically distantly related (see for example van Oppen et al. 2001; Fukami et al. 2004). To unravel the real systematic affinities of Antiphatharia, the use of an integrated systematic approach and when possible, genomic tools, is essential.
The Red Sea Antipatharia fauna: new perspectives
We expanded the known biodiversity of antipatharians in the Red Sea through our study, increasing the reported nominal genera from the region from 4 to 11 (Supplementary Table S1). Prior to our study, only representatives of the family Antipathidae, i.e. Stichopathes (Risk et al. 2009; Qurban et al. 2014; Morgulis et al. 2022), Antipathes (Brook 1889; Jones et al. 2000; Risk et al. 2009; Ghandourah and Alorfi 2019; Morgulis et al. 2022) and Cirrhipathes (Brook 1889; Cooper 1903; Herler 2007) were reported from the Red Sea. Additionally, Chimienti et al. (2022) recently reported members of the Schizopathidae, with the newly described species Bathypathes thermophila. Our study confirms the presence of the genera Antipathes, Cirrhipathes, Stichopathes and Bathypathes in the basin. Moreover, thanks to unprecedented sampling efforts below SCUBA diving depths, for the first time we retrieved Allopathes and Pseudocirrhipathes within the Antipathidae, the genera Acanthopathes, Asteriopathes, Aphanipathes and Tetrapathes for the family Aphanipathidae and the genus Myriopathes within the Myriopathidae.
The assessment of the geographical distribution of taxa is fundamental to the investigation of isolation, speciation and migration events (Wiens and Donoghue 2004; Bowen et al. 2013) in evolutionary biology. The findings of this study revealed that of the 11 clades that comprised Red Sea sequences, some were widespread throughout the Gulf of Aqaba, and the northern and central Red Sea, whereas others were restricted to a given region (Table 2). Six clades retrieved from the concatenated dataset potentially represent Red Sea endemics, pending confirmation through additional morphomolecular studies. Indeed, several nominal species of black corals have not been assessed genetically, therefore the identification of Red Sea taxa without proximity to sequences deposited in GenBank based on markers used does not conclusively establish these taxa or lineages as new to science. The Red Sea is considered a marine biodiversity hotspot, hosting the highest rates of endemism for marine fauna in the Arabian and Indian Ocean regions (Sheppard et al. 1992; DiBattista et al. 2016a; Berumen et al. 2019). For example, 6.6% of the Red Sea Scleractinia are endemic to the basin (Berumen et al. 2019; Terraneo et al. 2019, 2021), whereas Veron et al. (2015) and Obura (2016) reported a level of endemism lower than 3% for all the other areas of the Indian Ocean. The evolutionary events that contributed to the origin of high biodiversity and endemism in the Red Sea are still debated. The peculiar abiotic conditions (temperature, salinity and nutrients), and a complex geological and paleoclimatic history of the region (DiBattista et al. 2016a) may have contributed to the isolation of the Red Sea fauna. This includes reef fish, scleractinian corals (DiBattista et al. 2013, 2016a; Berumen et al. 2019) and possibly also black corals.
Bathymetric distribution of black corals in the Saudi Arabian Red Sea
Although black corals are known to have higher abundance and diversity in deep waters (Tazioli et al. 2007), our Red Sea investigation of shallow, mesophotic and deep ecosystems revealed a high diversity of black corals occurring at mesophotic depths (30–200 m) (Table 3). This corroborates the findings of Bo et al. (2019), in which the authors related tropical and subtropical latitudes (Central–Western–Eastern Indo-Pacific regions and the Caribbean Sea) with mesophotic black coral assemblages, whereas temperate latitudes (the Northeast Atlantic and Mediterranean Sea) appeared to host black coral assemblages at depths greater than 150 m. In our study most of the recovered clades were depth specialists and only clade I – Antipathes, Cirrhipathes and Stichopathes, and clade VIII – Myriopathes, cf. Myriopathes, Plumapathes, Tanacetipathes, Antipathes and Stylopathes occurred throughout the depth gradient. Specifically, Red Sea specimens that nested in clades of the Aphanipathidae were exclusively encountered at mesophotic depths, with a single exception in clade VI – Acanthopathes, in which only two Acanthopathes specimens were sampled in low mesophotic waters. Another family typical of cold, deep waters is Schizopathidae. As already highlighted by Chimienti et al. (2022), B. thermophila is the only member of this family known to live in relatively warm deep waters. Interestingly, the families Antipathidae and Myriopathidae do not appear to be depth specialists (Table 3). In the Antipathidae, except for clade III – Antipathes that inhabits the mesophotic, all the other clades were depth generalist. The genus Myriopathes occurred from shallow to deep waters (4–412 m) (Table 3), although Myriopathidae members typically inhabit shallow-mesophotic ecosystem (Bo et al. 2019). This is evidence of how the distribution expands to the deep sea in a basin such as the Red Sea that is characterised by extreme conditions and deep waters warmer than in any other existing ocean basin. The extreme temperature gradients, seasonal temperature fluctuations exceeding 10°C (Berumen et al. 2019) and deep water temperature stabilised at 21°C may have allowed thermosensitive taxa to expand bathymetric ranges in the Red Sea.
Conclusions
Our study represents a stepping stone towards a better understanding of antipatharian biodiversity, distribution and evolutionary relationships. The phylogenetic reconstructions presented confirmed that the families Antipathidae and Aphanipathidae are polyphyletic and therefore do not represent natural groups. Through first time analysis of the mitochondrial DNA of Allopathes, Pseudocirrhipathes and Tetrapathes we strongly encourage a taxonomic revision of these families. Genetic evidence provided in this work highlighted the unsuitability of relying solely on traditional morphological characters in delimiting boundaries within the order Antipatharia at different taxonomic levels, as variable and/or plastic skeletal characters do not correspond to evolutionary relationships. Indeed, several morphologies typical of different nominal genera were spread among separate clades. This is the first study that investigated the morphological and molecular diversity of black corals in the Red Sea and several undescribed taxa are expected to be found among the mesophotic and deep water samples. However, only a small fraction of the basin has been investigated in this work, and the south and western coasts of the Red Sea must be included in future studies to adequately assess the biodiversity of antipatharians in the basin. Moreover, many important areas outside the basin are missing, where the poorly studied south-western Indian Ocean would be the natural biogeographical fit to complement this study.
Supplementary material
Supplementary Table S1 lists the specimens analysed in this study and used in phylogenetic analyses. Metadata, family and genus-level identification and corresponding GenBank accession numbers for each marker are provided for each specimen. Supplementary Table S2 provides the list and details of the primers used for sequences newly produced in this study. The ‘Morphological results’ section provides a detailed description of the morphological characters used to identify the specimens. Supplementary Fig. S1, S2, S3 and S4 illustrate in situ, polyp and spine images of the analysed Antipathidae, Aphanipathidae, Myriopathidae and Schizopathidae genera respectively. Supplementary Fig. S5 shows the BI analysis based on mitochondrial region trnW-IGR-nad2. Supplementary Fig. S6 reports the BI analysis based on mitochondrial region nad5-IGR-nad1. Supplementary Fig. S7 provides the BI analysis based on mitochondrial region cox3-IGR-cox1. Supplementary material is available online.
Data availability
The sequences that support this study are available in GenBank, with corresponding accession numbers listed in Supplementary Table S1.
Declaration of funding
This work was supported and founded by NEOM and KAUST (award FCC/1/1973-50-01 and baseline research funds to F. Benzoni).
Acknowledgements
We kindly thank NEOM, particularly Thamer Habis, Razan Khamis, Peter Mackelworth, Paul Marshall, Justin Mynar and Giacomo Palavacini for organising, coordinating and facilitating the Red Sea Expedition in 2020. We thank OceanX and the crew of OceanXplorer for operational and logistical support for the duration of this expedition. Specifically, we would like to acknowledge the dive support, ROV and Sub teams for data acquisition, sample collection and support of scientific operations on board OceanXplorer. We thank OceanX Media for documenting and communicating this work to the public. We are grateful to Christina Alcon for support in the DNA extraction and amplification, KAUST Sanger Sequencing Core Lab for helping with sequencing and Alessandro Genovese for facilitating the use of the SEM at KAUST Imaging Core Lab. We also thank the anonymous reviewers and editor of the journal, Dr Allen Collins, for useful suggestions that helped improve the manuscript.
References
Arrigoni R, Benzoni F, Terraneo TI, Caragnano A, Berumen ML (2016) Recent origin and semi-permeable species boundaries in the scleractinian coral genus Stylophora from the Red Sea. Scientific Reports 6(1), 34612.
| Crossref | Google Scholar | PubMed |
Arrigoni R, Berumen ML, Huang D, Terraneo TI, Benzoni F (2017) Cyphastrea (Cnidaria: Scleractinia: Merulinidae) in the Red Sea: phylogeny and a new reef coral species. Invertebrate Systematics 31(2), 141-156.
| Crossref | Google Scholar |
Arrigoni R, Huang D, Berumen ML, Budd AF, Montano S, Richards ZT, et al. (2021) Integrative systematics of the scleractinian coral genera Caulastraea, Erythrastrea and Oulophyllia. Zoologica Scripta 50(4), 509-527.
| Crossref | Google Scholar |
Barrett NJ, Hogan RI, Allcock AL, Molodtsova T, Hopkins K, Wheeler AJ, Yesson C (2020) Phylogenetics and mitogenome organisation in black corals (Anthozoa: Hexacorallia: Antipatharia): an order-wide survey inferred from complete mitochondrial genomes. Frontiers in Marine Science 7, 440.
| Crossref | Google Scholar |
Berumen ML, Arrigoni R, Bouwmeester J, Terraneo TI, Benzoni F (2019) Corals of the Red Sea. In ‘Coral Reefs of the Red Sea’. (Eds CR Voolstra, ML Berumen) pp. 123–155. (Springer: Cham, Switzerland) 10.1007/978-3-030-05802-9
Bo M, Barucca M, Biscotti MA, Canapa A, Lapian HFN, Olmo E, Bavestrello G (2009) Description of Pseudocirrhipathes (Cnidaria: Anthozoa: Hexacorallia: Antipathidae), a new genus of whip black corals from the Indo-Pacific. Italian Journal of Zoology 76(4), 392-402.
| Crossref | Google Scholar |
Bo M, Bavestrello G, Barucca M, Makapedua DM, Poliseno A, Forconi M, Olmo E, Canapa A (2012) Morphological and molecular characterization of the problematic whip black coral genus Stichopathes (Hexacorallia: Antipatharia) from Indonesia (North Sulawesi, Celebes Sea). Zoological Journal of the Linnean Society 166, 1-13.
| Crossref | Google Scholar |
Bo M, Barucca M, Biscotti MA, Brugler MR, Canapa A, Canese S, Lo Iacono C, Bavestrello G (2018) Phylogenetic relationships of Mediterranean black corals (Cnidaria : Anthozoa : Hexacorallia) and implications for classification within the order Antipatharia. Invertebrate Systematics 32(5), 1102-1110.
| Crossref | Google Scholar |
Bo M, Montgomery AD, Opresko DM, Wagner D, Bavestrello G (2019) Antipatharians of the mesophotic zone: four case studies. In ‘Mesophotic coral ecosystems.’ (Eds Y Loya, KA Puglise, TC Bridge) Coral reefs of the worls series (n. 12). pp. 683–708. (Springer: NY) 10.1007/978-3-319-92735
Bouwmeester J, Benzoni F, Baird AH, Berumen ML (2015) Cyphastrea kausti sp. n. (Cnidaria, Anthozoa, Scleractinia), a new species of reef coral from the Red Sea. ZooKeys 496, 1-13.
| Crossref | Google Scholar | PubMed |
Bowen BW, Rocha LA, Toonen RJ, Karl SA, the ToBo Laboratory (2013) The origins of tropical marine biodiversity. Trends in Ecology & Evolution 28(6), 359-366.
| Crossref | Google Scholar | PubMed |
Brook G (1889) The Voyage of HMS Challenger. Zoology. Report on the Antipatharia collected by HMS Challenger during the years 1873–1876. The Voyage of HMS Challenger LXXX, 1-222.
| Google Scholar |
Brugler MR, Opresko DM, France SC (2013) The evolutionary history of Antipatharia (Cnidaria: Anthozoa: Hexacorallia) as inferred from mitochondrial and nuclear DNA: implications for black coral taxonomy and systematics. Zoological Journal of the Linnean Society 169(2), 351-361.
| Crossref | Google Scholar |
Budd AF, Fukami H, Smith ND, Knowlton N (2012) Taxonomic classification of the reef coral family Mussidae (Cnidaria: Anthozoa: Scleractinia). Zoological Journal of the Linnean Society 166(3), 465-529.
| Crossref | Google Scholar |
Cairns SD (2007) Deep-water corals: an overview with special reference to diversity and distribution of deep-water scleractinian corals. Bullettin of Marine Science 81, 311-322.
| Google Scholar |
Chery N, Parra K, Evankow A, Stein D, Distel D, Appiah-madson H, Ross R, Sanon E, Alomari N, Johnson R, Vasovic A, Horowitz A, Popa HL, Short B, Vasquez DM, Rodriguez E, Opresko DM, Brugler MR (2018) Partnering with Ocean Genome Legacy to advance our understanding of black corals (Order Antipatharia). In ‘15th Deep-Sea Biology Symposium’, 9–14 September 2018, Monterey, CA, USA. Available at https://2328be37‐4b2b‐4774‐894d‐916f74520ece.filesusr.com/ugd/5fcd02_e4a81ea0a872469196f20467f9f9ee4c.pdf [Poster presentation]
Chimienti G, De Padova D, Mossa M, Mastrototaro F (2020) A mesophotic black coral forest in the Adriatic Sea. Scientific Reports 10(1), 8504.
| Crossref | Google Scholar | PubMed |
Chimienti G, Terraneo TI, Vicario S, Marchese F, Purkis SJ, Abdulla Eweida A, Rodrigue M, Benzoni F (2022) A new species of Bathypathes (Cnidaria, Anthozoa, Antipatharia, Schizopathidae) from the Red Sea and its phylogenetic position. Zookeys 1116, 1-22.
| Crossref | Google Scholar | PubMed |
Cowman PF, Quattrini AM, Bridge TCL, Watkins-Colwell GJ, Fadli N, Grinblat M, Roberts TE, McFadden CS, Miller DJ, Baird AH (2020) An enhanced target-enrichment bait set for Hexacorallia provides phylogenomic resolution of the staghorn corals (Acroporidae) and close relatives. Molecular Phylogenetics and Evolution 153, 106944.
| Crossref | Google Scholar |
Darriba D, Taboada GL, Doallo R, Posada D (2012) JModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9(8), 772.
| Crossref | Google Scholar | PubMed |
DiBattista JD, Berumen ML, Gaither MR, Rocha LA, Eble JA, Choat JH, Craig MT, Skillings DJ, Bowen BW (2013) After continents divide: comparative phylogeography of reef fishes from the Red Sea and Indian Ocean. Journal of Biogeography 40, 1170-1181.
| Crossref | Google Scholar |
DiBattista JD, Howard Choat J, Gaither MR, Hobbs JPA, Lozano-Cortés DF, Myers RF, Paulay G, Rocha LA, Toonen RJ, Westneat MW, Berumen ML (2016a) On the origin of endemic species in the Red Sea. Journal of Biogeography 43(1), 13-30.
| Crossref | Google Scholar |
DiBattista JD, Roberts MB, Bouwmeester J, Bowen BW, Coker DJ, Lozano-Cortés DF, Howard Choat J, Gaither MR, Hobbs JPA, Khalil MT, Kochzius M, Myers RF, Paulay G, Robitzch VSN, Saenz-Agudelo P, Salas E, Sinclair-Taylor TH, Toonen RJ, Westneat MW, Williams ST, Berumen ML (2016b) A review of contemporary patterns of endemism for shallow water reef fauna in the Red Sea. Journal of Biogeography 43(3), 423-439.
| Crossref | Google Scholar |
Fukami H, Budd AF, Paulay G, Solé-Cava A, Chen CA, Iwao K, Knowlton N (2004) Conventional taxonomy obscures deep divergence between Pacific and Atlantic corals. Nature 427, 832-835.
| Crossref | Google Scholar | PubMed |
Fukami H, Chen CA, Budd AF, Collins A, Wallace C, Chuang YY, Chen C, Dai CF, Iwao K, Sheppard C, Knowlton N (2008) Mitochondrial and nuclear genes suggest that stony corals are monophyletic but most families of stony corals are not (Order Scleractinia, Class Anthozoa, Phylum Cnidaria). PLoS One 3, e3222.
| Crossref | Google Scholar | PubMed |
Ghandourah MA, Alorfi HS (2019) New cytotoxic fatty acid esters from the Black coral, Antipathes dichotoma. Tropical Journal of Pharmaceutical Research 18(1), 69.
| Crossref | Google Scholar |
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52(5), 696-704.
| Crossref | Google Scholar | PubMed |
Haverkort-Yeh RD, McFadden CS, Benayahu Y, Berumen M, Halász A, Toonen RJ (2013) A taxonomic survey of Saudi Arabian Red Sea octocorals (Cnidaria: Alcyonacea). Marine Biodiversity 43, 279-291.
| Crossref | Google Scholar |
Herler J (2007) Microhabitats and ecomorphology of coral- and coral rock-associated gobiid fish (Teleostei: Gobiidae) in the northern Red Sea. Marine Ecology 28, 82-94.
| Crossref | Google Scholar |
Horowitz J, Brugler MR, Bridge TCL, Cowman PF (2020) Morphological and molecular description of a new genus and species of black coral (Cnidaria: Anthozoa: Hexacorallia: Antipatharia: Antipathidae: Blastopathes) from Papua New Guinea. Zootaxa 4821(3), 553-569.
| Crossref | Google Scholar | PubMed |
Horowitz J, Opresko D, Molodtsova TN, Beaman RJ, Cowman PF, Bridge TC (2022) Five new species of black coral (Anthozoa; Antipatharia) from the Great Barrier Reef and Coral Sea, Australia. Zootaxa 5213(1), 1-35.
| Crossref | Google Scholar | PubMed |
Huang D, Benzoni F, Fukami H, Knowlton N, Smith ND, Budd AF (2014) Taxonomic classification of the reef coral families Merulinidae, Montastraeidae and Diploastraeidae (Cnidaria: Anthozoa: Scleractinia). Zoological Journal of the Linnean Society 171, 277-355.
| Crossref | Google Scholar |
Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17(8), 754-755.
| Crossref | Google Scholar | PubMed |
Jones DS, Hewitt MA, Sampey A (2000) A checklist of the Cirripedia of the south China Sea. The Raffles Bulletin of Zoology 8(Supplement), 233-307.
| Google Scholar |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30(4), 772-780.
| Crossref | Google Scholar | PubMed |
Kitahara MV, Cairns SD, Stolarski J, Blair D, Miller DJ (2010) A comprehensive phylogenetic analysis of the Scleractinia (Cnidaria, Anthozoa) based on mitochondrial CO1 sequence data. PLoS One 5(7), e11490.
| Crossref | Google Scholar | PubMed |
Kitahara MV, Fukami H, Benzoni F, Huang D (2016). The new systematics of Scleractinia: integrating molecular and morphological evidence. In ‘The Cnidaria, past, present and future’. (Eds S Goffredo, Z Dubinsky) pp. 41–59. (Springer: Cham, Switzerland) 10.1007/978-3-319-31305-4_4
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34(3), 772-773.
| Crossref | Google Scholar | PubMed |
Lapian HFN (2009) Biodiversity study of black coral (Order: Antipatharia) collected from Manado, Indonesia based on rDNA internal transcribed spacer (ITS) sequences analysis. Biodiversitas Journal of Biological Diversity 10(1), 1-5.
| Crossref | Google Scholar |
Lapian HFN, Barucca M, Bavestrello G, Biscotti MA, Bo M, Canapa A, Tazioli S, Olmo E (2007) A systematic study of some black corals species (Antipatharia, Hexacorallia) based on rDNA internal transcribed spacers sequences. Marine Biology 151(2), 785-792.
| Crossref | Google Scholar |
Lü T, Zhan Z, Xu K (2021) Morphology and molecular phylogeny of three black corals (Antipatharia, Schizopathidae) from seamounts in the Western Pacific Ocean, with description of a new species. Journal of Oceanology and Limnology 39(5), 1740-1757.
| Crossref | Google Scholar |
Macisaac KG, Best M, Brugler MR, Kenchington ELR, Anstey LJ, Jordan T (2013) Telopathes magna gen. nov., spec. nov. (Cnidaria: Anthozoa: Antipatharia: Schizopathidae) from deep waters off Atlantic Canada and the first molecular phylogeny of the deep-sea family Schizopathidae. Zootaxa 3700(2), 237-258.
| Crossref | Google Scholar | PubMed |
McFadden CS, Haverkort-Yeh R, Reynolds AM, Halàsz A, Quattrini AM, Forsman ZH, Benayahu Y, Toonen RJ (2017) Species boundaries in the absence of morphological, ecological or geographical differentiation in the Red Sea octocoral genus Ovabunda (Alcyonacea: Xeniidae). Molecular Phylogenetics and Evolution 112, 174-184.
| Crossref | Google Scholar | PubMed |
McFadden CS, Van Ofwegen LP, Quattrini AM (2022) Revisionary systematics of Octocorallia (Cnidaria: Anthozoa) guided by phylogenomics. Bulletin of the Society of Systematic Biologists 1(3), 8735.
| Crossref | Google Scholar |
Miller MA, Pfeiffer W, Schwartz T (2011) The CIPRES science gateway. In ‘TG ‘11: Proceedings of the 2011 TeraGrid Conference on Extreme Digital Discovery’, 18–21 July 2011, Salt Lake City, UT, USA. Article 41. (Association for Computing Machinery: New York, NY, USA) doi: 10.1145/2016741.2016785
Molodtsova TN, Budaeva N (2007) Modifications of corallum morphology in black corals as an effect of associated fauna. Bulletin of Marine Science 81(3), 469-480.
| Google Scholar |
Molodtsova TN, Opresko DM (2017) Black corals (Anthozoa: Antipatharia) of the Clarion–Clipperton Fracture Zone. Marine Biodiversity 47(2), 349-365.
| Crossref | Google Scholar |
Morgulis M, Martinez S, Almuly R, Einbinder S, Zaslansky P, Mass T (2022) Black corals (Antipatharia) of the northern Red Sea: ancient predators of the mesophotic reef. Marine Ecology Progress Series 688, 33-47.
| Crossref | Google Scholar |
Obura DO (2016) An Indian Ocean centre of origin revisited: Palaeogene and Neogene influences defining a biogeographic realm. Journal of Biogeography 43, 229-242.
| Crossref | Google Scholar |
Okamura SI, Habe T (1976) The strange habit of Rhizochilus teramachii Kuroda. Venus 35(2), 91-92.
| Google Scholar |
Opresko DM (2001) Revision of the Antipatharia (Cnidaria: Anthozoa). Part I. Establishment of a new family, Myriopathidae. Zoologische Mededelingen 75, 343-370.
| Google Scholar |
Opresko DM (2002) Revision of the Antipatharia (Cnidaria: Anthozoa). Part II. Schizopathidae. Zoologische Mededelingen 76(22), 411-442.
| Google Scholar |
Opresko DM (2004) Revision of the Antipatharia (Cnidaria: Anthozoa). Part IV. Establishment of a new family, Aphanipathidae. Zoologische Mededelingen 78, 209-240.
| Google Scholar |
Opresko DM, Cairns S (1994) Description of the new genus Allopathes (Cnidaria: Antipatharia) and its type species Cirripathes desbonni. Proceedings of the Biological Society of Washington 107(1), 185-192.
| Google Scholar |
Opresko DM, Goldman SL, Johnson R, Parra K, Nuttall M, Schmahl GP, Brugler MR (2020) Morphological and molecular characterization of a new species of black coral from Elvers Bank, north-western Gulf of Mexico (Cnidaria: Anthozoa: Hexacorallia: Antipatharia: Aphanipathidae: Distichopathes). Journal of the Marine Biological Association of the United Kingdom 100(4), 559-566.
| Crossref | Google Scholar |
Opresko DM, Bo M, Stein DP, Evankow A, Distel DL, Brugler MR (2021) Description of two new genera and two new species of antipatharian corals in the family Aphanipathidae (Cnidaria: Anthozoa: Antipatharia). Zootaxa 4966(2), 161-174.
| Crossref | Google Scholar | PubMed |
Quattrini AM, Faircloth BC, Dueñas LF, Bridge TCL, Brugler MR, Calixto‐Botía IF, DeLeo DM, Forêt S, Herrera S, Lee SMY, Miller DJ, Prada C, Rádis‐Baptista G, Ramírez‐Portilla C, Sánchez JA, Rodríguez E, McFadden CS (2017) Universal target‐enrichment baits for anthozoan (Cnidaria) phylogenomics: new approaches to long‐standing problems. Molecular Ecology Resources 18(2), 281-295.
| Crossref | Google Scholar |
Qurban MA, Krishnakumar PK, Joydas TV, Manikandan KP, Ashraf TTM, Quadri SI, Wafar M, Qasem A, Cairns SD (2014) In situ observation of deep water corals in the northern Red Sea waters of Saudi Arabia. Deep-Sea Research Part I: Oceanographic Research Papers 89, 35-43.
| Crossref | Google Scholar |
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67, 901-904.
| Crossref | Google Scholar | PubMed |
Rees WJ (1969) A brief survey of the symbiotic associations of Cnidaria with Mollusca. Journal of Molluscan Studies 37, 213-231.
| Crossref | Google Scholar |
Richmond RH, Hunter CL (1990) Reproduction and recruitment of corals: comparisons among the Caribbean, the Tropical Pacific, and the Red Sea. Marine Ecology Progress Series 60, 185-203.
| Crossref | Google Scholar |
Risk MJ, Sherwood OA, Nairn R, Gibbons C (2009) Tracking the record of sewage discharge off Jeddah, Saudi Arabia, since 1950, using stable isotope records from antipatharians. Marine Ecology Progress Series 397, 219-226.
| Crossref | Google Scholar |
Roark EB, Guilderson TP, Dunbar RB, Fallon SJ, Mucciarone DA (2009) Extreme longevity in proteinaceous deep-sea corals. Proceedings of the National Academy of Sciences of the United States of America 106(13), 5204-5208.
| Crossref | Google Scholar | PubMed |
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9), 1312-1313.
| Crossref | Google Scholar | PubMed |
Stolarski J, Kitahara MV, Miller DJ, Cairns SD, Mazur M, Meibom A (2011) The ancient evolutionary origins of Scleractinia revealed by azooxanthellate corals. BMC Evolutionary Biology 11, 316.
| Crossref | Google Scholar | PubMed |
Tazioli S, Bo M, Boyer M, Rotinsulu H, Bavestrello G (2007) Ecological observations of some common antipatharian corals in the marine park of Bunaken (North Sulawesi, Indonesia). Zoological Studies 46(2), 227-241.
| Google Scholar |
Terrana L, Flot JF, Eeckhaut I (2021) ITS1 variation among Stichopathes cf. maldivensis (Hexacorallia: Antipatharia) whip black corals unveils conspecificity and population connectivity at local and global scales across the Indo-Pacific. Coral Reefs 40(2), 521-533.
| Crossref | Google Scholar |
Terraneo TI, Berumen ML, Arrigoni R, Waheed Z, Bouwmeester J, Caragnano A, Stefani F, Benzoni F (2014) Pachyseris inattesa sp. n. (Cnidaria, Anthozoa, Scleractinia): a new reef coral species from the Red Sea and its phylogenetic relationships. ZooKeys 433, 1-30.
| Crossref | Google Scholar | PubMed |
Terraneo TI, Benzoni F, Arrigoni R, Berumen ML (2016) Species delimitation in the coral genus Goniopora (Scleractinia, Poritidae) from the Saudi Arabian Red Sea. Molecular Phylogenetics and Evolution 102, 278-294.
| Crossref | Google Scholar | PubMed |
Terraneo TI, Benzoni F, Baird AH, Arrigoni R, Berumen ML (2019) Morphology and molecules reveal two new species of Porites (Scleractinia, Poritidae) from the Red Sea and the Gulf of Aden. Systematics and Biodiversity 17(5), 491-508.
| Crossref | Google Scholar |
Terraneo TI, Benzoni F, Arrigoni R, Baird AH, Mariappan KG, Forsman ZH, Wooster MK, Bouwmeester J, Marshell A, Berumen ML (2021) Phylogenomics of Porites from the Arabian Peninsula. Molecular Phylogenetics and Evolution 161, 107173.
| Crossref | Google Scholar | PubMed |
Totton AK (1923) Coelenterata, Part III, Antipatharia (and their Cirripede commensals). British Antarctic (Terra Nova) Expedition, 1910–1913. Natural History Reports, Zoology 5, 97-120.
| Google Scholar |
van Oppen MJH, McDonald BJ, Willis BL, Miller DJ (2001) The evolutionary history of the coral genus Acropora (Scleractinia, Cnidaria) based on a mitochondrial and a nuclear marker: reticulation, incomplete lineage sorting or morphological convergence? Molecular Biology and Evolution 18, 1315-1329.
| Crossref | Google Scholar | PubMed |
Veron J, Stafford-Smith M, DeVantier L, Turak E (2015) Overview of distribution patterns of zooxanthellate Scleractinia. Frontiers in Marine Science 1, 81.
| Crossref | Google Scholar |
Wagner D (2015a) A taxonomic survey of the shallow-water (<150 m) black corals (Cnidaria: Antipatharia) of the Hawaiian Islands. Frontiers in Marine Science 2, 24.
| Crossref | Google Scholar |
Wagner D (2015b) The spatial distribution of shallow-water (<150 m) black corals (Cnidaria: Antipatharia) in the Hawaiian archipelago. Marine Biodiversity Records 8, e54.
| Crossref | Google Scholar |
Wagner D, Brugler MR, Opresko DM, France SC, Montgomery AD, Toonen RJ (2010) Using morphometrics, in situ observations and genetic characters to distinguish among commercially valuable Hawaiian black coral species, a redescription of Antipathes grandis Verrill, 1928 (Antipatharia: Antipathidae). Invertebrate Systematics 24, 271-290.
| Crossref | Google Scholar |
Wagner D, Luck DG, Toonen RJ (2012) The Biology and Ecology of Black Corals (Cnidaria: Anthozoa: Hexacorallia: Antipatharia). In ‘Advances in Marine Biology’. (Ed. M Lesser) Vol. 63, pp. 67–132. (Academic Press) 10.1016/B978-0-12-394282-1.00002-8
Warner GF (1981) Species descriptions and ecological observations of black corals (Antipatharia) from Trinidad. Bulletin of Marine Science 31(1), 147-163.
| Google Scholar |
Wiens JJ, Donoghue MJ (2004) Historical biogeography, ecology and species richness. Trends in Ecology & Evolution 19, 639-644.
| Crossref | Google Scholar | PubMed |


