Identification and expression characterisation of SbERECTA family genes in Sorghum bicolor
Jia Cheng Zheng A B C , Jie Yu C B , Ting Liu C B , Xin Wang B , Qiu Wen Zhan B , Jie Qin Li B , Zhao Shi Xu A C and You Zhi Ma A C
A B C , Jie Yu C B , Ting Liu C B , Xin Wang B , Qiu Wen Zhan B , Jie Qin Li B , Zhao Shi Xu A C and You Zhi Ma A C
A Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS)/National Key Facility for Crop Gene Resources and Genetic Improvement, Key Laboratory of Biology and Genetic Improvement of Triticeae Crops, Ministry of Agriculture, Beijing 10081, P.R. China.
B College of Agronomy, Anhui Science and Technology University, Fengyang, Anhui, 233100, P.R. China.
C Corresponding authors. Email: xuzhaoshi@caas.cn; mayouzhi@caas.cn
Crop and Pasture Science 72(2) 125-135 https://doi.org/10.1071/CP20434
Submitted: 29 October 2020 Accepted: 7 January 2021 Published: 16 February 2021
Journal Compilation © CSIRO 2021 Open Access CC BY
Abstract
ERECTAs are receptor-like kinases that regulate plant biomass and stress resistance. In this study, the wheat (Triticum aestivum) TaERECTA gene was used as a probe to identify the SbERECTA family genes (SbERs) in the sorghum (Sorghum bicolor) genome, analyse their subcellular localisation and characterise their expression. Results showed that the two SbER members, SbER10 with three copies (SbER10_X1, SbER10_X2, and SbER10_X3) and SbER4 with two copies (SbER4_X1 and SbER4_X2), were found on chromosomes 10 and 4 of sorghum, respectively. SbER10 had the highest expression level in the pedicel tissue and showed a remarkable response under treatment with abscisic acid, brassinolide, gibberellin and indole-3-acetic acid. SbER10_X1, functioning on the cell membrane and chloroplast, exhibited abundant transcript in only a few sorghum varieties that are grown in mountainous areas and receive strong light, heat, and water supply. Expression of SbER10_X1 was significantly and positively correlated with plant biomass of 32 sorghum germplasm resources. These results indicate that SbER10 genes have an important regulatory role in sorghum growth, and increasing SbER10 transcription level offers a potential strategic target for breeding or biotechnological approaches to enhance sorghum biomass and environmental adaptability.
Keywords: correlation analysis, ERECTA family, expression characteristics, SbER, sorghum.
Introduction
Sorghum bicolor L., one of the key C4 crops, is used to produce food, forage and energy, and in brewing. It provides food and resources for more than 500 million people in arid and semiarid areas in Africa and Asia (Abdel-Ghany et al. 2020; Kimani et al. 2020). Global population growth and increases in per capita income intensify human demand for food and energy. Therefore, crop breeding is needed, drawing on traditional methods, functional gene screening, genome editing, and other technical means, to obtain new varieties with drought resistance, pest resistance, and high light efficiency. The sorghum genome contains many useful genes related to high yield, stress resistance, and high light efficiency (McCormick et al. 2018). Development and use of sorghum functional genes is a potential strategy to ensure global food security, provide energy, and promote the development of animal husbandry.
ERECTA (ER) is a receptor-like kinase (RLK) that regulates plant photosynthesis and transpiration efficiency, increases biomass, and improves plant stress resistance (Masle et al. 2005; van Zanten et al. 2009). Transgenic maize (Zea mays) with ZmER shows increased organ size, enhanced transpiration efficiency and drought resistance (Guo et al. 2011). After the wheat (Triticum aestivum) TaER gene was silenced, leaf stomatal density and conductance of the mutant plant increased (Zheng et al. 2019). The AtER gene (Arabidopsis thaliana) was overexpressed in tomatoes and rice, leading to the increased thermostability of transgenic lines (Shen et al. 2015). Interaction of ER and BAK1 genes can improve the resistance of A. thaliana to the necrotrophic fungus Plectosphaerella cucumerina BMM (Jordá et al. 2016). These results indicate that the ER family has broad application prospects in regulating plant development and stress resistance.
In sorghum, SbER2-1 (equivalent to SbER4_X1 in this study) was isolated and introduced into maize to increase drought resistance and net photosynthetic rate (Li et al. 2019). However, SbERECTA has two members (SbER4 and SbER10), and SbER10 was not specifically investigated and introduced into sorghum for improvement of forage biomass. In this study, the evolutionary relationship of the SbER family in dicotyledonous and monocotyledonous plants was clarified by analysing the characteristics of sorghum SbER family members and cis-regulatory elements. The expression pattern of the SbER10 genes under hormone induction was examined, and SbER10_X1 was isolated to detect its expression variation in 32 sorghum germplasm resources of China. This work provides a theoretical basis for using SbER10_X1 to improve sorghum production potential and stress resistance.
Materials and methods
Identification of SbER family in the sorghum genome
Wheat TaER gene (JQ599261.2) was used to search in the Phytozome v12.1 database; two SbER gene tags of sorghum (Sobic.004G317200 and Sobic.010G077000) were found and aligned against the NCBI database to obtain the SbER nucleotide and protein sequences, and predict the SbER full CDS coding frame and chromosome position information. The PROSITE database was used to analyse the amino acid composition, molecular weight, and isoelectric point of SbER. The GSDS database was used for predicting the exons of ER family genes, PlantCARE database for the cis-regulatory elements of the SbER promoter, and Plant-mPLoc database for the characteristics of SbER subcellular localisation. Supplementary Material Table S1 lists all databases used and their URLs (available at the journal’s website).
Analysis of protein sequence and phylogenetic tree
PROSITE and SMART databases were used to predict the core conserved regions and functional domains of the SbER family. The ER family members of monocotyledonous and dicotyledonous plants were obtained in the NCBI database, and amino acid sequences with >80% similarity were downloaded. MEGA 5.0 software was used for alignment (Tamura et al. 2013). A phylogenetic tree was generated with the neighbour-joining method under the threshold of 1000 replications for bootstrap (Tamura et al. 2013).
Prokaryotic expression and western blot analysis of SbER10 family
The common Ser/Thr kinase segment of the SbER10 genes was amplified with primers of SbER10-KF and SbER10-KR (Table S2), using the PCR reaction system (50 μL): 25 μL 2 × PCR buffer, 10 μL dNTP (2 mM), 1.5 μL Primer-F (10 μM), 1.5 μL Primer-R (10 μM), 1 μL KOD FX (1.0 U μL–1, KFX-101, Toyobo, China), 5 μL cDNA as template (the peduncle of sorghum variety Tx623B), 6 μL ddH2O. The PCR procedure was: 94°C for 2 min, 40 cycles (98°C for 10 s, 61°C for 30 s for renaturation, 68°C for 1 min for extension), 68°C for 10 min.
PCR products were introduced into pET-32a-c(+) vector via restriction site (Table S2), transformed into Escherichia coli (DH5α), and then cultured at 37°C until OD600 0.6–0.8. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added into the culture with a final concentrations of 0, 0.4 and 0.8 mM. The cells cultured at 37°C for 3 h were harvested and split by repeated freezing and thawing with lysozyme. The supernatant (~50 μg) was run on SDS–PAGE and transferred onto nitrocellulose filter membrane (NC membrane) by wet transblot apparatus (Bio-Rad, USA). Primary and secondary antibodies were anti-histidine (His) (mouse monoclonal antibody) and IgG antibody (horseradish peroxidase IgG). Protein marker was pre-dyed protein maker II (MP203, Tiangen, China). Immune complexes were visualised with scanner system (Canon, Japan).
Gene isolation and subcellular localisation of SbER10_X1
SbER10_X1 (XM_002437978.2) had an open reading frame of 2949 bp, encoding a protein of 983 amino acids (Table 1). The specific primers (SbER10_X1-2F/SbER10_X1-2R in Table S2) were designed to isolate the SbER10_X1 gene from the peduncle cDNA of sorghum variety Tx623B by using the KOD FX enzyme. The PCR reaction system and procedure were as above, except for 64°C for 30 s for renaturation and 68°C for 4 min for extension.

|
The code fragment of SbER10_X1 (without the stop codon for development of fusion protein in Supplementary materials Fig. S1) was amplified with primers GFP-SbER10_X1-F1 and GFP-SbER10-X1-R1 (Table S2), using the PCR procedures and reaction system as above, except for 62°C for 30 s for renaturation and 68°C for 3 min for extension. The PCR fragment was fused into the N-terminus of green fluorescent protein (GFP) containing the promoter of CaMV35S as 35S::SbER10_X1-GFP, and the 35S::GFP vector was used as the control (Shi et al. 2018), The fusion construct was confirmed by DNA sequencing. Vectors 35S::GFP and 35S::SbER10_X1-GFP were introduced into wheat mesophyll protoplasts to estimate their transient expression. The transformed cells were incubated in darkness at 22°C for 18–20 h, and then photographed under a confocal laser scanning microscope (LSM700; CarlZeiss, Germany).
Plant materials and treatments
For detection of expression profiles, sorghum Tx623B and 32 sorghum germplasm resources were planted in the experimental field of Anhui Science and Technology University in 2018 (32°52′?, 117°33′E; 43 m elevation). For expression profiles analysis at different growth stages, the latest expanded leaves of Tx623B were collected at seedling (stage 2), jointing (stage 3), and booting (stage 4); and flag leaves were collected at heading (stage 5), flowering (stage 6), filling (stage 7), and maturity (stage 8). For tissue specific expression, tissue samples of root (stage 3), stem (stage 4), leaf and leaf sheath (stage 5), panicle, pedicel, anther and ovary (stage 6), and kernel (stage 7) were collected. Details of the sampling period can be found in Fig. S2.
In 32 germplasm resources, the flag leaves were collected at only stage 5, in addition to investigation of photosynthetic index and agronomic traits. Nine plants of each of the 32 germplasm resources were evaluated as replications of each trait. Table S3 displays the source and names of 32 sorghum germplasm resources.
For hormone induction, sorghum variety Tx623B was planted in flowerpots (35 by 35 cm). Each flowerpot was sown with 35 seeds pre-germinated for 4 days and placed in a light incubator for growth (humidity 60%, 23°C/20°C day/night, 16 h/8 h light/dark culture, light intensity 525 μmol s–1 m–2). After 15 days of growth, the seedlings were collected including the roots, rinsed, dried on filter paper for instant drying, and then placed in hormone solution with deionised water (control) for recovery growth. Hormone solutions were abscisic acid (ABA) (100 μM), brassinolide (BR) (0.75 μM), gibberellin (GA3) (30 mM), and the auxin indole-3-acetic acid (IAA) (10 μM) (Zheng and Hu 2016). Samples (mixture with organs of roots, stems and leaves) were taken at different times: 0, 1, 2, 4, 6, 12, 24, 48, and 60 h. Three individual plants were selected as biological replicates each time.
All samples were immediately frozen in liquid nitrogen after collection and stored at –80°C for later use.
RNA extraction, RT-qPCR and data analysis
Total RNA of sorghum samples was extracted using RNAprep Pure Kit (Tiangen Biochemical Technology, China), and reverse transcribed to obtain cDNA samples by using PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Japan). Five cDNA sequences of SbER10 and SbER4 genes were aligned to design specific primers for SbER10 expression, selecting regions with high similarity among the three copies of SbER10 genes but with diversity between SbER10 and SbER4. The primers of SbER10 genes (SbER10-2QF/SbER10-2QR) and reference genes (SbActin-F/SbActin-R) are listed in Table S2. The SbER10 expression level was detected by using RT-qPCR methodology and the ABI Prism 7500 system (Applied Biosystems, USA) (Liu et al. 2013).
The RT-qPCR data were analysed in accordance with procedure of Zheng et al. (2015) and Zheng and Hu (2016). Error analysis was conducted with the SPSS Statistics Software version 18.0 (IBM, USA) based on the biological replicates of three individual plants. Correlation analysis between SbER10 expressions and measured agronomy traits was performed with SPSS 18.0 by using the Pearson product moment correlation test.
Results
Characteristics of the sorghum SbER gene family
The SbER family has two members, SbER10 (LOC8077321) and SbER4 (LOC8081937), located on sorghum chromosomes 10 and 4, respectively. SbER10 had three copies, namely, SbER10_X1 (XM_002437978.2), SbER10_X2 (XM_021448730.1) and SbER10_X3 (XM_021448731.1), and SbER4 had two copies, namely, SbER4_X1 (XM_021458593.1) and SbER4_X2 (XM_021458594.1) (Table 1). Among the five copies, four had 27 exons, except for SbER10_X3, in which exon 21 was deleted, and all members were predicted to be transmembrane proteins. In exon 26 of SbER4 and SbER10 genes, SbER4_X2 encoded one fewer alanine (Ala) than SbER4_X1, whereas SbER10_X2 encoded one fewer lysine (Lys) than SbER10_X1. SbER10_X3, with a deleted exon, encoded 24 fewer amino acids (Fig. 1).
SbER protein functional domain and phylogenetic relationship
The SbER family had typical characteristics of RLKs, including the N-terminal signal peptide area, leucine repeat region (LRR), transmembrane region, and C-terminal serine/threonine kinase region (Fig. S3). In particular, the amino acid sequences of the SbER family and other species in the LRR region and kinase region were relatively conserved, whereas the amino acids in the N-terminal signal peptide region and transmembrane region were different (Fig. S4).
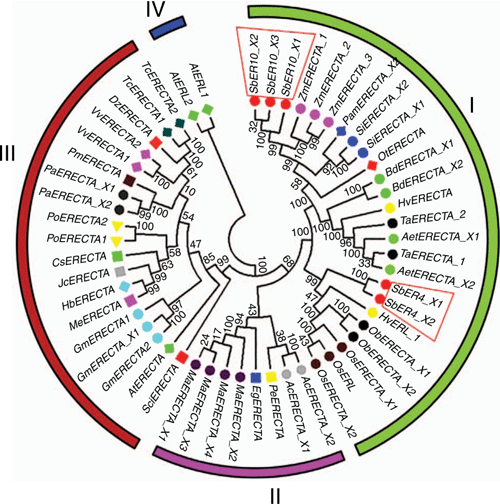
The ER proteins of common monocotyledonous and dicotyledonous plants can be clearly divided into four categories (Fig. 2). The first category is Gramineae crops, which can be divided into two subgroups: sorghum SbER10 family, wheat, barley, maize, millet and other crops constitute one subgroup; and SbER4 family, barley HvERL1 and rice ER family constitute the other subgroup. The second category consists of monocotyledonous plants such as pineapple, banana and Phalaenopsis. The third category contains dicotyledonous plants including the ER family of soybeans, grapes, sesame, poplar, cocoa and A. thaliana. The fourth category includes only AtERL1 and AtERL2, indicating that the A. thaliana AtERL has the farthest evolutionary relationship. SbER10_X1, SbER10_X2 and SbER10_X3 have the closest genetic relationship with maize, and SbER4_X1 and SbER4_X2 have a close relationship with rice, implying that the evolutionary relationship of SbER10 system is close to modern field xerophytes.
|
SbER gene structure and cis-regulatory elements
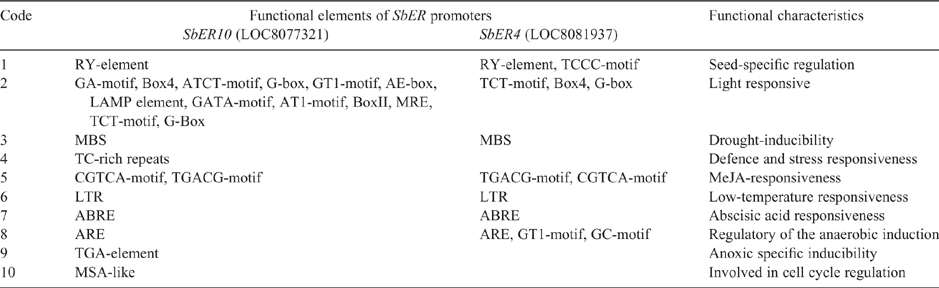
The cis-regulatory elements of the SbER family promoter regulate plant seed-specific development, light response, drought, low temperature induction, and signal responses mediated by methyl jasmonate and ABA (Table 2). Therefore, the SbER family may have a role in regulating plant resistance. In addition, SbER10 regulates the plant’s defence response to anaerobic induction reaction and the cell division cycle, suggesting that its regulatory function may be stronger than that of SbER4. Therefore, SbER10_X1 was used to analyse the ER gene family structure of Gramineae crops and published soybean, poplar, grape and A. thaliana. Figure 3 shows that the ER family nucleotide sequence can also be divided into monocotyledonous plants, dicotyledonous plants and the AtERL family. The selected members of the ER family all contained 27 exons, of which exons 25, 26 and 27 were large and mainly encode the threonine/serine kinase domain. Exon 1 of the dicotyledon ER family was large, and exon 1 of the monocotyledon was small. Exon 1 encoded the N-terminal signal peptide region or part of the LRR repeat region. AtERL family genes were relatively short and had separate branches. From this finding, it was speculated that the ER genes of different species have various regulatory functions in receiving upstream signals that are transmitted into the cell, owing to the different elements of the N-terminal signal peptide region.
|
Expression pattern of SbER10 gene family in different organs and at developmental stages
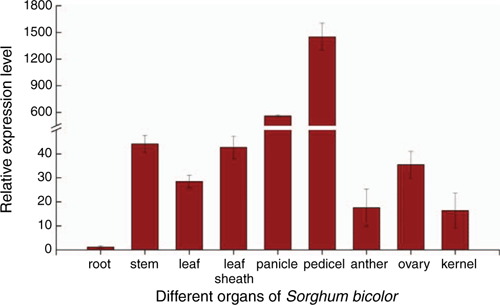
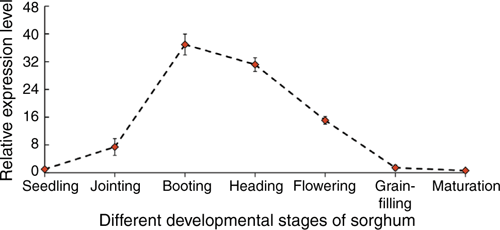
In different organs of sorghum Tx623B (Fig. 4), the SbER10 family was highly expressed in aboveground organs but hardly expressed in roots. With the expression level of root organs as a reference, SbER10 expression in the pedicels was the highest, at 1500 times, followed by that in the panicles. The expression levels in young stems, leaf sheaths and ovary were close and >40 times that of roots, and the expression level in anthers and kernels was lower at only ~16 times. Of the different developmental stages (Fig. 5), SbER10 showed the highest expression level at booting stage, 37 times higher than at seedling stage, followed by heading and flowering stages, but low expression at grain-filling and maturation stage. This finding indicates great variations in the functional mechanism of SbER10 when regulating the development of different sorghum organs during growth stages.
|
|
Response pattern of SbER10 gene family under hormone treatments
For the study of treatment with hormones ABA, BR, GA3 and IAA, the expression level of SbER10 genes was stable in control samples but significantly (P < 0.01) increased in the treated samples, thus showing a strong response (Fig. 6). After 2 h of ABA treatment, the expression level of SbER10 gradually increased, with the highest expression 14 times that of the control at 48 h (Fig. 6a). After 2 h of BR treatment, the expression level of SbER10 increased rapidly, with the highest expression 18 and 17 times that of the control at 12 and 24 h, respectively (Fig. 6b). After 2 h of GA3 treatment, the expression level of SbER10 increased slowly, with highest expression 14 times that of that control at 12 h, and then decreased rapidly (Fig. 6c). After IAA treatment, the SbER10 genes showed a positive response, with highest expression level 36 times that of the control at 2 h, and the expression level gradually decreased after 2 h (Fig. 6d). This finding indicates that SbER10 can actively respond to hormone induction and may regulate physiological processes related to sorghum development and stress resistance.
SbER10_kinase prokaryotic expression
For the study of treatment with IPTG at constant temperature (37°C) for 3 h, the target protein (pET-32a-c(+)-SbER10_kinase) without induction by IPTG slightly accumulated compared with the control (pET-32a-c(+) empty vector) in the prokaryotic system. After induction by IPTG (0.4 and 0.8 mM), the target protein accumulated in large amounts, with a size of ~45 kDa. After the His tag was removed for 14 kDa, the size of SbER10_kinase was close to 31 kDa, which is consistent with the predicted size of the target protein (Fig. 7a). Western blot analysis was conducted to confirm the reliability of the target protein; Fig. 7b shows that no band appeared after hybridisation of the empty vector pET-32a-c(+) protein, and the hybrid bands of the target protein without IPTG induction were weak. After IPTG induction, the target protein hybrid band pattern was evident, indicating that the SbER10_kinase sequence can completely encode the target protein for translation.
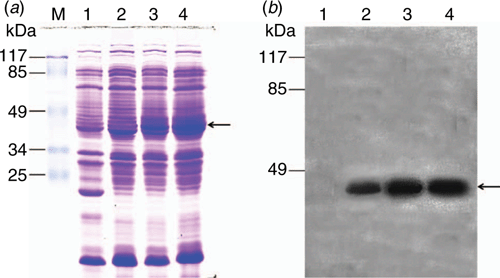
|
SbER10_X1 clone and subcellular localisation
SbER10_X1 gene fragment with size 3376 bp was isolated using the pedicel of sorghum Tx623B as material (Fig. S5). The 35S:SbER10_X1-GFP fusion vector was transferred into wheat leaf protoplasts and cultured using PEG to clarify the role of the SbER10_X1 protein in cells. Observation showed that the fluorescent signal of the SbER10_X1-GFP fusion protein was located on the cell membrane and chloroplast of wheat mesophyll protoplasts (Fig. 8). Meanwhile, the control pCAM35-GFP was located on the cell membrane, cytoplasm, and nucleus, indicating that SbER10_X1 mainly functions on the cell membrane and chloroplast. This finding is consistent with the above structural characteristics of predicted SbER10 as a transmembrane protein.
Correlation of expression of SbER10_X1 with photosynthetic index and agronomic traits
Thirty-two sorghum germplasm resources in different provinces of China were selected to identify the expression level of SbER10_X1. The results showed that the expression level was high in only a few sorghums and low in most materials (Fig. 9a). The resources can be clearly divided into three groups, with highly significant (P < 0.01) difference among the three categories. The first group contained three germplasm resources (No. 8, 22 and 29) and showed the highest expression of SbER10_X1 with an average of 10.9. The photosynthetic index and agronomic traits were greatly different among the 32 sorghum germplasm resources, and negative linear correlations were found for SbER10_X1 expression with transpiration rate (E), stomatal conductance (gs) and stomatal number per unit (SN), whereas positive linear correlations were detected for SbER10_X1 expression with 1000-kernel weight (TKW), main stem panicle grain yield (MSPGY), main stem diameter (MSD), main stem fresh weight (MSFW) and main stem drought weight (MSDW) (Table 3). Regression analyses confirmed that SbER10_X1 expression was significantly (P < 0.05) and negatively correlated with E and gs, and highly significantly (P < 0.01) and positively correlated with MSFW and MSDW (Fig. 9b). This finding shows that only a few varieties during sorghum natural evolution have high expression levels of SbER10_X1, probably leading to the variation in photosynthetic index and biomass.
Discussion
The characteristics of gene families are important to the study of their functions. The accuracy and reliability of analysis of the evolutionary characteristics of gene families depend on the genome sequences. Sorghum has two members of the ER family (SbER10 and SbER4) located on chromosomes 10 and 4, respectively. The SbER10 gene sequence in the sorghum genome was long with three spliceosomes, whereas the SbER4 gene sequence was small with two spliceosomes. The number of exons varied in the three copies of SbER10, and this finding is different from previous studies (Li et al. 2019). The ER family also has two members in rice, wheat, cotton, tobacco and other crops, and each member has different splice bodies, resulting in uneven distribution of introns and exons across the genome (Liu et al. 2019). In eukaryotes, the acquisition or loss of introns is one of the evolutionary mechanisms of a gene family (Roy and Penny 2007). Any difference in the number of introns will affect the expression levels of the target genes. When the AtER gene of A. thaliana lacks introns, the amount of target protein decreases 500–900 times (Karve et al. 2011). When the extracellular region of soybean GmER reduces the number of LRR tandems (exons decrease), shading treatment can increase the hypocotyl length, leaf area and petiole length (Du et al. 2018). Different splice bodies of SbER genes in sorghum probably cause functional differences, and need to be further investigated.
The ER gene family is a member of the RLK gene family and codes for a transmembrane protein that senses external stimuli, activates the expression of intracellular signalling factors, and regulates the physiological response of cells (Shpak et al. 2004; Zheng and Hu 2016;). Prior to the emergence of monocotyledonous and dicotyledonous plants, the ER family evolved into two large families of ER and ERL. Multiple copies of the ER and ERL families of monocotyledonous and dicotyledonous plants have gradually formed with the occurrence of gene replication events (Liu et al. 2019). In this work, the ER family can be clearly divided into three categories: monocotyledonous, dicotyledonous and Arabidopsis ERL. Among them, SbER10 has a close evolution relationship with maize, and SbER4 has a close relationship with rice. The SbER family and ER family of dicotyledonous and monocotyledonous plants have large differences in amino acid residues in the N-terminal signal peptide region and transmembrane region. The most important function of the ER gene is phosphorylation. The difference in the amino acid position of the transmembrane region affects the phosphorylation of RLKs; the N-terminal extension region is an integral part of kinase folding and is essential for kinase activity (Kosentka et al. 2017). The absence of the kinase region does not affect the increase in the elongation of stalk and pedicel cells but can cause the A. thaliana inflorescence to be compact and the silique to shrink. Moreover, this gene has different molecular mechanisms for stomatal development and cell elongation (Shpak et al. 2003; Villagarcia et al. 2012). After 3 h of IPTG induction, SbER10_kinase protein is completely expressed in prokaryotic cells. This finding provides theoretical guidance for the next synthesis of SbER-specific antibodies.
The ER family participates in light-induced subtropical growth (van Zanten et al. 2010), improves the drought resistance of maize (Li et al. 2019), and regulates rice blast non-host resistance (Takahashi et al. 2016). Combined with ABA and methyl jasmonate, the gene family regulates the quantitative resistant traits of A. thaliana (Häffner et al. 2014), and inhibits cell division and promotes cell elongation (Qu et al. 2017). In this study, the promoter of the SbER family contains ABA (ABRE), drought, low temperature, methyl jasmonate, anaerobic induction and other core elements, implying that the gene improves plant stress resistance. However, the mechanism of low temperature response and anaerobic induction reaction has not been reported. For the light response mechanism, ER genes are involved in mediating hypocotyl elongation, shortening the cycle of the plant biological clock, and reducing the range of leaf surface movement under the interaction of blue and dark light (van Zanten et al. 2009). Those authors also reported that ER affects the carboxylation rate and photoelectron transfer ability of ribulose diphosphate carboxylase and increases the photosynthetic ability of A. thaliana (van Zanten et al. 2009). SbER has four cis-acting elements that are all involved in the photoreaction, and the SbER10_X1 protein is located on the cell membrane and chloroplast. This finding indicates that this gene is involved in plant photosynthesis and has great application potential for improving sorghum photosynthesis and increasing biological yield.
The ER family is involved in signal regulation networks mediated by IAA (Qu et al. 2017), BR (Wang et al. 2017), GA3 (Ragni et al. 2011) and ABA (Häffner et al. 2014). SbER2-1 can improve the drought resistance and water use efficiency of maize (Li et al. 2019), and VvER (in grape) has a positive response to hormone, drought and pathogenic bacteria treatment (Liu et al. 2019). This study found that SbER10 genes actively respond to hormone induction. This finding is consistent with previous research results and suggests that SbER has similar functions in plant growth and development and stress resistance regulation.
In A. thaliana and wheat, the ER gene is hardly expressed in plant roots (Shpak et al. 2004; Zheng and Hu 2016), and the expression level of SbER10 in sorghum roots is low. The expression levels in tissues such as young panicles and pedicels are extremely high, indicating that SbER mainly regulates the development of young tissues and organs above ground. Among different growth stages, SbER has the highest expression level in the new leaves at booting stage and has low expression at the seedling stage and after heading. This finding is different from the tissue expression specificity of wheat TaER gene (Zheng and Hu 2016), probably because germinal leaves develop from the seed embryo at the germination stage of wheat, whereas the new leaves of sorghum develop from the growth cone of stem tissues after jointing stage. Transcriptional level of SbER was high in young tissues, displaying abundant transcript at booting stage and a reduced trend with leaves senescence process.
Of the 32 sorghum germplasm resources of different regions of China used to assess variation in SbER10_X1 expression, analysis found that SbER10_X1 is abundant in only a few varieties, most of which come from mountainous areas with sufficient light, heat and water resources. In other regions, SbER10_X1 expression is relatively low, indicating that differences in the natural environment greatly influence the SbER transcription level in sorghum. SbER10_X1 expression was positively correlated with MSFW, MSDW, MSD, MSPGY and TKW, but negatively correlated with E, gs and SN; this concurs with previous studies (Zheng at al. 2015). However, the strongest correlation of SbER10_X1 expression was with MSFW and MSDW, which greatly contribute to the biomass of forage sorghum. The link between SbER expression with photosynthetic index and agronomic traits confers another strategic target for breeding or biotechnological approaches to increase sorghum biomass.
Conclusions
SbERECTA (SbERs) family has two members, SbER10 with three copies (SbER10_X1, SbER10_X2, and SbER10_X3) and SbER4 with two copies (SbER4_X1 and SbER4_X2), in sorghum genome. SbER10 has a close evolutionary relationship with maize, and SbER4 with rice. The promoter of the SbER family contains ABA, low temperature, anaerobic induction and other core elements, and SbER10 genes actively respond to hormone induction. SbER10_X1, functioning on the cell membrane and chloroplast, showed abundant transcript in only a few mountain varieties experiencing strong light, heat and water supply among 32 sorghum germplasm resources. SbER10_X1 expression was significantly and positively correlated with plant biomass, providing a strategic target for forage sorghum breeding.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work was financially supported by the National Key Research and Development Program of China (2018YFD0300902, 2017YFD0301301), Outstanding Talents Program of University (gxyq2020039), the National Natural Science Foundation of China (31971993), and Talents Introduced Fund of Anhui Science and Technology University (NXYJ201604).
References
Abdel-Ghany SE, Ullah F, Ben-Hur A, Reddy ASN (2020) Transcriptome analysis of drought-resistant and drought-sensitive sorghum (Sorghum bicolor) genotypes in response to PEG-induced drought stress. International Journal of Molecular Sciences 21, 772| Transcriptome analysis of drought-resistant and drought-sensitive sorghum (Sorghum bicolor) genotypes in response to PEG-induced drought stress.Crossref | GoogleScholarGoogle Scholar |
Du JB, Li Y, Sun X, Yu L, Jiang HK, Cao QL, Shang J, Sun MY, Liu Y, Shu K, Liu J, Yong TW, Liu WG, Yang F, Wang XC, Liu CY, Yang WY (2018) Characterization of a splice variant of soybean ERECTA devoid of an intracellular kinase domain in response to shade stress. Journal of Genetics 97, 1353–1361.
| Characterization of a splice variant of soybean ERECTA devoid of an intracellular kinase domain in response to shade stress.Crossref | GoogleScholarGoogle Scholar |
Guo M, Rupe M, Simmons C, Sivasankar S (2011) The Maize ERECTA genes for improving plant growth, transpiration, efficiency and drought tolerance in crop plants. United States Patent Application Publication 2011/0035844, A1.
Häffner E, Karlovsky P, Splivallo R, Traczewska A, Diederichsen E (2014) ERECTA, salicylic acid, abscisic acid, and jasmonic acid modulate quantitative disease resistance of Arabidopsis thaliana to Verticillium longisporum. BMC Plant Biology 14, 85
| ERECTA, salicylic acid, abscisic acid, and jasmonic acid modulate quantitative disease resistance of Arabidopsis thaliana to Verticillium longisporum.Crossref | GoogleScholarGoogle Scholar | 24690463PubMed |
Jordá L, Sopena-Torres S, Escudero V, Nunez-Corcuera B, Delgado-Cerezo M, Torii KU, Molina A (2016) ERECTA and BAK1 receptor like kinases interact to regulate immune responses in Arabidopsis. Frontiers in Plant Science 7, 897
| ERECTA and BAK1 receptor like kinases interact to regulate immune responses in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 27446127PubMed |
Karve R, Liu WS, Willet SG, Torii KU, Shpak ED (2011) The presence of multiple introns is essential for ERECTA expression in Arabidopsis. RNA 17, 1907–1921.
| The presence of multiple introns is essential for ERECTA expression in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 21880780PubMed |
Kimani W, Zhang LM, Wu XY, Hao HQ, Jing HC (2020) Genome-wide association study reveals that different pathways contribute to grain quality variation in sorghum (Sorghum bicolor). BMC Genomics 21, 112
| Genome-wide association study reveals that different pathways contribute to grain quality variation in sorghum (Sorghum bicolor).Crossref | GoogleScholarGoogle Scholar | 32005168PubMed |
Kosentka PZ, Zhang L, Simon YA, Satpathy B, Maradiaga R, Mitoubsi O, Shpak ED (2017) Identification of critical functional residues of receptor-like kinase ERECTA. Journal of Experimental Botany 68, 1507–1518.
| Identification of critical functional residues of receptor-like kinase ERECTA.Crossref | GoogleScholarGoogle Scholar | 28207053PubMed |
Li HS, Han XD, Liu XX, Zhou MY, Ren W, Zhao BB, Ju CL, Liu Y, Zhao JR (2019) A leucine-rich repeat-receptor-like kinase gene SbER2-1 from sorghum (Sorghum bicolor L.) confers drought tolerance in maize. BMC Genomics 20, 737
| A leucine-rich repeat-receptor-like kinase gene SbER2-1 from sorghum (Sorghum bicolor L.) confers drought tolerance in maize.Crossref | GoogleScholarGoogle Scholar |
Liu P, Xu ZS, Lu PP, Hu D, Chen M, Li LC, Ma YZ (2013) A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. Journal of Experimental Botany 64, 2915–2927.
| A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 23682116PubMed |
Liu M, Li WP, Min Z, Cheng XH, Fang YL (2019) Identification and expression analysis of ERECTA family genes in grape (Vitis vinifera L.). Genes & Genomics 41, 723–735.
| Identification and expression analysis of ERECTA family genes in grape (Vitis vinifera L.).Crossref | GoogleScholarGoogle Scholar |
Masle J, Gilmore SR, Farquhar GD (2005) The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 436, 866–870.
| The ERECTA gene regulates plant transpiration efficiency in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 16007076PubMed |
McCormick RF, Truong SK, Sreedasyam A, Jenkins J, Shu SQ, Sims D, Kennedy M, Amirebrahimi M, Weers BD, McKinley B, Mattison A, Morishige DT, Grimwood J, Schmutz J, Mullet JE (2018) The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. The Plant Journal 93, 338–354.
| The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization.Crossref | GoogleScholarGoogle Scholar | 29161754PubMed |
Qu XY, Zhao Z, Tian ZX (2017) ERECTA regulates cell elongation by activating auxin biosynthesis in Arabidopsis thaliana. Frontiers in Plant Science 8, 1688
| ERECTA regulates cell elongation by activating auxin biosynthesis in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar |
Ragni L, Nieminen K, Pacheco-Villalobos D, Sibout R, Schwechheimer C, Hardtke CS (2011) Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion. The Plant Cell 23, 1322–1336.
| Mobile gibberellin directly stimulates Arabidopsis hypocotyl xylem expansion.Crossref | GoogleScholarGoogle Scholar | 21498678PubMed |
Roy SW, Penny D (2007) Patterns of intron loss and gain in plants: intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Molecular Biology and Evolution 24, 171–181.
| Patterns of intron loss and gain in plants: intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana.Crossref | GoogleScholarGoogle Scholar | 17065597PubMed |
Shen H, Zhong XB, Zhao FF, Wang YM, Yan BX, Li Q, Chen GY, Mao BZ, Wang JJ, Li YS, Xiao GY, He YK, Xiao H, Li JM, He ZH (2015) Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato. Nature Biotechnology 33, 996–1003.
| Overexpression of receptor-like kinase ERECTA improves thermotolerance in rice and tomato.Crossref | GoogleScholarGoogle Scholar | 26280413PubMed |
Shi WY, Du YT, Ma J, Min DH, Jin LG, Chen J, Chen M, Zhou YB, Ma YZ, Xu ZS, Zhang XH (2018) The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean. International Journal of Molecular Sciences 19, 4087
| The WRKY transcription factor GmWRKY12 confers drought and salt tolerance in soybean.Crossref | GoogleScholarGoogle Scholar |
Shpak ED, Lakeman MB, Torii KU (2003) Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape. The Plant Cell 15, 1095–1110.
| Dominant-negative receptor uncovers redundancy in the Arabidopsis ERECTA leucine-rich repeat receptor-like kinase signaling pathway that regulates organ shape.Crossref | GoogleScholarGoogle Scholar | 12724536PubMed |
Shpak ED, Berthiaume CT, Hill EJ, Torii U (2004) Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation. Development 131, 1491–1501.
| Synergistic interaction of three ERECTA-family receptor-like kinases controls Arabidopsis organ growth and flower development by promoting cell proliferation.Crossref | GoogleScholarGoogle Scholar | 14985254PubMed |
Takahashi T, Shibuya H, Ishikawa A (2016) ERECTA contributes to non-host resistance to Magnaporthe oryzae in Arabidopsis. Bioscience, Biotechnology, and Biochemistry 80, 1390–1392.
| ERECTA contributes to non-host resistance to Magnaporthe oryzae in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 26924213PubMed |
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729.
| MEGA6: Molecular evolutionary genetics analysis version 6.0.Crossref | GoogleScholarGoogle Scholar | 24132122PubMed |
van Zanten M, Snoek LB, Proveniers MCG, Peeters AJM (2009) The many functions of ERECTA. Trends in Plant Science 14, 214–218.
| The many functions of ERECTA.Crossref | GoogleScholarGoogle Scholar | 19303350PubMed |
van Zanten M, Snoek LB, Eck-Stouten EV, Proveniers MCG, Torii KU, Voesenek LACJ, Millenaar FF, Peeters AJM (2010) ERECTA controls low light intensity-induced differential petiole growth independent of phytochrome B and cryptochrome 2 action in Arabidopsis thaliana. Plant Signaling & Behavior 5, 284–286.
| ERECTA controls low light intensity-induced differential petiole growth independent of phytochrome B and cryptochrome 2 action in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar |
Villagarcia H, Morin A, Shpak ED, Khodakovskaya MV (2012) Modification of tomato growth by expression of truncated ERECTA protein from Arabidopsis thaliana. Journal of Experimental Botany 63, 6493–6504.
| Modification of tomato growth by expression of truncated ERECTA protein from Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 23096000PubMed |
Wang DX, Yang CJ, Wang HJ, Wu ZH, Jiang JJ, Liu JJ, He ZN, Chang F, Ma H, Wan XL (2017) BKI1 regulates plant architecture through coordinated inhibition of the brassinosteroid and ERECTA signaling pathways in Arabidopsis. Molecular Plant 10, 297–308.
| BKI1 regulates plant architecture through coordinated inhibition of the brassinosteroid and ERECTA signaling pathways in Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
Zheng JC, Hu YG (2016) TaERECTA response to phytohormone, Mg2+ stress and dehydration and its association with stomatal density in bread wheat. Cereal Research Communications 44, 206–216.
| TaERECTA response to phytohormone, Mg2+ stress and dehydration and its association with stomatal density in bread wheat.Crossref | GoogleScholarGoogle Scholar |
Zheng JC, Yang ZY, Madgwick PJ, Carmo-Silva E, Parry MAJ, Hu YG (2015) TaER expression is associated with transpiration efficiency traits and yield in bread wheat. PLoS One 10, e0128415
| TaER expression is associated with transpiration efficiency traits and yield in bread wheat.Crossref | GoogleScholarGoogle Scholar |
Zheng JC, Liu T, Li JQ, Wang X, Li WY, Xu F, Zha QW (2019) Virus-induced gene silencing of TaERECTA increases stomatal density in bread wheat. Cereal Research Communications 47, 67–77.
| Virus-induced gene silencing of TaERECTA increases stomatal density in bread wheat.Crossref | GoogleScholarGoogle Scholar |
† These authors contributed equally to this work.








