Untargeted Metabolomic Analyses Reveal Chemical Complexity of Dioecious Cannabis Flowers
Matthew T. Welling A , Myrna A. Deseo A B , Antony Bacic A B and Monika S. Doblin A B C
A , Myrna A. Deseo A B , Antony Bacic A B and Monika S. Doblin A B C
A La Trobe Institute for Agriculture and Food, Department of Animal, Plant and Soil Sciences, School of Life Sciences, AgriBio Building, La Trobe University, Bundoora, Vic. 3086, Australia.
B Australian Research Council Research Hub for Medicinal Agriculture, AgriBio Building, La Trobe University, Bundoora, Vic. 3086, Australia.
C Corresponding author. Email: m.doblin@latrobe.edu.au
Australian Journal of Chemistry 74(6) 463-479 https://doi.org/10.1071/CH21033
Submitted: 30 January 2021 Accepted: 26 April 2021 Published: 21 May 2021
Journal Compilation © CSIRO 2021 Open Access CC BY-NC-ND
Abstract
Cannabis is a mostly dioecious multi-use flowering plant genus. Sexual dimorphism is an important characteristic in Cannabis-based commercial production systems, which has consequences for fibre, seed, and the yield of secondary metabolites, such as phytocannabinoid and terpenes for therapeutic uses. Beyond the obvious morphological differences between male and female plants, metabolic variation among dioecious flowers is largely undefined. Here, we report a pilot metabolomic study comparing staminate (male) and pistillate (female) unisexual flowers. Enrichment of the α-linolenic acid pathway and consensus evaluation of the jasmonic acid (JA) related compound 12-oxo-phytodienoicacid (OPDA) among differentially abundant metabolites suggests that oxylipin signalling is associated with secondary metabolism and sex expression in female flowers. Several putative phytocannabinoid-like compounds were observed to be upregulated in female flowers, but full identification was not possible due to the limitation of available databases. Targeted analysis of 14 phytocannabinoids using certified reference standards (cannabidiolic acid (CBDA), cannabidiol (CBD), Δ9-tetrahydrocannabinolic acid A (Δ9-THCAA), Δ9-tetrahydrocannabinol (Δ9-THC), cannabichromenic acid (CBCA), cannabichromene (CBC), cannabigerolic acid (CBGA), cannabigerol (CBG), cannabinolic acid (CBNA), cannabinol (CBN), cannabidivarinic acid (CBDVA), cannabidivarin (CBDV), tetrahydrocannabivarinic acid (THCVA), and tetrahydrocannabivarin (THCV)) showed a higher total phytocannabinoid content in female flowers compared with the male flowers, as expected. In summary, the development of a phytocannabinoid-specific accurate-mass MSn fragmentation spectral library and gene pool representative metabolome has the potential to improve small molecule compound annotation and accelerate understanding of metabolic variation underlying phenotypic diversity in Cannabis.
Keywords: liquid chromatography, mass spectrometry, phytochemistry, metabolism, Cannabis, cannabinoids, untargeted metabolomics, flowering.
Introduction
Cannabis, a member of the family Cannabaceae, is a highly heterozygote multi-use anemophilous (wind pollinated) flowering plant genus, which is believed to have originated in Central Asia.[1] Taxonomic assignment and speciation of Cannabis is unresolved.[2] Despite this ambiguity, a monotypic status is widely supported on the basis that physiological and genetic barriers to gene flow are underreported.[3] Domestication of Cannabis over prolonged periods of time has led to its global dispersal. Fossil evidence in the form of pollen deposits and seed remains suggest widespread use across Eurasia dating back several thousand years.[1,4] In addition to traditional uses for food and fibre, plants also have recreational and medicinal value due to their secondary metabolites, particularly phytocannabinoids and terpenes.
Cannabis plants are prolific producers of secondary metabolites, with their repertoire spanning several chemical classes including terpenoids, flavonoids, stilbenoids, alkaloids and phenolic amides.[5] However, it is the group of isoprenylated resorcinyl polyketides, commonly referred to as phytocannabinoids, that Cannabis is most recognised for on account of their exclusivity to this taxon and their recreational and potential therapeutic uses.[6] The phytocannabinoid Δ9-tetrahydrocannabinol (THC), which is a partial agonist of the human endocannabinoid CB1 receptor,[7] is primarily responsible for the narcotic status of this plant. A large proportion of the more than 100 phytocannabinoid constituents are non-intoxicating, with a subset representing molecules of potential therapeutic importance. These include cannabidiol (CBD) which is the active ingredient in Epidiolex, a United States Food and Drug Administration (FDA) approved prescription medicine for the treatment of intractable seizures in childhood epileptic disorders.[8] Terpenes which are also produced by Cannabis are thought to contribute to phytocannabinoid ligand activity, although evidence for the so-called ‘entourage effect’ when extended to terpenoids is largely anecdotal.[9]
An arbitrary level of THC content, which can vary by jurisdiction, is used to demarcate crop use, with a low THC content in female flowers congruent with industrial hemp end-uses.[10] Plants typically segregate into one of three chemotypes based on THC/CBD ratio, although this system of classification does not account for variation of other phytocannabinoid constituents such as those with altered alkyl groups which can be more abundant than either THC or CBD.[11] The biosynthesis of phytocannabinoids and other secondary metabolites, such as terpenoids, is concentrated within specialised capitate stalked glandular trichomes.[12] These are hair-like structures which are abundant on the epidermal surfaces of floral tissues of female inflorescences.[13] Unisexual female inflorescences are highly branched compound racemes that are comprised of mono-photometric structures consisting of an axillary shoot, solitary pistillate flowers, subtending bracts, and reduced leaves.[14] In contrast, male inflorescences have sparse leaf coverage and consist largely of pendulous panicles.[15] Staminate flowers have a five-sepal enclosed androecium, which, on opening, allows for longitudinal release of pollen grains (Fig. 1a).[15]

|
Being mainly dioecious, Cannabis segregates into distinct female and male plants, although monecious phenotypes with bisexual flowers or inflorescences bearing separate male and female flowers occur to a lesser extent.[15,16] Reproductive commitment occurs as early as the emergence of the fourth leaflet pair.[15] Sex expression can be influenced by temperature and photoperiod, although not all genotypes are strictly obligate in this regard and sensitivity to photoperiodic induction can vary considerably amongst germplasm.[17] Sex is determined by heteromorphic X and Y chromosomes.[18] Males are the heterogametic sex (XY) while females are the homogametic (XX) sex.[18] However, sexual phenotype can be modified in well differentiated male as well as female plants by various chemical and phytohormonal applications, such as silver thiosulfate, gibberellic acid, and ethephon.[19–21]
Sexual dimorphism is a critical factor in Cannabis-based production systems and is essential for genetic improvement of germplasm.[22] For phytocannabinoid production, genetically female plants are grown in the absence of pollen. Male and hermaphroditic plants have less floral biomass, reduced phytocannabinoid yield, and also negatively impact female inflorescence quality via fertilisation and initiation of seed development.[16] Prolonged virginity in female plants during flowering also aids in the formation of congested pistillate inflorescences which improves phytocannabinoid yield.[16,23] Conversely, monoecy can be desirable for the cultivation of industrial hemp. Not only does this reduce crop heterogeneity, but also mitigates early flowering and senescence of unisexual male plants which can complicate mechanical harvesting of both stems and seed. However, the monoecious state can be intergenerationally unstable and sexual phenotypic extremes can vary from predominantly male to predominantly female.[24]
Despite the commercial importance of sexual dimorphism in Cannabis, few studies have comprehensively examined chemical phenotypes of male and female flowers. While comparisons have been made between unisexual flowers, these have either been targeted towards a small number of phytocannabinoids or have been applied to a narrow subset of the gene pool.[25–28] Ultra-high-performance liquid chromatography–electrospray ionisation–high-resolution mass spectrometry (UHPLC-ESI-HRMS) with data-dependent acquisition is an effective analytical technique for untargeted chemical profiling of plant constituents, as it facilitates chromatographic separation of complex matrices and simultaneous acquisition of HRMS and MSn spectral data.[29] Here, we performed a pilot study comparing the floral tissues of three male and three female industrial hemp accessions to establish the veracity of the methodological approach and the minimal sampling requirements for a large-scale germplasm comparison of Cannabis chemotypes. Both a targeted approach with consideration of 14 phytocannabinoid constituents as well as a non-targeted approach using UHPLC-ESI-HRMS methodology was performed with the aim of bridging the gap in understanding the metabolic variation underlying sexual phenotype in Cannabis.
Results
Metabolic Variation Among Cannabis Flowers
A panel of six industrial hemp accessions were grown from seed in a controlled environment room. As is common practice for indoor-grown Cannabis, seedlings were grown initially under a long-day photoperiod to promote vegetative growth and then under a short-day photoperiod to initiate flowering. The time it took plants to reach flower maturity varied among accessions. Female plants used for analysis were harvested ~4 (accession 11) and ~5 (accessions 06 and 07) weeks after exposure to a short-day photoperiod when ~95 % of the stigma on pistillate (hereafter referred to as female) flowers were browned and shrivelled, while male plant individuals were harvested after a period of ~5 (accession 20) and ~7 weeks (accession 12 and 14) following photoperiod induction when ~95 % staminate (hereafter referred to as male) flowers had opened and expelled pollen (Table 1). No consistent differences in height were observed between male and female plants within sample groups, with female and male plants varying from 1.4 m (accessions 06 and 12) to ~2 m (accessions 07 and 14). To enrich for sex-specific flowers and capture within-plant metabolic variation, foliage leaves, seeds, and/or stems were removed from inflorescences before analysis and floral tissues were sampled from the apical and basal inflorescence of each male and female plant.

|
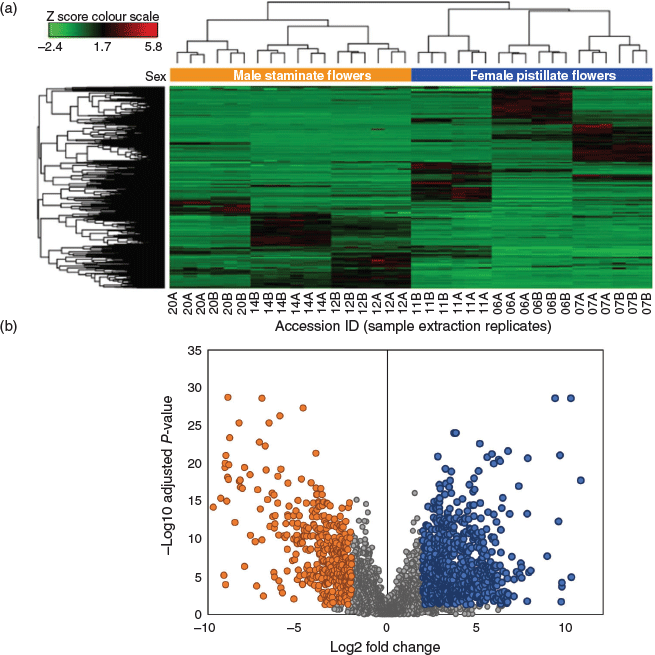
Untargeted metabolic analysis was performed using UHPLC-ESI-HRMS and data-dependent MS2 acquisition. Preliminary method development involving Cannabis floral tissues showed an increase in the number of features and coverage of the metabolome in positive ion mode as compared with negative ion mode (data not shown) and so untargeted analysis and quantification of unknown compounds was performed in positive ion mode. The representative total ion chromatogram (TIC) from the six accessions showed a high level of dissimilarity (Fig. 2), indicating that the floral samples had diverse chemical profiles. Analysis of male and female floral tissues across all accessions generated 74 151 features that accounted for 5161 putative compounds distinguishable by retention time and molecular weight. The compound list was filtered by eliminating those that fall within the group with low abundant chromatographic peak areas < 50 000 and those lacking MS2 fragmentation spectra. After filtering, 2599 compounds/metabolites remained in the list (Supplementary Material, Table S1). Principal component analysis (PCA) of the 2599 putative metabolites showed a clear separation of male and female flower samples, with the first two principal components (PCs) accounting for 37.5 % (PC1) and 18.1 % (PC2) of the variability in the dataset (Fig. 1b and Supplementary Material, Fig. S1). Processing of putative metabolites by hierarchical clustering analysis (HCA) grouped technical (extraction) replicates by accession and by inflorescence position (apical and basal; descending developmental maturity) as well as by male and female flowers. Floral samples from the six accessions formed two major clades representative of male and female flowers, suggesting that unisexual flowers have distinct metabolomes (Fig. 3a).

|
|
Analysis of Differentially Abundant Metabolites (DAMs) of Cannabis Floral Tissues
Analysis of DAMs between male and female floral samples indicated a greater number of metabolites were upregulated in the female floral tissues as compared with the male (Fig. 3b). Of the 2599 putative metabolites, 861 were upregulated while 626 were downregulated in female versus male flowers (P-value of < 0.05, log2 fold change ≥ 1.5) (Fig. 3b). A large proportion of putative metabolite abundances (~43 %) were shared between male and female flowers (shaded in grey in Fig. 3b). Metabolites common to both unisexual flowers were dismissed and only DAMs were investigated further.
To gain an overall understanding of metabolic variation between female and male floral samples, and to interpret the biological significance associated with these tissues, DAMs were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/). More pathways were impacted in downregulated DAMs as compared with upregulated DAMs (Fig. 4 and Supplementary Table S2). Secondary metabolism and polyunsaturated fatty acid (PUFA) metabolism pathways were significantly enriched in both up- and downregulated DAMs (adj. P-value < 0.001) (Fig. 4). In the downregulated DAMs, the following pathways were enriched (adj. P-value < 0.001): arachidonic acid metabolism; α-linolenic acid metabolism; linoleic acid metabolism; flavone and flavonol biosynthesis; tropane, piperidine, and pyridine alkaloid biosynthesis; and phenylpropanoid biosynthesis (Fig. 4a). The α-linolenic acid metabolism pathway was also significantly enriched among upregulated DAMs, along with sesquiterpenoid biosynthesis pathway as well as limonene and pinene degradation pathways (adj. P-value < 0.001) (Fig. 4b). In addition to PUFA and secondary metabolism pathways, amino acid biosynthesis and metabolism pathways were also enriched in downregulated DAMs (adj. P-value < 0.05), with phenylalanine metabolism being significantly enriched in the downregulated DAMs (adj. P-value < 0.001) (Fig. 4a).

|
Annotation of Female versus Male Flower DAMs
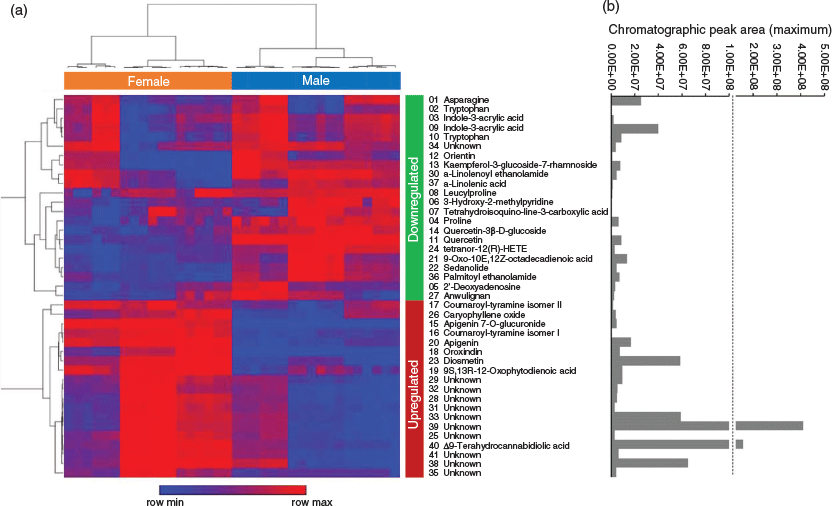
Annotations of unknown floral tissue metabolites were assigned using Compound Discoverer software (CD) from consensus evaluation based on elemental composition prediction, spectral library (mzCloud) and database (ChemSpider) searches, as well as the database ranking algorithmic tool mzLogic. Of the 626 downregulated and 861 upregulated putative compounds (1487 compounds in total), 41 compounds that represented 2.8 % of the total number of DAMs were assigned annotations based on the consensus evaluation. There were 19 compounds annotated in the upregulated group (Table 2), with 22 compounds in the downregulated group (Table 3).
Of the 19 upregulated compounds in female versus male floral tissues, 10 compounds were initially annotated as the cannabidiol (CBD) hydroxyquinone HU‐331 (compounds 25, 28, 29, 31, 32, 33, 35, 38, 39, and 41 in Table 2). One compound in the downregulated group (compound 34 in Table 3), was also initially annotated as HU-331. These 11 HU-331-annotated compounds all had a calculated molecular mass of 328.2039 based on MS data, which matches the molecular formula of C21H28O3. The different retention times of putative HU-331-annotated compounds ranged from 9.1 to 14.6 min and indicated that these compounds are molecular isomers.
Fragment Ion Search (FISh) coverage score, an algorithmic measurement of the percentage of MS2 ions which match in silico fragmentation patterns based on literature-defined chemical reactions from the HighChem Fragmentation Library, was applied to the HU-331 annotated compounds in an attempt to further discriminate the compounds from one another. FISh coverage scores of HU-331-like compounds varied from 38.46 % to 94.74 % (Tables 2 and 3), with higher FISh scores being more supportive of the annotated chemical structure. Compound 32 with retention time of 11.4 min gave the highest FISh score of 94.7 % among the 11 similarly annotated compounds, indicating that this compound is likely to be HU-331. The MS2 spectrum of compound 32 (Supplementary Material, Appendix S1) gave a base peak of m/z 329.2108 that corresponded to the [M + H]+ ion. Upon comparison of its MS2 spectrum to that of HU-331 from the mzCloud reference library, it became apparent that the fragmentation patterns and their calculated elemental composition did not match, indicating that compound 32 could be a structural isomer of HU-331. The MS2 spectra of the other compounds with the same pseudo-molecular ions were also extracted (Supplementary Material, Appendix S1) and compared with that of HU-331 (Supplementary Material, Appendix S2). Compounds 28, 29, 31, 32, and 33 showed the presence of fragment ion m/z 311.201, which corresponds to a loss of a water molecule and is indicative of the presence of a hydroxy substituent in the molecule. Compound 25 that eluted at 9.1 min showed a different MS2 spectrum compared with the other compounds, suggesting that this compound could be of a different class to that of the other 10 unknown compounds. Compound 35 at 12.8 min also showed greater variation in the fragment ions which also differentiated this compound from the others. Compounds 38, 39, 41, and 34 all showed a base peak at m/z 229.086 that matched the fragment ion of HU-331 with a chemical formula of C14H13O3+ for the benzoquinone moiety with alkene sidechain as shown in the mzCloud reference library (Supplementary Material, Appendix S2). The four compounds 38, 39, 41, and 34, and compounds 28, 29, 31, 32, and 33 showed some similarity of fragment ions with that of HU-331 and most probably represent two groups of hydroxy-benzoquinones with different substituents.
Compound 40, corresponding to the molecular formula C22H30O4 and [M + H]+ ion at m/z 358.21434, was incorrectly annotated as cannabidiolic acid by the CD software. Despite being identified as CBDA across multiple annotation sources, this compound eluted 2.9 min after the CBDA reference standard and had a low FISh coverage score of 42.96 % (Table 2). This compound was an upregulated DAM in the female flowers and was subsequently identified as Δ9-tetrahydrocannabinolic acid A (Δ9-THCAA), with the nominal mass (Δ ppm < 5) as well as retention time matching that of the Δ9-THCAA reference standard at 14.6 min.
Analogous with the KEGG pathway enrichment analysis, several lipid and flavonoid metabolites were identified through consensus evaluation from DAMs of the female versus male floral tissues. Lipid constituents with FISh coverage scores ≥ 75 % included α-linolenic acid, 9-oxo-octadecadienoic acid (ODE), tetranor-12(R)-HETE in the downregulated group, and 12-oxo-phytodienoic acid in the upregulated group (Fig. 5 and Tables 2 and 3). Two PUFA-derived endocannabinoid-like acylethanolamides, palmitoyl ethanolamide and α-linolenoyl ethanolamide, were also observed in the downregulated DAMs, with FISh coverage scores of these metabolites being greater than 90 %. The flavonoids apigenin, apigenin 7-O-glucuronide, diosmetin, and oroxindin were identified in the upregulated group (Fig. 5 and Table 2), while kaempferol-3-glucoside-7-rhamnoside, orientin, quercetin, and quercetin-3-β-D-glucoside were identified in the downregulated group (Fig. 5 and Table 3). Of these, only quercetin, oroxindin, and diosmetin had FISh coverage scores ≥ 75 % (Tables 2 and 3).
|
Quantitative Analysis of Phytocannabinoids
Fourteen phytocannabinoids, for which certified reference standards are available, were determined quantitatively from the male and female floral tissues of six Cannabis accessions and the results are summarised in Table 4. Given that the plants used for metabolomic analysis were derived from industrial hemp varieties, it was anticipated that CBDA/CBD, which are commonly associated with such germplasm, would be the predominant phytocannabinoids among the floral samples. CBDA concentration was observed to be highest in accessions 06 and 11 (554.53 to 798.10 mg/100 g), and its decarboxylated form CBD was highest in accession 11 (901.29 mg/100 g in the apical floral tissue), which was evident in the representative TICs of the accessions in Fig. 2 (compounds 42 and 43, respectively). Surprisingly, the profile of accession 07 was high in Δ9-THCAA (1254.30 and 653.86 mg/100 g for apical and basal floral tissues, respectively) and Δ9-THC (1481.90 and 1350.29 mg/100 g for apical and basal floral tissues, respectively) compared with other accessions and these compounds were the predominant phytocannabinoids in the profile of this plant. The total THC content of two individual plants derived from accessions 06 and 07 exceed the limit set under the provisions of the Drugs, Poisons and Controlled Substances Act 1981. These plants were subsequently destroyed during harvest and were not used for further propagation.

|
Another indication of the chemical diversity of plants used for comparative metabolomic analysis was the unusual chemotype of the female plant from accession 06 which showed elevated levels of what are typically minor alkyl phytocannabinoid homologues. CBDVA and THCVA, which differ from CBDA and Δ9-THCAA, respectively, by the number of carbon atoms on their alkyl side chains, exhibited variable concentration among the accessions with accession 06 having the highest concentration. As expected, a similar trend was observed for the corresponding decarboxylated forms, with CBDV and THCV highest in accession 06. CBNA and CBN, which are oxidation artefacts of Δ9-THCAA and THC, respectively,[6] had relatively low concentrations among the floral samples examined. Interestingly, the concentration of the juvenile cannabinoid CBCA was 5–20-fold higher in the female plants from accessions 06, 07, and 11 (431.52 to 719.86 mg/100 g) compared with the male plants from accessions 12, 14, and 20 (19.60 to 126.31 mg/100 g). CBGA, which is the precursor of CBDA, Δ9-THCAA, and CBCA,[6] exhibited a concentration range of > 30 fold (0.71 to 32.35 mg/100 g) for the six accessions.
The decarboxylated phytocannabinoids are generated from their respective carboxylated forms either in planta, post-harvest (upon exposure to, for example, light and/or heat), or both. The floral samples exhibited varying levels of decarboxylation among the phytocannabinoid species examined (e.g. CBD/CBDA and THC/THCA), although a higher Δ9-THC concentration than its acid form (Δ9-THCAA) is evident for all floral tissue samples. A similar trend of higher CBD concentration can be observed within accessions 11, 12, 14, and 20 but the reverse is true for accessions 06 and 07.
In addition to variation in the relative abundance of phytocannabinoids among male and female plants, variation in the total cannabinoid content was also evident from the targeted analysis of phytocannabinoids (Fig. 6a) To assess the relative proportion of each phytocannabinoid species in the samples, the concentration of the acid and decarboxylated forms were combined (e.g. CBD + CBDA, Δ9-THCAA + Δ9-THC, etc; Fig. 6b). The analysis of the apical and basal floral tissues from the six accessions generally did not exhibit a large variation in the phytocannabinoid concentration within the accession, although a trend of slightly higher levels in the apical inflorescence can be observed, especially among the female floral samples. Accession 06 gave the highest total phytocannabinoid concentration (5427.69 and 4964.72 mg/100 g for apical and basal floral tissues, respectively) with CBDVs and THCVs as the more dominant phytocannabinoids within the accession. The total phytocannabinoid concentration for the other accessions varied with accession 07 > accession 11 > accession 20 > accession 14 > accession 12. From these data it can be observed that the average total concentration of phytocannabinoids from floral tissues of female plants (accessions 06, 07, and 11) are higher compared with those of the male plants (accessions 12, 14, and 20) as shown in Fig. 6c.

|
Discussion
The Metabolome of Cannabis Flowers
While the use of genomic and transcriptomic resources to elucidate molecular mechanisms underlying trait diversity in Cannabis has accelerated in recent years,[30] our understanding of the abundance and diversity of chemical constituents defining chemotypic groups is lacking.[29] Metabolomic information can provide greater resolution of underlying biological processes than is possible with other omics-based analyses as metabolites represent the endpoint of cellular process and are therefore more proximal to the phenotype or chemotype under investigation.[31] Identifying a set of metabolites that distinguishes samples from two or more groups is reliant on both a comparison of a sufficient number of samples to obtain statistically meaningful information as well as on the ability of these samples to adequately capture biological variation so as to avoid spurious associations.[31,32] In the absence of pilot data to inform experimental design, extracting relevant and interpretable information from metabolic datasets remains challenging.[31,33] To maximise group variation, biological replicates used in the comparison of male and female Cannabis flowers were selected at the level of accession (variety). Multivariant analysis showed that the biological replicates of each accession clustered in distinct spaces indicating that the impact of sexual phenotype was substantial and specific, despite the limited sample size (Fig. 1b and Fig. 3a).
Another obstacle which complicates interpretation of metabolomic variation between chemotypes is identifying the vast catalogue of small molecules present within samples.[29,34] Annotation assignments of unknown floral metabolites was performed in Compound Discoverer software by consensus evaluation of elemental composition prediction, database, and MS2 spectral library searches. These integrated approaches increase the level of confidence in the annotation of metabolites from the available HRMS information, although this level of stringency ultimately reduces the number of reportable compounds. Of the 2599 compounds identified across floral samples, only 211 or ~8 % of all compounds had a full match with the vendor-specific spectral library (mzCloud), compared with 936 compounds which matched ChemSpider database searches and 2382 compounds with predicted elemental compositions matching theoretical isotope patterns. While there are varying levels of identification rigour which can be applied to the reporting of compounds from untargeted metabolomic analyses (varying from unambiguous identification to compound classes and unclassified metabolites),[35] a major bottleneck in the confidence of compound annotation is matching MS2 fragments with library spectra fragments. Development of customised MS2 spectral libraries, such as those applied to Cannabis sativa var. Kompolti for phenolic compound identification,[29] coupled with the acquisition of high-quality HRMS fragmentation data is anticipated to improve annotation of unknown compounds and allow for more detailed profiling of phenotypic variation in Cannabis. Application of these spectral libraries within untargeted metabolomic analysis pipelines will also be a critical step in understanding the complexity of botanical formulations derived from Cannabis.
Phytocannabinoid Variation Among Floral Samples
Our analysis among six accessions showed that cannabinoid contents were consistently higher in the female samples as compared with male flowers (Fig. 6c). Previous analyses of cannabinoid contents among unisexual plants yielded inconsistent results, with comparisons either showing no differences between male and female inflorescences or showing a higher cannabinoid content in female floral tissues.[25–28] This could be attributed to several factors.
One possible source of variation contributing to inconsistences in phytocannabinoid content among unisexual flowers could relate to the sampling of floral tissues. Male inflorescences are largely comprised of staminate flowers and have a low abundance of vegetative tissue, with glandular trichomes, which are the major site for phytocannabinoid synthesis and accumulation, concentrated on the surface of anthers.[12,15,36] In contrast, female inflorescences have a more complex highly branched heterogeneous structure where trichome abundance is concentrated on a subset of vegetative tissues, such as the perigonal bracts, surrounding flowers (see Fig. 1a).[14,36]
Previous comparisons between unisexual flowers have not discriminated between inflorescence component tissues such as low-phytocannabinoid containing foliage leaves and those directly encasing the carpel and pistillate flowers, while others have homogenised inflorescences bearing immature fruit (seed).[26–28] These details are especially relevant given the variation in female plant architecture and inflorescence structure reported among sub-taxa, such as variation in branching, internode length, and bract abundance.[14] Congruent with the cluster analysis, four of the six accessions showed a higher cannabinoid content in the apical inflorescence as compared with basal inflorescence samples (Fig. 6a), indicating that the age and tissue-type of the plant material sampled will also impact comparative analyses among unisexual flowers. As with other aspects of chemotype identified in female plants, such as proportion and content of phytocannabinoids,[11,37] genotype-specific variation could also be contributing to patterns of cannabinoid content among unisexual flowers.
DAMs Associated with Sexual Phenotype
While incorrectly annotated as CBDA by Compound Discoverer software due to the absence of Δ9-THCAA MS2 fragmentation data in the available spectral libraries, independent identification of one of the predominant phytocannabinoids segregating between male and female floral samples indicates that the DAMs identified from untargeted metabolomic analysis are biologically meaningful and specific to the sexual phenotypes under investigation (Fig. 2 and Fig. 5). In addition to identifying phytocannabinoid variation, our untargeted analysis of unisexual flowers also revealed several metabolites known to be involved in pollen sterility to be among the male-specific DAMs.
Consistent with other analyses of male flowers and pollen-producing tissues,[38–40] several pathways relating to PUFA metabolism as well as flavone and flavonol biosynthesis were enriched in the downregulated (male-specific) DAMs (Fig. 4). It is well established that flavonoids play a key role in pollen viability.[38] Flavonols, as opposed to other flavonoids produced in the anthers of the flower, such as quercetin and kaempferol, promote pollen fertility and pollen tube growth in vitro.[38] Analysis of transgenic plants with reduced chalcone synthase gene expression have resulted in abnormal anthers with impaired pollen tube growth and low germination efficiency in Petunia and Solanum, respectively.[39,40] In agreement with previous HPLC‐DAD‐MSn analysis comparing quercetin di‐C‐hexoside between male and female Cannabis sativa var. Spontanea flowers,[25] our analysis shows quercetin to be predominantly present in the male sexual phenotype with only trace levels detected in female floral samples (Fig. 5). While the mechanism underlying flavonoid accumulation and pollen fertility remain unclear, quercetin and possibly other flavones identified to be upregulated in male floral tissues may participate in reactive oxygen species (ROS) homeostasis in anthers, thereby limiting ROS-facilitated pollen abortion.[41] As with the flavonoid pathways, over-representation of linoleic and α-linolenic pathways in pollen producing tissues is not uncommon. Stearoyl-ACP desaturase, ER desaturases (FAD3, FAD2), and FATB thioesterases involved in the termination of C16 fatty acids are differentially expressed in male flowers, and linolenic acid as well as linoleic acid are the predominant constituents in the lipid fractions of pollen.[42–44]
While the α-linolenic acid metabolism pathway was impacted by DAMs specific to both male and female flowers, this pathway could be utilised differently between unisexual flowers, with the untargeted analysis of floral tissues indicating that the jasmonate (JA) pathway may play a role in flower development and secondary metabolism in female plants. Among the upregulated differential metabolites, one fatty acyl octadecanoid with the molecular formula C18H28O3 and [M + H]+ ion at m/z 292.2039 was identified via consensus evaluation as 12-oxo-phytodienoic acid (OPDA) (Fig. 5 and Table 2). OPDA is a linolenic acid-derived oxylipin signalling molecule and a direct precursor to JA.[45] JA, jasmonoyl-l-isoleucine, and methyl JA, collectively known as jasmonates, regulate gene expression governing plant developmental, physiological, and defence processes and are directly implicated in flower development, seed maturation, and induction of plant defence compounds such as various terpenoids.[46,47]
Despite OPDA being biosynthetically related to JA, it has a distinct role in signalling and gene transcription independent of other jasmonates. Of relevance to sexual phenotype in Arabidopsis are upregulated OPDA-specific genes associated with auxin (At3g09870, At5g35735) and abscisic acid (ABA) (At5g13200) hormonal responses.[46] OPDA is the dominant oxylipin in seed coat tissues of various plant species, and female-sterility in JA-insensitive mutants can be rescued by wound-induction synthesis of OPDA, indicating OPDA-specific regulation of embryo development.[45,48,49] OPDA also shows synergistic effects with ABA, increases ABA INSENSITIVE5 protein abundance, and its concentration is affected by ABA via the proposed action of PLASTID LIPASE2 and 3.[50] In Cannabis, ABA can fully inhibit GA induction of male flowers in female plants and indole-3-acetic acid (IAA) induction of female flowers in male plants.[20,51] Two compounds corresponding to the molecular formula C11H9NO2 and [M + H]+ ion at m/z 187.0633 were identified as indole-3-acrylic acid (IAcrA) in the downregulated DAMs (Table 3 and Fig. 5), although the function of this auxin is primarily associated with plant growth metabolic processes.[52]
Irrespective of florogenesis, the upregulation of jasmonates, such as OPDA, in the high phytocannabinoid yielding female floral samples may indicate a possible role for oxylipin signalling in Cannabis secondary metabolism. Congruent with this hypothesis is the enrichment of monoterpenoid and sesquiterpenoid pathways as well as the annotation of sesquiterpene caryophyllene oxide in the female flowers (Fig. 4b and Table 2). Changes in THC, CBD, and carotenoid content have been reported in Cannabis vegetative tissues treated with between 1 and 100 μM JA,[53] while application of ABA increases THC content in female and male floral tissues.[54] There are four stereoisomers of OPDA, with the natural cis isomer (9S,13S)-OPDA being relevant in JA biosynthesis. The OPDA upregulated in female flowers was identified across annotation sources as the trans isomer (9S,13R)-OPDA (Table 2). However, it is not possible to discriminate these isomers with the LC stationary phase used in this study. This is the first report of the upregulation of OPDA in Cannabis flowers and given that APA- and auxin-related genes are directly implicated in sex expression,[20,51,55] this compound may represent a molecule of interest for further investigation.
Unknown Phytocannabinoid-Like DAMs in Cannabis Flowers
Eleven compounds were identified as molecular isomers of HU‐331 corresponding to the molecular formula C21H28O3 and [M + H]+ ion at m/z 329.2115; 10 of which were upregulated in the female floral samples (Fig. 5 and Table 2). In addition to differing by retention time, the MS2 spectra of these compounds showed differences in their fragmentation patterns and suggested three different groups or classes with compounds 25 and 35 differing from each other and the other molecular isomers. The other compounds 28, 29, 31, 32, 33, 34, 38, 39, and 41 showed several fragment ions that were similar to the fragment ions of HU-331, although some of the elemental compositions did not match that of HU-331 when calculated within 5 ppm of the observed masses. However, the MS2 spectra of these nine compounds indicated that these could be structural isomers of a hydroxybenzoquinone-type compound with different substituents.
Some benzoquinones can be synthesised non-enzymatically from phytocannabinoids. Examples are HU-331,[56] HU-336,[57] HU-345,[57] and VCE-003[58] from the oxidation of CBD, Δ8-THC, CBN, and CBG, respectively. Upregulation of multiple phytocannabinoid derivatives may be suggestive of a greater flux through the phytocannabinoid pathway in female flowers as compared with the male flowers which is consistent with the higher phytocannabinoid content identified in the female floral samples. HU-331 represents a potential anti-cancer drug which inhibits ATPase activity of topoisomerase II at micromolar concentrations.[59,60] Similarly, as with VCE-003, various HU-331 analogues act as perixome proliferator activated receptors (PPAR)γ agonists and are predicted to have neuroprotective activities.[61] HU-331 is also produced in mammals by hepatic microsomal enzymes where it forms adducts with glutathione.[62] While it is unclear if these benzoquinone compounds could be an autoxidation artefact, a subset may represent bona fide natural products worth further investigation when considering the therapeutic potential of quinone cannabinoid derivativities.[59–61]
In addition to the classical isoprenylated resorcinyl polyketide phytocannabinoids, other interesting PUFA-derived cannabimimetic-like compounds were found as DAMs across female versus male floral samples. N-Acyl-ethanolamines are ubiquitous chemical signalling lipids which can potentiate responses of the mammalian endocannabinoid anandamide signalling pathway.[63,64] Two such compounds, α-linolenoyl ethanolamide and palmitoyl ethanolamide, were found to be upregulated in male flowers (Table 3 and Fig. 5). While it is uncertain if the concentrations present would modify phytocannabinoid ligand activity, the presence of these compounds in male flowers is nonetheless intriguing.
Conclusion
This pilot study sought to both substantiate the methodological pipeline for tissue/organ comparative metabolome analyses by UHPLC-ESI-HRMS and to examine metabolic variation between unisexual flowers of the dioecious plant genus Cannabis. Our data indicate that this approach was successful in identifying several metabolites associated with sexual phenotype.
The chemical complexity of female flowers is evident from the PCA of floral tissues, with the metabolomes of the three female accessions showing a high level of dissimilarity (Fig. 1b). A large proportion of this metabolic variation is not currently accounted for in the manufacture of crude herbal formulations, artisanal extracts, and magistral preparations intended for human consumption.[34] While medicinal chemotypes of Cannabis are most commonly maintained through clonal propagation under controlled environment conditions,[65,66] seed-grown industrial hemp sub-taxa are increasingly being used as feedstock for either nutraceutical or therapeutic-based products.[67] Given the outcrossing and highly heterozygote nature of Cannabis plants in general, this could generate significant chemical variation which may have consequences for the quality, safety, and efficacy of Cannabis preparations.
Notwithstanding the targeted measurement of 14 major and minor cannabinoids, we identified several putative unknown phytocannabinoid-like metabolites upregulated in female flowers but were unable to unequivocally assign annotations to these compounds. The annotation of metabolites was limited by the availability of reference compounds to compare observed MS1 and MS2 information for correct annotation, thus highlighting the need to establish a comprehensive reference library or database for Cannabis metabolites. Development of phytocannabinoid-specific accurate-mass MSn fragmentation spectral libraries coupled with nuclear magnetic resonance analysis will improve the confidence of compound structure annotation and mitigate challenges associated with the discrimination of multiple cannabinoid species and their constituent isomers and stereoisomers within complex botanical matrices.[6,68] Moreover, the completion of a well annotated gene pool-representative Cannabis metabolome compiled from various plant tissues is predicted to improve small molecule and pathway analysis, thereby allowing comprehensive evaluation of metabolic variation underlying trait diversity. These resources will ultimately pave the way for contemporary multi-omics-based breeding strategies capable of facilitating metabolic engineering and genetic improvement of Cannabis for a variety of end-uses, therapeutic or industrial.
Experimental
Genetic Materials and Cultivation of Industrial Hemp Cannabis Germplasm
All research activities, including the procurement and cultivation of industrial hemp Cannabis germplasm, were carried out in accordance with Part IVA of the Drugs, Poisons and Controlled Substances Act 1981, and under an authorisation issued by Agriculture Victoria, Department of Jobs, Precincts and Regions (DJPR), Victorian State Government, Australia.
Seed from six industrial hemp Cannabis sativa L. accessions (varieties) were kindly provided gratis under permit from University and commercial sources within Australia (Table 1). Seeds were individually sown into seedling trays containing soil media at a depth of 1.5 cm and sprayed with reverse osmosis (RO) water daily until germination. Seedlings were transplanted into 400 mL pots and then transferred into 8 L pots at the emergence of the first (at ~1 week after sowing) and fourth leaflet pair (at ~3 weeks after sowing), respectively. Soil media consisted of one-part perlite, one-part peat moss, and one-part vermiculite as well as dolomite at a concentration of 1.1 g L−1. Plants were grown in a controlled environment room (CER) at 24°C with 55 % humidity. The photoperiod was initially set to a long-day cycle (18/6 h light/dark) to promote vegetative growth and plants were watered daily using RO water supplemented with CANNA Aqua Vega nutrient solution (40 mL CANNA Aqua Vega 1 and 40 mL CANNA Aqua Vega 2 in 10 L RO water). After 14 days, a shorter-day photoperiod (12/12 h light/dark) was maintained to initiate flowering and from this period, plants were watered daily with CANNA Classic Flores nutrient solution (40 mL Canna Flores A and 40 mL Canna Flores B in 10 L RO water) until harvest. Plants were harvested at maturation when ~95 % of the stigma on female flowers were browned and shrivelled or when ~95 % of male flowers had opened and expelled pollen. For each accession, floral tissue was taken from the apical (top 30 cm) and basal (bottom 30 cm) inflorescence and 1st, 2nd, and 3rd order foliage leaves were manually removed from floral samples before drying. All plants were destroyed during sample collection and were not used for further propagation.
Reagents and Standards
LC-MS grade methanol (Lichrosolv) used for extraction was purchased from Merck (Sigma Aldrich, NSW, Australia). Water with 0.1 % (v/v) formic acid (Optima LC-MS grade, mobile phase A), acetonitrile with 0.1 % (v/v) formic acid (Optima LCMS grade, mobile phase B), and Pierce FlexMix calibration solution were purchased from Thermo Fisher Scientific Inc. (Scoresby, Vic., Australia).
Cannabinoid certified reference standards in 1.0 mg mL−1 solution in ampoules were purchased from Novachem Pty Ltd (Heidelberg West, Vic., Australia) as distributor for Cerilliant Corporation (Round Rock, Texas, USA). Cannabidiol (CBD), Δ9-tetrahydrocannabinol (Δ9-THC), cannabinol (CBN), cannabichromene (CBC), cannabigerol (CBG), cannabidivarin (CBDV), and tetrahydrocannabivarin (THCV) were in methanol. Cannabidiolic acid (CBDA), Δ9-tetrahydrocannabinolic acid A (Δ9-THCAA), cannabinolic acid (CBNA), cannabichromenic acid (CBCA), cannabigerolic acid (CBGA), cannabidivarinic acid (CBDVA), and tetrahydrocannabivarinic acid (THCVA) were in acetonitrile. A mixed standard of CBD, Δ9-THC, CBN, CBC, CBDV, and THCV (Standard Set 1) was prepared at 200 µg mL−1 in methanol and diluted to prepare the working standards at 0.1, 0.5, 1, 5, 10, 25, 50, and 100 µg mL−1. A mixed standard of CBDA, Δ9-THCAA, CBNA, CBCA, CBDVA, and THCVA (Standard Set 2) was prepared at 150 µg mL−1 in acetonitrile and diluted to prepare the working standards at 0.1, 0.5, 1, 5, 25, 50, 75, and 100 µg mL−1. CBG (Standard Set 3) and CBGA (Standard Set 4) were prepared separately at 200 µg mL−1 in acetonitrile and methanol, respectively, and diluted to 0.1, 0.5, 1, 5, 25, 50, 75, and 100 µg mL−1. All standard solutions were stored at –80°C.
Sample Preparation
Male and female floral samples were dried in a Thermo Scientific Heratherm gravity convection oven at 40°C for 96 h. Seeds and stalks were manually separated using a coarse sieve and discarded. For male inflorescences, panicles comprising of staminate flowers and ≥ 4th order reduced leaves were retained, while for female inflorescences, phytomere tissues comprising of pistillate flowers, perigonal bracts, bracts, and ≥ 4th order reduced leaves remained. Inflorescence material was homogenised in a Geno/Grinder 2010 (Spex SamplePrep, Metuchen, NJ, USA) at 1500 rpm for 2 × 30 s intervals in 15 mL short polycarbonate vials with stainless steel grinding balls. Fifty milligrams (50 mg) of finely ground floral tissue was weighed into an Eppendorf Safe-Lock microcentrifuge tube and extracted in 5 mL of LCMS-grade methanol with vortex mixing (1 min) and sonication at room temperature for 20 min. The mixture was allowed to sit for a few minutes and then filtered through a 0.45 μm polytetrafluoroethylene (PTFE) syringe filter. Each sample extract was diluted 10 × with methanol. Aliquots of the filtrate were transferred to 2 mL amber HPLC vials and stored at –80°C before analysis. Three extraction replicates were performed on each apical and basal floral tissue sample, totalling six extraction (technical) replicates per accession.
Ultra-High-Performance Liquid Chromatography–High-Resolution Mass Spectrometry (UHPLC-HRMS) Analysis
UHPLC-HRMS separation, detection, and analysis of diluted and undiluted extracts was carried out on a Thermo Fisher Vanquish Flex UHPLC system with solvent degasser, quaternary pump, temperature-controlled sampler/auto injector (maintained at 5°C), temperature-regulated column compartment maintained at 30°C, and photodiode array detector (DAD) coupled to an Orbitrap ID-X Tribrid high resolution mass spectrometer (Thermo Fisher Scientific Inc., MA, USA). Separation was carried out on a Phenomenex Kinetex C18 column, 1.7 μm, 150 mm × 2.1 mm (Phenomenex Australia Pty Ltd, NSW, Australia). Mobile phases used were water with 0.1 % (v/v) formic acid (mobile phase A) and acetonitrile with 0.1 % (v/v) formic acid (mobile phase B) with 0.3 mL min−1 flow rate using the following gradient program: 10 % B, 0–2 min; 10 to 40 % B, 2–3 min; 40 % B, 3–5 min; 40 to 80 % B, 5–6 min; 80 % B, 6–9 min; 80 to 90 % B, 9–11 min; 90 to 100 % B, 11–12 min; 100 % B, 12–15 min; 100 to 10 % B, 15–16 min; and 10 % B from 16–20 min. Injection volume was 3 μL.
MS analysis was carried out using an electrospray ionisation (ESI) interface in the positive ion mode. Resolution was 120 000 in full scan mode and 15 000 in data dependent MS2 with HCD activation, with both MS and MS2 using the orbitrap as the mass analyser and a mass range of m/z 100 to 1000. Sheath and auxiliary gases were nitrogen at 50 and 10 arbitrary units (a.u.), respectively. ESI spray voltage was 3.5 kV, and the capillary and vaporiser temperatures were 325 and 350°C, respectively. Prior to data acquisition, the MS was calibrated with Pierce FlexMix calibration solution and the internal mass calibrant fluoranthene (Easy-IC) was activated during data acquisition for real-time mass calibration. Software for data acquisition was Xcalibur 4.4.
MS Data Processing and Statistical Analysis
Full MS data were processed using Trace Finder 4.1 (Thermo Fisher Scientific Inc., MA, USA) for quantitative analysis. Peak identity was determined by comparison of retention time with reference standards and integration of extracted ion peaks. The concentration of each analyte was determined by interpolation from standard calibration curves generated in MS Excel and results were expressed as average concentration ± standard deviation (s.d.).
The high-resolution accurate mass (HRAM) data were further processed using Thermo Scientific Compound Discoverer 3.1 software. The following parameters were used for aligning retention times: alignment model = adaptive curve; maximum shift [min] = 2; mass tolerance [ppm] = 5. The following parameters were used for compound detection: mass tolerance [ppm] = 5; intensity tolerance [%] = 30; S/N threshold = 3; min peak intensity = 50000; Ions = [M + H]+1, [M + H – H2O]+1; Base Ions = [M + H]+1, [M – H]−1; min element counts = C H; max. element counts = C100 H300 Br5 Cl10 K5 N20 Na5 O50 P10 S10. To eliminate drift and correct for batch effects, peak area was normalised linearly using the area under the peak of the quality control (QC) sample. QC samples were developed from pooled male and female floral tissue samples and these were measured every nine injections. QC-corrected chromatographic peak area data was used for statistical analysis. For principle component analysis (PCA), PC1 and PC2 data points were centred and scaled. Euclidean distance was the distance measure used for hierarchical cluster analysis (HCA). To detect differential metabolites between male and female floral tissues, volcano plot analysis using Student’s t-test was performed, with adjusted P-values calculated using the Benjamini–Hochberg correction for the false-discovery rate.
Compound Annotation
Small-molecule identification was performed using Thermo Scientific Compound Discoverer 3.1 software, with annotation of putative compounds achieved using consensus evaluation from calculated elemental composition and searches using HRAM MSn spectral library mzCloud and compound database ChemSpider along with fragment ion searching (FISh) to predict in silico fragmentation and ranking (mzLogic) algorithmic tools. For mzCloud searches, the following parameters were used: compound classes = all; precursor mass tolerance [ppm] = 10; FT fragment mass tolerance [ppm] = 10; IT fragment mass tolerance [Da] = 0.4; Library = auto processed, reference; post processing = recalibrated; max # results = 10; annotate matching fragments = true. DDA search parameters were as follows: identity search = cosine; match activation type = true; match activation energy = match with tolerance; activation energy tolerance = 20; apply intensity threshold = true; similarity search = confidence forward; match factor threshold = 60. ChemSpider searches included the following databases: Cambridge Structural Database; Carotenoids Database; DrugBank; KEGG; LipidMAPS; NIST Chemistry WebBook; Phenol-Explorer; PlantCyc; PubMed. Searches were conducted by Formula and Mass using the following parameters: mass tolerance [ppm] = 5 ppm; max. # of results per compound = 100; max. # of predicted compositions searched per compound = 3. mzLogic was applied with the following parameters: FT fragment mass tolerance [ppm] = 10; IT fragment mass tolerance [Da] = 0.4; max. # compounds = 0; max. # mzCloud similarity results to consider per compound = 10; match factor threshold = 30. FISh scoring settings used were high accuracy mass tolerance = 5 ppm; low accuracy mass tolerance = 20 ppm; S/N threshold = 3; max depth = 5.
Metabolic Pathways Enrichment Analysis
To interpret the biological significance associated with metabolite changes between female vs male floral tissues, Kyoto Encyclopedia of Genes and Genomes (KEGG, website: https://www.genome.jp/kegg/pathway.html) pathway overrepresentation (enrichment) analysis was performed on putative differential metabolites using the Metabolites Biological Role (MBRole) server.[69] KEGG compound IDs which served as input for enrichment analysis were retrieved from Compound Discoverer software (v 3.11.12). At the time of analysis, neither a metabolome nor a reference set of compounds for Cannabis sativa was available and so the background set was based on all compounds in the database associated with KEGG pathway annotations. Enrichment p-values were adjusted for multiple testing using the Benjamini–Hochberg correction for the false-discovery rate.[70]
Supplementary Material
The list of putative compounds identified from male and female flowers, KEGG enrichment of differentially regulated metabolic pathways, and principal component analysis loadings and variances plots of metabolite abundances, as well as MS2 spectral data of HU-331 and unknown compounds are available on the Journal’s website. Table S1: List of 2599 putative compounds identified from male and female flowers; Table S2: KEGG enrichment of differentially regulated metabolic pathways in female v. male floral tissues; Table S3: Consensus annotation of 41 DAMs; Fig. S1: Principal component analysis loadings and variances plots of metabolite abundances from male and female floral tissue samples; Appendix S1: MS2 spectral data of 11 compounds that were initially annotated as HU-331; and Appendix S2: MS2 spectral data of HU-331.
Conflicts of Interest
The authors declare no conflicts of interest.
Declaration of Funding
MSD, AB, and MTW acknowledge the support of the Australian Research Council (ARC) Linkage Program grant (LP160101317). AB, MSD, and MAD acknowledge the support of the ARC Research Hub for Medicinal Agriculture (IH180100006). Cann Group Limited are a partner organisation of the ARC linkage program grant LP160101317 and the ARC Research Hub for Medicinal Agriculture (IH180100006). The authors also acknowledge generous resource allocations from an Ian Potter Foundation grant (#31110299) and the ARC LIEF scheme grant (LE200100117) towards the purchase of the mass spectrometers. AB and MSD were supported by La Trobe University through the La Trobe Institute for Agriculture and Food (LIAF).
References
[1] J. M. McPartland, W. Hegman, T. Long, Veg. Hist. Archaeobot. 2019, 28, 691.| Crossref | GoogleScholarGoogle Scholar |
[2] E. Small, Bot. Rev. 2015, 81, 306.
| Crossref | GoogleScholarGoogle Scholar |
[3] E. Small, A. Cronquist, Taxon 1976, 25, 405.
| Crossref | GoogleScholarGoogle Scholar |
[4] R. H. W. Bradshaw, P. Coxon, J. R. A. Greig, A. R. Hall, New Phytol. 1981, 89, 503.
| Crossref | GoogleScholarGoogle Scholar |
[5] I. J. Flores-Sanchez, R. Verpoorte, Phytochem. Rev. 2008, 7, 615.
| Crossref | GoogleScholarGoogle Scholar |
[6] L. O. Hanuš, S. M. Meyer, E. Muñoz, O. Taglialatela-Scafati, G. Appendino, Nat. Prod. Rep. 2016, 33, 1357.
| Crossref | GoogleScholarGoogle Scholar | 27722705PubMed |
[7] E. D. French, Neurosci. Lett. 1997, 226, 159.
| Crossref | GoogleScholarGoogle Scholar | 9175591PubMed |
[8] O. Devinsky, J. H. Cross, L. Laux, E. Marsh, I. Miller, R. Nabbout, et al. N. Engl. J. Med. 2017, 376, 2011.
| Crossref | GoogleScholarGoogle Scholar | 28538134PubMed |
[9] M. Heblinski, M. Santiago, C. Fletcher, J. Stuart, M. Connor, I. S. McGregor, et al. Cannabis Cannabinoid Res. 2020, 5, 305.
| Crossref | GoogleScholarGoogle Scholar | 33376801PubMed |
[10] C. Staginnus, S. Zörntlein, E. de Meijer, J. Forensic Sci. 2014, 59, 919.
| Crossref | GoogleScholarGoogle Scholar | 24579739PubMed |
[11] M. T. Welling, L. Liu, C. A. Raymond, T. Kretzschmar, O. Ansari, G. J. King, Sci. Rep. 2019, 9, 11421.
| Crossref | GoogleScholarGoogle Scholar | 31388099PubMed |
[12] S. J. Livingston, T. D. Quilichini, J. K. Booth, D. C. Wong, K. H. Rensing, J. Laflamme‐Yonkman, et al. Plant J. 2020, 101, 37.
| Crossref | GoogleScholarGoogle Scholar | 31469934PubMed |
[13] N. Happyana, S. Agnolet, R. Muntendam, A. Van Dam, B. Schneider, O. Kayser, Phytochemistry 2013, 87, 51.
| Crossref | GoogleScholarGoogle Scholar | 23280038PubMed |
[14] B. Spitzer-Rimon, S. Duchin, N. Bernstein, R. Kamenetsky, Front. Plant Sci. 2019, 10, 350.
| Crossref | GoogleScholarGoogle Scholar | 31001293PubMed |
[15] V. C. Moliterni, L. Cattivelli, P. Ranalli, G. Mandolino, Euphytica 2004, 140, 95.
| Crossref | GoogleScholarGoogle Scholar |
[16] Z. K. Punja, J. E. Holmes, Front. Plant Sci. 2020, 11, 718.
| Crossref | GoogleScholarGoogle Scholar | 32670310PubMed |
[17] J. Petit, E. M. Salentijn, M.-J. Paulo, C. Thouminot, B. J. van Dinter, G. Magagnini, et al. Front. Plant Sci. 2020, 11, 102.
| Crossref | GoogleScholarGoogle Scholar | 32153610PubMed |
[18] M. G. Divashuk, O. S. Alexandrov, O. V. Razumova, I. V. Kirov, G. I. Karlov, PLoS One 2014, 9, e85118.
| Crossref | GoogleScholarGoogle Scholar | 24465491PubMed |
[19] J. D. Lubell, M. H. Brand, Horttechnology 2018, 28, 743.
| Crossref | GoogleScholarGoogle Scholar |
[20] H. M. Ram, V. S. Jaiswal, Planta 1972, 105, 263.
| Crossref | GoogleScholarGoogle Scholar |
[21] N. Sriram, H. M. Ram, Curr. Sci. 1984, 53, 735.
[22] G. Mandolino, A. Carboni, Euphytica 2004, 140, 107.
| Crossref | GoogleScholarGoogle Scholar |
[23] E. Small, S. G. Naraine, Genet. Resour. Crop Evol. 2016, 63, 339.
| Crossref | GoogleScholarGoogle Scholar |
[24] A.-M. Faux, X. Draye, R. Lambert, R. d’Andrimont, P. Raulier, P. Bertin, Eur. J. Agron. 2013, 47, 11.
| Crossref | GoogleScholarGoogle Scholar |
[25] D. U. Nagy, K. Cianfaglione, F. Maggi, S. Sut, S. Dall’Acqua, Chem. Biodivers. 2019, 16, e1800562.
| Crossref | GoogleScholarGoogle Scholar | 30548994PubMed |
[26] A. Ohlsson, C. Abou-Chaar, S. Agurell, I. Nilsson, K. Olofsson, F. Sandberg, Bull. Narc. 1971, 23, 29.
[27] R. Latta, B. Eaton, Econ. Bot. 1975, 29, 153.
| Crossref | GoogleScholarGoogle Scholar |
[28] H. Kushima, Y. Shoyama, I. Nishioka, Chem. Pharm. Bull. 1980, 28, 594.
| Crossref | GoogleScholarGoogle Scholar |
[29] A. Cerrato, G. Cannazza, A. L. Capriotti, C. Citti, G. La Barbera, A. Laganà, et al. Talanta 2020, 209, 120573.
| Crossref | GoogleScholarGoogle Scholar | 31892008PubMed |
[30] B. Hurgobin, M. Tamiru‐Oli, M. T. Welling, M. S. Doblin, A. Bacic, J. Whelan, et al. New Phytol. 2021, 230, 73.
| Crossref | GoogleScholarGoogle Scholar | 33283274PubMed |
[31] G. Nyamundanda, I. C. Gormley, Y. Fan, W. M. Gallagher, L. Brennan, BMC Bioinformatics 2013, 14, 338.
| Crossref | GoogleScholarGoogle Scholar | 24261687PubMed |
[32] D. Dudzik, C. Barbas-Bernardos, A. García, C. Barbas, J. Pharm. Biomed. Anal. 2018, 147, 149.
| Crossref | GoogleScholarGoogle Scholar | 28823764PubMed |
[33] B. Worley, R. Powers, Curr. Metabolomics 2013, 1, 92.
| 26078916PubMed |
[34] C. Citti, U. M. Battisti, D. Braghiroli, G. Ciccarella, M. Schmid, M. A. Vandelli, et al. Phytochem. Anal. 2018, 29, 144.
| Crossref | GoogleScholarGoogle Scholar | 28915313PubMed |
[35] L. W. Sumner, A. Amberg, D. Barrett, M. H. Beale, R. Beger, C. A. Daykin, et al. Metabolomics 2007, 3, 211.
| Crossref | GoogleScholarGoogle Scholar | 24039616PubMed |
[36] P. Dayanandan, P. B. Kaufman, Am. J. Bot. 1976, 63, 578.
| Crossref | GoogleScholarGoogle Scholar |
[37] E. P. M. de Meijer, K. M. Hammond, Euphytica 2016, 210, 291.
| Crossref | GoogleScholarGoogle Scholar |
[38] B. Ylstra, A. Touraev, R. M. B. Moreno, E. Stöger, A. J. Van Tunen, O. Vicente, et al. Plant Physiol. 1992, 100, 902.
| Crossref | GoogleScholarGoogle Scholar | 16653074PubMed |
[39] L. Taylor, R. Jorgensen, J. Hered. 1992, 83, 11.
| Crossref | GoogleScholarGoogle Scholar |
[40] E. G. Schijlen, C. R. de Vos, S. Martens, H. H. Jonker, F. M. Rosin, J. W. Molthoff, et al. Plant Physiol. 2007, 144, 1520.
| Crossref | GoogleScholarGoogle Scholar | 17478633PubMed |
[41] X. Ding, X. Wang, Q. Li, L. Yu, Q. Song, J. Gai, et al. Int. J. Mol. Sci. 2019, 20, 2869.
| Crossref | GoogleScholarGoogle Scholar |
[42] A. P. Brown, J. T. Kroon, D. Swarbreck, M. Febrer, T. R. Larson, I. A. Graham, et al. PLoS One 2012, 7, e30100.
| Crossref | GoogleScholarGoogle Scholar | 23284689PubMed |
[43] N. E. Rothnie, M. V. Palmer, D. G. Burke, J. P. Sang, E. P. Hilliard, P. A. Salisbury, et al. Phytochemistry 1987, 26, 1895.
| Crossref | GoogleScholarGoogle Scholar |
[44] F. Opute, Phytochemistry 1975, 14, 1023.
| Crossref | GoogleScholarGoogle Scholar |
[45] A. Dave, I. A. Graham, Front. Plant Sci. 2012, 3, 42.
| Crossref | GoogleScholarGoogle Scholar | 22645585PubMed |
[46] N. Taki, Y. Sasaki-Sekimoto, T. Obayashi, A. Kikuta, K. Kobayashi, T. Ainai, et al. Plant Physiol. 2005, 139, 1268.
| Crossref | GoogleScholarGoogle Scholar | 16258017PubMed |
[47] A. Moreno-Rodríguez, R. Santos-Castro, J. Vázquez-Medrano, R. E. Quintanar-Zúñiga, F. A. García-García, L. B. Hernández-Portilla, et al. Nat. Prod. Res. 2020, 34, 1942.
| Crossref | GoogleScholarGoogle Scholar | 30724587PubMed |
[48] H. Enomoto, T. Sensu, K. Sato, F. Sato, T. Paxton, E. Yumoto, et al. Sci. Rep. 2017, 7, 42977.
| Crossref | GoogleScholarGoogle Scholar | 28211480PubMed |
[49] S. Goetz, A. Hellwege, I. Stenzel, C. Kutter, V. Hauptmann, S. Forner, et al. Plant Physiol. 2012, 158, 1715.
| Crossref | GoogleScholarGoogle Scholar | 22337921PubMed |
[50] K. Wang, Q. Guo, J. E. Froehlich, H. L. Hersh, A. Zienkiewicz, G. A. Howe, et al. Plant Cell 2018, 30, 1006.
| Crossref | GoogleScholarGoogle Scholar | 29666162PubMed |
[51] E. Galoch, Acta Soc. Bot. Pol. 1978, 47, 153.
| Crossref | GoogleScholarGoogle Scholar |
[52] M. Hofinger, Arch. Int. Physiol. Biochim. 1969, 77, 225.
| Crossref | GoogleScholarGoogle Scholar | 4184292PubMed |
[53] F. Salari, H. Mansori, Journal of Plant Process and Function 2013, 1, 51.
[54] H. Mansouri, Z. Asrar, J. Szopa, Plant Growth Regul. 2009, 58, 269.
| Crossref | GoogleScholarGoogle Scholar |
[55] A. M. Adal, K. Doshi, L. Holbrook, S. S. Mahmoud, Planta 2021, 253, 17.
| Crossref | GoogleScholarGoogle Scholar | 33392743PubMed |
[56] R. Mechoulam, Z. Ben-Zvi, Y. Gaoni, Tetrahedron 1968, 24, 5615.
| Crossref | GoogleScholarGoogle Scholar | 5732891PubMed |
[57] N. M. Kogan, R. Rabinowitz, P. Levi, D. Gibson, P. Sandor, M. Schlesinger, et al. J. Med. Chem. 2004, 47, 3800.
| Crossref | GoogleScholarGoogle Scholar | 15239658PubMed |
[58] J. Díaz-Alonso, J. Paraíso-Luna, C. Navarrete, C. Del Río, I. Cantarero, B. Palomares, et al. Sci. Rep. 2016, 6, 29789.
| Crossref | GoogleScholarGoogle Scholar | 27430371PubMed |
[59] K. M. Regal, S. L. Mercer, J. E. Deweese, Chem. Res. Toxicol. 2014, 27, 2044.
| Crossref | GoogleScholarGoogle Scholar | 25409338PubMed |
[60] J. T. Wilson, C. A. Fief, K. D. Jackson, S. L. Mercer, J. E. Deweese, Chem. Res. Toxicol. 2018, 31, 137.
| Crossref | GoogleScholarGoogle Scholar | 29272108PubMed |
[61] R. Mechoulam, L. Hanuš, Chem. Phys. Lipids 2002, 121, 35.
| Crossref | GoogleScholarGoogle Scholar | 12505688PubMed |
[62] L. M. Bornheim, M. P. Grillo, Chem. Res. Toxicol. 1998, 11, 1209.
| Crossref | GoogleScholarGoogle Scholar | 9778318PubMed |
[63] E. Palazzo, L. Luongo, F. Guida, V. de Novellis, S. Boccella, C. Cristiano, et al. Neuropsychiatry 2019, 9, 2035.
[64] S. Petrosino, V. Di Marzo, Br. J. Pharmacol. 2017, 174, 1349.
| Crossref | GoogleScholarGoogle Scholar | 27539936PubMed |
[65] O. Aizpurua-Olaizola, U. Soydaner, E. Öztürk, D. Schibano, Y. Simsir, P. Navarro, et al. J. Nat. Prod. 2016, 79, 324.
| Crossref | GoogleScholarGoogle Scholar | 26836472PubMed |
[66] B. De Backer, K. Maebe, A. G. Verstraete, C. Charlier, J. Forensic Sci. 2012, 57, 918.
| Crossref | GoogleScholarGoogle Scholar | 22390363PubMed |
[67] S. Chandra, H. Lata, M. A. ElSohly, L. A. Walker, D. Potter, Epilepsy Behav. 2017, 70, 302.
| Crossref | GoogleScholarGoogle Scholar | 28202406PubMed |
[68] C. Citti, F. Russo, S. Sgrò, A. Gallo, A. Zanotto, F. Forni, et al. Anal. Bioanal. Chem. 2020, 412, 4009.
| Crossref | GoogleScholarGoogle Scholar | 32285185PubMed |
[69] J. López-Ibáñez, F. Pazos, M. Chagoyen, Nucleic Acids Res. 2016, 44, W201.
| Crossref | GoogleScholarGoogle Scholar | 27084944PubMed |
[70] Y. Benjamini, Y. Hochberg, J. R. Stat. Soc. B 1995, 57, 289.
| Crossref | GoogleScholarGoogle Scholar |




