First report of a ‘Candidatus Phytoplasma cynodontis’-related strain (group 16SrXIV) associated with white leaf disease of Dichanthium annulatum in India
G. P. Rao A C , S. Mall A , M. Singh A and C. Marcone BA Sugarcane Research Station, Kunraghat, Gorakhpur 273 008, Uttar Pradesh, India.
B Dipartimento di Scienze Farmaceutiche, Università degli Studi di Salerno, Via Ponte Don Melillo, I-84084 Fisciano (Salerno), Italy.
C Corresponding author. Email: gprao_gor@rediffmail.com
Australasian Plant Disease Notes 4(1) 56-58 https://doi.org/10.1071/DN09023
Submitted: 14 October 2008 Accepted: 17 April 2009 Published: 15 June 2009
Abstract
A phytoplasma has been detected in a formerly undescribed white leaf disease of Dichanthium annulatum (Kleberg’s bluestem) in India. By sequence and phylogenetic analyses of polymerase chain reaction-amplified rDNA sequences, the detected phytoplasma proved to be closely related to Bermuda grass white leaf agent (‘Candidatus Phytoplasma cynodontis’) which is a member of the 16SrXIV group.
Dichanthium annulatum (Kleberg’s bluestem, also named marvel grass or Delhi grass in India) is a perennial, stoloniferous grass of the Poaceae family, native to south-eastern Asia, which is common throughout the plains and hills of India up to 1660 m as well as in tropical and North Africa extending east through South-east Asia to China, New Guinea, Australia and Fiji. Dichanthium annulatum is regarded as a highly esteemed fodder grass, especially in India. During the summer of 2008, a white leaf disease of D. annulatum was observed at one location at Gorakhpur, Uttar Pradesh, India. Diseased plants were growing along the roadsides of the research farm and university campus in Gorakhpur districts. The most striking symptoms of the white leaf disease affecting D. annulatum were excessive chlorosis, bushy growth, small leaves and stunting of the plants (Fig. 1). Disease incidence ranged from 2 to 10%. Because the symptoms observed in India were similar to those previously described for white leaf diseases affecting other graminaceous plants (Jung et al. 2003; Marcone et al. 2004), symptomatic D. annulatum plants were examined for phytoplasmal infections employing the highly sensitive polymerase chain reaction (PCR) technology. The detected phytoplasma was identified and characterised using sequence and phylogenetic analyses of PCR-amplified ribosomal DNA (rDNA).

|
For DNA isolation, young shoots including leaves were taken from diseased D. annulatum plants showing typical white leaf symptoms and from non-symptomatic plants of the same species. Total DNA was extracted from ~1 g of tissue employing a phytoplasma-enrichment procedure as described previously (Ahrens and Seemüller 1992). Phytoplasma DNA was amplified using a nested PCR assay. The first amplification was with the universal phytoplasma pair P1/P6 (Deng and Hiruki 1991), and the second with the universal phytoplasma primer pair R16F2n/R2 (Gundersen and Lee 1996). All 10 symptomatic D. annulatum plants examined tested phytoplasma-positive, whereas no visible PCR products could be obtained in template DNA isolated from three non-symptomatic plants. The R16F2n/R16R2 PCR products (~1245 bp in length which include most of the 16S rRNA gene) from two plants were separated by electrophoresis in 1.5% agarose gel, excised from the gel and eluted using the QIAquick gel extraction kit (Qiagen, Hilden, Germany). DNA fragments were either sequenced directly or cloned before sequencing. Sequences were then assembled and edited using DNASTAR’s LaserGene software (DNASTAR) and consensus sequences were generated. Sequence alignments were performed by using CLUSTAL version 5, of the same software. The sequences obtained in the present work proved to be identical and have been deposited in GenBank database under the accession number FJ348654.
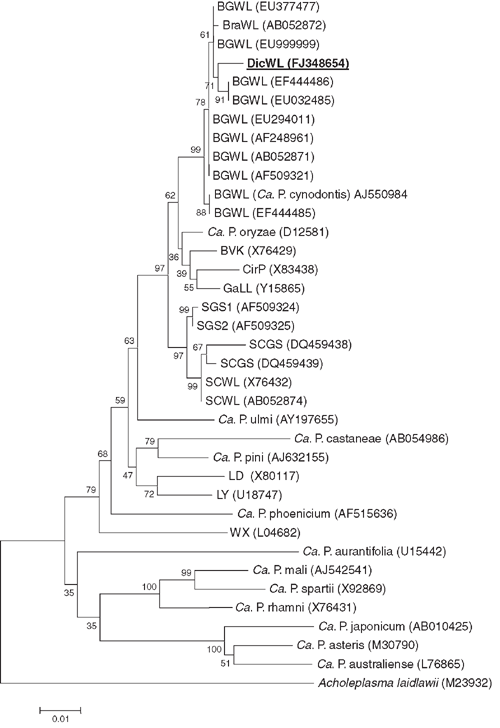
Phylogenetic and molecular evolutionary analyses were conducted using the neighbour-joining program of the genetic analysis software Molecular Evolutionary Genetics Analysis (MEGA), version 4 (Tamura et al. 2007). The data were resampled 1000 times and the bootstrap percentage values are given at the nodes of the tree. Phylogenetic distances were calculated by pairwise comparison. The phylogenetic relatedness of the 16S sequences of the phytoplasma detected in D. annulatum in India to other phytoplasmas is depicted in Fig. 2. The phytoplasma from D. annulatum in India clustered together with BGWL phytoplasma strains. Nucleotide sequence analysis revealed that the phytoplasma detected in diseased D. annulatum in India, is closely related to strains of the Bermuda grass white leaf (BGWL) agent (‘Candidatus Phytoplasma cynodontis’) whose 16S rDNA sequences are available in the GenBank database. The sequence similarity of the D. annulatum-infecting agent to the Malaysian strain of BGWL (GenBank accession number EU294011) is 98.1% whereas the Chinese strain of BGWL (GenBank accession number EU999999) is 99.1%.
|
The D. annulatum-infecting phytoplasma is a member of the same subclade as the BGWL group, or 16SrXIV group (Marcone et al. 2004). Flanked to this cluster were members from other phytoplasma groups such as ‘Candidatus Phytoplasma oryzae’, the phytoplasma obtained from the leafhopper Psammotettix cephalothes (strain BVK) and the cirsium phyllody (CirP) agent. The name dichanthium white leaf (DicWL) phytoplasma is proposed for this BGWL phytoplasma-related agent. As the 16S rDNA sequence similarity is greater than 97.5%, DicWL phytoplasma should be considered as part of the Candidatus species ‘Candidatus Phytoplasma cynodontis’. However, further databases on other molecular markers, cross inoculation experiments and vector transmission specificity are needed to confirm that they are the same taxonomic entity (IRPCM Phytoplasma/Spiroplasma Working Team – Phytoplasma taxonomy group 2004). To our knowledge, this is the first report on the occurrence of a phytoplasmal disease of D. annulatum.
Ahrens U, Seemüller E
(1992) Detection of DNA of plant pathogenic mycoplasmalike organisms by a polymerase chain reaction that amplifies a sequence of the 16S rRNA gene. Phytopathology 82, 828–832.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Deng S, Hiruki C
(1991) Amplification of 16S rRNA genes from culturable and nonculturable mollicutes. Journal of Microbiological Methods 14, 53–61.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Gundersen DE, Lee I-M
(1996) Ultrasensitive detection of phytoplasmas by nested-PCR assays using two universal primer pairs. Phytopathologia Mediterranea 35, 144–151.
|
CAS |

IRPCM Phytoplasma/Spiroplasma Working Team – Phytoplasma taxonomy group
(2004) ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. International Journal of Systematic and Evolutionary Microbiology 54, 1243–1255.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Jung H-Y,
Sawayanagi T,
Wongkaew P,
Kakizawa S,
Nishigawa H,
Wei W,
Oshima K,
Miyata S-I,
Ugaki M,
Hibi T, Namba S
(2003) ‘Candidatus Phytoplasma oryzae’, a novel phytoplasma taxon associated with rice yellow dwarf disease. International Journal of Systematic and Evolutionary Microbiology 53, 1925–1929.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Marcone C,
Schneider B, Seemüller E
(2004) ‘Candidatus Phytoplasma cynodontis’, the phytoplasma associated with Bermuda grass white leaf disease. International Journal of Systematic and Evolutionary Microbiology 54, 1077–1082.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Tamura K,
Dudley J,
Nei M, Kumar S
(2007) MEGA 4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |



