First report of Squash leaf curl China virus in Pakistan
M. Tahir A D , M. S. Haider B and R. W. Briddon CA NUST Centre of Virology and Immunology, National University of Science and Technology, Tamiz ud Din Road, Lalkurti, Rawalpindi, Pakistan.
B School of Biological Sciences, University of the Punjab, Lahore, Pakistan.
C National Institute for Biotechnology and Genetic Engineering, Jhang Road, Faisalabad, Pakistan.
D Corresponding author. Email: gullsbs@yahoo.com
Australasian Plant Disease Notes 5(1) 21-24 https://doi.org/10.1071/DN10009
Submitted: 5 January 2010 Accepted: 22 February 2010 Published: 22 March 2010
Abstract
Leaf samples of Cucurbita pepo plants showing yellow mosaic symptoms were collected from Lahore, Pakistan. Full-length clones of a bipartite begomovirus were isolated and sequenced in their entirety. The sequences showed the highest levels of nucleotide sequence identity (98.4% for the DNA-A and 89.6% for the DNA-B) to the ‘Indian’ strain of Squash leaf curl China virus (SLCCNV).
The Geminiviridae is a family of circular, single-stranded DNA viruses with characteristic quasi-icosahedral particles (Zhang et al. 2001; Böttcher et al. 2004). The viruses in this family are divided across four genera that are distinguished on the basis of host range, insect vector and genome arrangement (Fauquet et al. 2008). Economically the most important geminiviruses are in the genus Begomovirus. Begomoviruses are transmitted exclusively by the whitefly Bemisia tabaci (Jones 2003). In the Old World the majority of begomoviruses have monopartite genomes, although a small number have genomes consisting of two components, of approximately equal size, which are known as DNA-A and DNA-B. The DNA-A encodes all factors required for virus replication, overcoming host defences, insect transmission and control of gene expression, while DNA-B encodes factors required for inter- and intra-cellular movement in host plants (Hanley-Bowdoin et al. 1999). The majority of the monopartite begomoviruses instead associate with a newly identified class of single-stranded DNA satellites termed betasatellites (Briddon and Stanley 2006).
Several cucurbits are important vegetable crops grown across the Indian subcontinent. Cucurbits throughout Pakistan are affected by several phytopathogenic RNA-containing viruses (Ali et al. 2004). However, little is known about the nature of geminiviruses which affect cucurbit crops in Pakistan. Here we characterise a virus affecting squash in Lahore and show it to be an isolate of the begomovirus Squash leaf curl China virus (SLCCNV), the first time this species has been identified in Pakistan.
Leaf samples of squash (Cucurbita pepo) showing yellow mosaic symptoms were collected in areas around Lahore, Pakistan in 2005 (Fig. 1). Samples were maintained at −80°C until DNA extraction. Total nucleic acids were extracted from leaf samples by the cetyl trimethyl ammonium bromide (CTAB) method (Doyle and Doyle 1990). A partial clone of the begomovirus was obtained by polymerase chain reaction (PCR) amplification with begomovirus group-specific primers (Mansoor et al. 1998). Specific abutting primers (SONF 5′-GGGCCCCCATGAACTCTTTAAAGTG-3′, SONR 5′-GGGCCCAAAGGGACTGGCAATC-3′), for amplification of the full-length DNA-A component were designed to the sequence of the partial clone. The presence of a begomovirus DNA-B component was confirmed using the primer pair BC1F/BC1R (Hussain et al. 2004). Specific abutting primers (KTBF 5′-CTGCAGAGGTCACCTTGTCATTTCCTTC-3′, KTBR 5′-CTGCAGCATCATTTGTGAGCGCATATTC-3′), for the amplification of full-length DNA-B component were designed to the sequences of the partial clones.

|
Potential full-length amplification products were cloned into the pTZ57R/T vector (Fermentas, Burlington, ON, Canada). Sequences were determined by dideoxynucleotide chain-termination sequencing using GenomeLab Dye Terminator Cycle Sequencing kits (Beckman Coulter, Brea, CA, USA) on a Beckman Coulter automated sequencer (CEqn 8000).
Sequences were assembled and analysed using Lasergene (DNAStar Inc., Madison, WI, USA) software. Multiple sequence alignments and phylogenetic trees were produced using ClustalX (Thompson et al. 1997). Phylogenetic trees were manipulated and printed using Treeview (Page 1996). Pairwise sequence comparisons against all begomovirus genome (or DNA-A component) sequences available in the databases were run on-line using pairwise sequence comparison (PASC) software (http://www.ncbi.nlm.nih.gov/sutils/pasc/viridty.cgi).
The amplified DNA-A and DNA-B molecules were cloned and sequenced in their entirety, with no ambiguities remaining. The complete nucleotide sequence of the DNA-A component was determined to be 2756 bp in length (accession number AM286794). From analysis of the sequence the component is predicted to contain six open reading frames (ORFs) with an arrangement typical of the DNA-A components of the Old World begomoviruses, consisting of two ORFs in the virion-sense (encoding the coat protein [CP] and AV2 [of unknown function]) and four in the complementary-sense (encoding the replication associated protein [Rep], the transcriptional activator protein [TrAP], the replication enhancer protein [REn] and AC4 [of unknown function]). The features of this newly derived DNA-A component are summarised in Table 1.
Error: Incorrect filename or format (DN10009_T1.gif). Please check out
| Component | ORF | Start codon coordinates | Stop codon coordinates | Predicted size (no. of amino acids) | Predicted molecular weight (kDa) |
| DNA-A | AV2 | 121 | 459 | 112 | 13.0 |
| CP | 281 | 1051 | 256 | 29.4 | |
| Rep | 2603 | 1500 | 367 | 41.7 | |
| TrAP | 1597 | 1193 | 134 | 15.2 | |
| REn | 1458 | 1048 | 136 | 15.9 | |
| AC4 | 2446 | 2270 | 58 | 6.5 | |
| DNA-B | NSP | 582 (468)A | 1274 | 230 (268)A | 26.4 (30.8)A |
| MP | 2173 | 1328 | 281 | 31.3 |
AA stop codon at the N-terminus of the product (position 525) truncates the gene with respect to other isolates of SLCCNV. The figures in brackets are for the features ignoring the premature stop codon.
The complete nucleotide sequence of the DNA-B component was determined to be 2703 bp (accession number AM778959) and predicted to encode two ORFs (the nuclear shuttle protein [NSP] in the virion-sense and the movement protein [MP] in the complementary-sense; Table 1).
Comparison of the newly derived DNA-A nucleotide sequence using the PASC software showed it was most similar to isolates of SLCCNV with sequence identities between 89.95% and 98.33% and the highest being to Squash leaf curl China virus-India [India:Lucknow:Pumpkin] (SLCCNV-IN[IN:Luc:Pum]; accession number DQ026296; Singh et al. 2008). Based on the presently applicable species demarcation criteria for begomoviruses (89% nucleotide sequence identity; Fauquet et al. 2008) the virus infecting C. pepo is an isolate of SLCCNV. This species occurs as two geographically distinct strains, the ‘China’ and ‘India’ strains. The Pakistani isolate shows between 89.95% and 90.54% identity to the ‘China’ strain, whereas to the two isolates of the ‘India’ strain it is 95.11% and 98.33%. Based on the recently proposed criteria for distinguishing strains of begomoviruses (Fauquet et al. 2008) this indicates that the isolate from Pakistan is of the ‘India’ strain, for which we propose the isolate descriptor Squash leaf curl China virus-India [Pakistan:Lahore:2005] (SLCCNV-IN[PK:Lah:05]). This conclusion is supported by the grouping of the Pakistani isolate with isolates of the ‘India’ strain, and being distinct from isolates of the ‘China’ strain in phylogenetic analyses (Fig. 2).
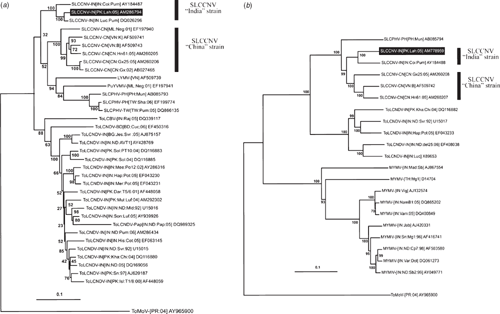
|
Similarly the complete nucleotide sequence of the DNA-B component of SLCCNV-IN[PK:Lah:05] showed the highest level of nucleotide sequence identity (89.6%) with SLCCNV-IN[IN:Coi:Pum] (AY184488). In comparisons to the DNA-B components of other isolates of SLCCNV, the NSP gene of SLCCNV-IN[PK:Lah:04] was truncated due to the presence of a premature stop codon at position 525. Reconstruction of the gene, by removal of the stop codon, yielded a gene encoding a predicted 268 amino acid product more typical of the other SLCCNV isolates. This is likely a naturally occurring mutation, although the possibility that it was introduced by the PCR amplification before cloning cannot be ruled out. A phylogenetic tree based on the complete DNA-B sequence and selected other sequences available in the databases, shows SLCCNV-IN[PK:Lah:05] to group with the other SLCCNV isolates and to be most closely related to the only other ‘India’ strain isolate for which a DNA-B sequence is publically available.
This is the first report of the occurrence of SLCCNV in Pakistan. This species occurs across a wide area from Pakistan and India, through southern China to Vietnam. However, the ‘India’ strain has thus far only been identified on the subcontinent; the China strain occurring in China and Vietnam (Revill et al. 2003). This suggests that the two strains are evolving independently, likely due to geographic isolation. Most probably this is due to the Himalayan mountain range preventing the free movement of the virus, and/or its insect vector, between India/Pakistan and the rest of Asia. The greater diversity of SLCCNV in southern China suggests that the virus may have originated there and was subsequently introduced to India/Pakistan, possibly by trade in agricultural products across the mountain range.
Acknowledgement
M. Tahir and M. S. Haider acknowledge the support of the School of Biological Sciences, University of the Punjab, in conducting this study. R. W. Briddon is supported by the Higher Education Commission (Pakistan) under the ‘‘Foreign Faculty Hiring’’ scheme.
Ali A,
Natsuaki T, Okuda S
(2004) Identification and molecular characterization of viruses infecting cucurbits in Pakistan. Journal of Phytopathology 152, 677–682.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Böttcher B,
Unseld S,
Ceulemans H,
Russell RB, Jeske H
(2004) Geminate structures of African cassava mosaic virus. Journal of Virology 78, 6758–6765.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Briddon RW, Stanley J
(2006) Sub-viral agents associated with plant-infecting single-stranded DNA viruses. Virology 344, 198–210.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Doyle JJ, Doyle JL
(1990) Isolation of plant DNA from fresh tissue. Focus (San Francisco, Calif.) 12, 13–15.

Fauquet CM,
Briddon RW,
Brown JK,
Moriones E,
Stanley J,
Zerbini M, Zhou X
(2008) Geminivirus strain demarcation and nomenclature. Archives of Virology 153, 783–821.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Hanley-Bowdoin L,
Settlage SB,
Orozco BM,
Nagar S, Robertson D
(1999) Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Critical Reviews in Plant Sciences 18, 71–106.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Hussain M,
Mansoor S,
Iram S,
Zafar Y, Briddon RW
(2004) First report of Tomato leaf curl New Delhi virus affecting chilli pepper in Pakistan. Plant Pathology 53, 794.
| Crossref | GoogleScholarGoogle Scholar |

Jones DR
(2003) Plant viruses transmitted by whiteflies. European Journal of Plant Pathology 109, 195–219.
| Crossref | GoogleScholarGoogle Scholar |

Mansoor S,
Hussain M,
Khan SH,
Bashir A,
Leghari AB,
Panwar GA,
Siddiqui WA,
Zafar Y, Malik KA
(1998) Polymerase chain reaction-based detection of cotton leaf curl and other whitefly-transmitted geminiviruses from Sindh. Pakistan Journal of Biological Sciences 1, 39–43.
| Crossref | GoogleScholarGoogle Scholar |

Page RDM
(1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12, 357–358.
|
CAS |
PubMed |

Revill PA,
Ha CV,
Porchun SC,
Vu MT, Dale JL
(2003) The complete nucleotide sequence of two distinct geminiviruses infecting cucurbits in Vietnam. Archives of Virology 148, 1523–1541.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Singh R,
Raj SK, Prasad V
(2008) Molecular characterization of a strain of Squash leaf curl China virus from north India. Journal of Phytopathology 156, 222–228.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Thompson JD,
Gibson TJ,
Plewniak F,
Jeanmougin F, Higgins DG
(1997) The Clustal X windows interface; flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Zhang W,
Olson NH,
Baker TS,
Faulkner L,
Agbandje-McKenna M,
Boulton MI,
Davies JW, McKenna R
(2001) Structure of the maize streak virus geminate particle. Virology 279, 471–477.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |



