Enhancing gut health and performance of grower-finisher pigs: assessing the efficacy of the mixture of microencapsulated essential oils and saponins
Sung J. Yu A , Andrew Morris B , Advait Kayal
A , Andrew Morris B , Advait Kayal  A , Thi Thu Hao Van
A , Thi Thu Hao Van  C , Dragana Stanley
C , Dragana Stanley  A and Yadav S. Bajagai
A and Yadav S. Bajagai  A *
A *
A
B
C
Abstract
The livestock industry demands alternative approaches to maintain animal health and productivity without using antimicrobials. Phytogenic feed additives, consisting of plant-derived compounds, offer promising benefits to grower-finisher pigs’ gut microbiota and overall health.
This study investigated the impact of essential oils and saponins as phytogenic feed additives on the gut microbiota and productivity of grower-finisher pigs.
Four hundred male Large White × Landrace pigs were randomly allocated into control and treatment groups and provided with a basal diet. The treatment group supplemented commercial phytogenic feed additives. Performance data, general health, cumulative mortality, and bodyweight were monitored. Intestinal samples from different gut locations and rectal swab samples were collected for microbiota analysis.
No significant difference in weight was observed between the two groups; however, the phytogenic feed additives notably affected gut microbiota. Greater microbial diversity and changes in genera abundance were evident. Campylobacter and Clostridium increased, while beneficial Lactobacillus increased, and Escherichia decreased. The supplemented group demonstrated more consistent growth, with higher minimum weights.
Phytogenic feed additives have a significant impact on the gut microbiota of grower-finisher pigs. A diverse microbial community and specific changes in genera abundance were observed, indicating the potential role of these additives in modulating gut health.
This study emphasises the potential advantages of using phytogenic feed additives to improve the health and productivity of livestock without the use of antibiotics.
Keywords: essential oils, grower-finisher pigs, gut microbiota, natural antimicrobial product, phytogenic product, post-weaning diarrhoea, saponins, swine, rosemary, thyme.
Introduction
Antimicrobial resistance (AMR) is acknowledged for its severe public health impacts, with growing attention worldwide. Before the crisis of AMR, antibiotics were used to treat many common bacterial infections and have improved the safety of numerous medical procedures for both humans and animals. The human data based on 471 million individuals recorded 4.95 million deaths related to drug-resistant bacterial infections and 1.27 million deaths associated with AMR (The Lancet 2022). In the USA, two million AMR-related cases were reported in humans in 2016, costing USD20 billion (O’Neill 2016). Humans and livestock share commensal and pathogenic bacteria, often using the same classes of antibiotics in veterinary and human medicine, contributing to an increase in AMR.
Antibiotics have been widely used in commercial livestock, mainly as growth promoters and for prophylactic purposes rather than to treat disease. Different classes of antibiotics change the microbial community in the gut, influencing the growth performance through alteration in the gut microbiota (Crisol-Martinez et al. 2017). However, the emergence of antibiotic resistance has jeopardised animal welfare, health, and productivity (Bengtsson and Wierup 2006). After the ban or voluntary discontinuation of antibiotics, swine industries have faced an increase in disease outbreaks, leading to reduced production performance. Livestock industries have promoted research in alternatives to antibiotics, such as prebiotics, probiotics, highly fermentable carbohydrate cereal grains, microbial transplants, and phytogenic products, to alleviate the economic loss and animal welfare.
Phytogenic feed additives (PFA) are the most successful and widely used antibiotic alternatives (Karaskova et al. 2015; Bartos et al. 2016). They are mostly comprised of an optimised combination of plant bioactive compounds established to reduce oxidative stress and improve gut health and growth and stimulate anti-inflammatory processes (Abdelli et al. 2021; Shehata et al. 2022; Yu et al. 2022), and control pathogens.
In commercial piggery, the piglets are weaned early before establishing a stable microbial population and immune systems (Konstantinov et al. 2006). This increases the susceptibility to bacterial post-weaning diarrhoea, resulting in substantial morbidity and mortality (Lalles et al. 2004). Moreover, the weaning process triggers rapid alterations in diet presentation and composition, as well as significant social and behavioural modifications, which affect gut fill, ingesta composition, and subsequent microbial shifts (Lalles et al. 2004; Shen et al. 2012; Kim et al. 2019).
The gut microbes play an important role in sustaining the immunological, nutritional, and physiological functions of the pig (Lee and Mazmanian 2010). Intestinal microbiota encodes most enzymes needed to disintegrate dietary fibre from feed because the pigs do not have the ability to produce such enzymes (Verschuren et al. 2018). The host microbiome is one of the major determinants of regulating physiology and maintaining the balance between health and disease (Bhat et al. 2022). Therefore, establishing robust microbiota in early life is imperative for pigs’ healthy growth (Kabat et al. 2014; Chen et al. 2018).
Rosemary extracts are well known for their biological and pharmacological activities as additives in foods and medicine. In livestock industries, supplementing feeds with rosemary extracts increases bodyweight gain in the broiler chickens (Mathlouthi et al. 2012) and presents antimicrobial activity against Streptococcus-associated disease in pigs (de Aguiar et al. 2018).
The essential oil extracted from thyme contains volatile components thymol and carvacrol, commonly used in cosmetics, food, and medicinal purposes (Ghasemi et al. 2020). Thyme has an antibacterial effect on Gram-negative and Gram-positive bacteria and possesses antifungal, antioxidant, anti-inflammatory, and antiviral activity (Sidiropoulou et al. 2022). In intensive piggery, thymol and carvacrol reduce waste and odour emissions (Varel 2002). For this reason, thymol has been a key ingredient of PFA.
Saponins are naturally occurring compounds found in a variety of fruits and plants as surface-active glycosides of triterpenes and possess biological and pharmacological activities. Dang and Kim (2020) supplemented saponins as feed additives and reported improved growth performance, nutrient digestibility, meat quality, and faecal gas emissions in finishing pigs.
The focus of this study was to explore how a PFA containing rosemary, thyme, and saponins affects the gut microbial composition in grower-finisher pigs. By comparing gut microbiota profiles between pigs supplemented with PFA and a control group, we aimed to identify key changes in microbial diversity and structure that could influence pigs’ overall health.
Materials and methods
Animal experiment
The experiment was conducted in the specifically constructed pigsty to study the impact on the productivity of the grower to finisher pigs with phytogenic supplements. In total, 400 pigs (Large White × Landrace, males, 16–20 kg, 8 weeks of age) were randomised and separated into two groups (control and treatment). Each group consisted of 200 pigs, and 50 pigs from both groups were randomly selected and tagged for identification throughout the production process. Ten of these identified pigs were then chosen to follow through the production cycle. Animals were provided with ad libitum access to water and feed. Basal diets were composed of barley, wheat, sorghum and other grains as energy sources and soya meal with meat and bone meal (MBM) as protein sources, which were designed by the company nutritionists to meet the production and animal welfare requirements. A standard trace minerals and vitamin premix, organic acids and prebiotics and probiotics were added to the feed to cover the vitamin and mineral requirements.
Treatment groups were supplemented with commercial PFA (Aromex Pro, Delacon Biotechnik GmbH) containing a blend of micro-encapsulated essential oils of rosemary and thyme, and saponins, to the basal diet at the recommended dosage of 200 g/1000 kg. The phytogenic supplements were administered to the animals 7 days after their arrival and supplementation continued for 9 weeks, from 8 to 17 weeks of age.
All animals were supplemented with amoxycillin and a blend of organic acids (Selko-pH, AusPac Ingredients, Tamworth, NSW, Australia) through water for the first 2 weeks on arrival to the farm, and with oxytetracycline and Selko-pH through water thenafter.
The study monitored various performance data, including general health, cumulative mortality, and bodyweight of the grower-finisher pigs. Individual body weights of 50 pigs from each group were recorded, and a total of 150 rectal swab samples were collected using a sterile cotton-swab applicator three times during the trial period, at the ages of 8, 14 and 17 weeks. The samples were kept on dry ice during collection and transportation and stored at −80°C until DNA extraction. Week 17 samples were obtained 1 day prior to the transportation of the animals to the abattoir.
Intestinal content samples for preliminary insights on microbiota profile in different gut sections were collected from the duodenum, jejunum, ileum, caecum and colon in the abattoir from 10 randomly selected animals in each group.
DNA extraction, 16S rRNA gene sequencing and data analysis
The DNA from intestinal-content and rectal-swab samples was extracted as previously described (Bauer et al. 2019). Briefly, the cells were lysed as per the improved lysis protocol specifically developed for digesta and faecal samples (Yu and Morrison 2004) and purified using the DNA mini spin column kits (Enzymax LLC, CAT# EZC101, Lexington, Kentucky, USA). Following the extraction, the quantity and quality of the DNA were measured with a NanoDrop One UV-Vis spectrophotometer (ThermoFisher Scientific, Wilmington, USA).
The V3–V4 region of the 16S rRNA gene was amplified using the dual index primer pairs (forward primer pro341F (5′-CCTACGGGNBGCASCAG-3′) and reverse primer pro805R (5′-GACTACNVGGGTATCTAATCC-3′) with index, heterogeneity spacer and Illumina sequencing linkers (Fadrosh et al. 2014). An AMPure XP Kit (Beckman Coulter, Brea, CA, USA) was used to purify the 16S amplicon library and it was sequenced with the Illumina Miseq platform (2 × 300 bp paired-end). A minimum Phred score of 25 across the length of 200 nucleotides of the forward read was used for downstream analysis.
The raw DNA sequences were demultiplexed using Cutadapt (Martin 2011), followed by analysis using quantitative insights into microbial ecology 2 (QIIME2) for microbiota analysis (Bolyen et al. 2019). Dada2 (Callahan et al. 2016) plugin with all recommended parameters was used for quality filtering, denoising, and chimera removal. SILVA v 138.1 database (Quast et al. 2013) was used as reference data to assign taxonomy. The data were analysed at amplicon sequence variant (ASV) and genus levels. The interpretation and analysis of the data were completed with a minimum of 3000 rarefied sequences per sample. For further downstream analysis and visualisation, R packages Phyloseq (McMurdie and Holmes 2013), Phylosmith (Smith 2019), Vegan (Dixon 2003) and Microeco (Liu et al. 2021) were used. Permutation multivariate ANOVA (PERMANOVA) and a paired multivariate ANOVA (Paired MANOVA) were completed using Primer-e v7 (Anderson et al. 2008).
Ethics approval
The Animal Ethics Committee of Central Queensland University approved the study under Approval number 0000023290. All animal manipulations were performed according to the Australian Code for the Care and Use of Animals for Scientific Purposes and reported in proportion to guidelines and regulations of Animal Research: Reporting of In Vivo Experiments (ARRIVE).
Results
Performance of animals
No significant improvements in weight gain were observed in the pigs that received phytogen (Phy) compared with the control group, and the mortality rate was similar between the two groups (Table 1, Table S1, Fig. 1).
Overall microbiota composition
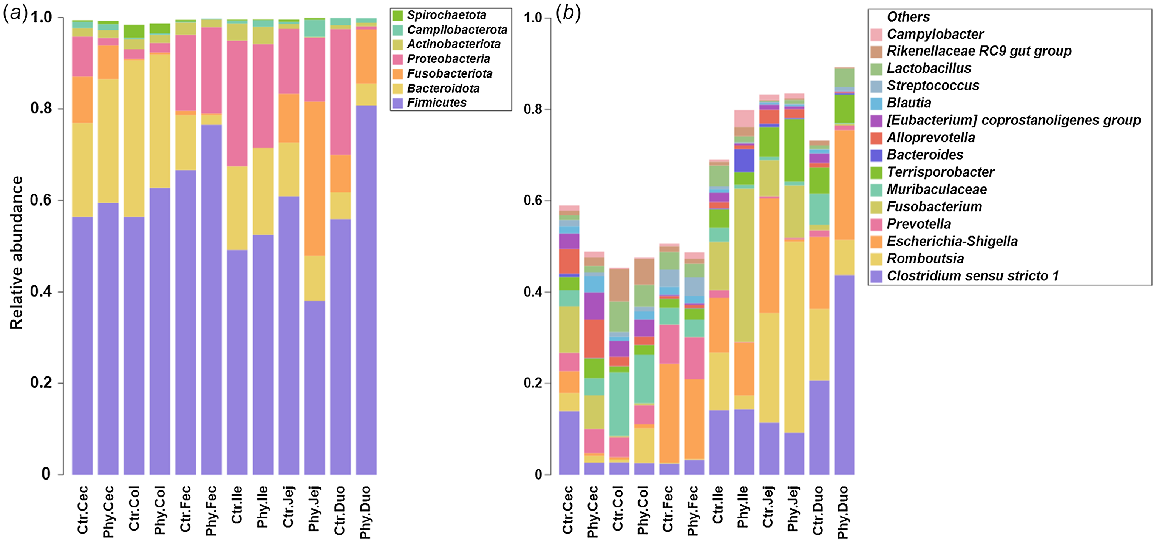
The microbiota composition at the phylum level in various gut regions, such as caecal, colonic, jejunal, duodenal, and ileal, and in rectal swab (faecal) is shown in the stacked bar chart (Fig. 2a). At phylum level, the most abundant phyla discovered in both groups were Firmicutes, Bacteroidota, Fusobacteriota, and Proteobacteria, with lesser amounts of Actinobacteriota, Spirochaetoata and Campilobacterota. Fig. 2b presents the most abundant genera in the gut for the two groups observed in the duodenum, jejunum, ileum, caecum and colon, and in rectal swabs (faecal). In the caecum of Phy-treated animals, the genera Clostridium sensu stricto 1 and Escherichia–Shigella showed a noticeable reduction. In the colon, the genus Romboutsia was enriched in the Phy-treated group compared with the control, while the majority of rectal-swab samples did not show apparent variation in the microbial population.
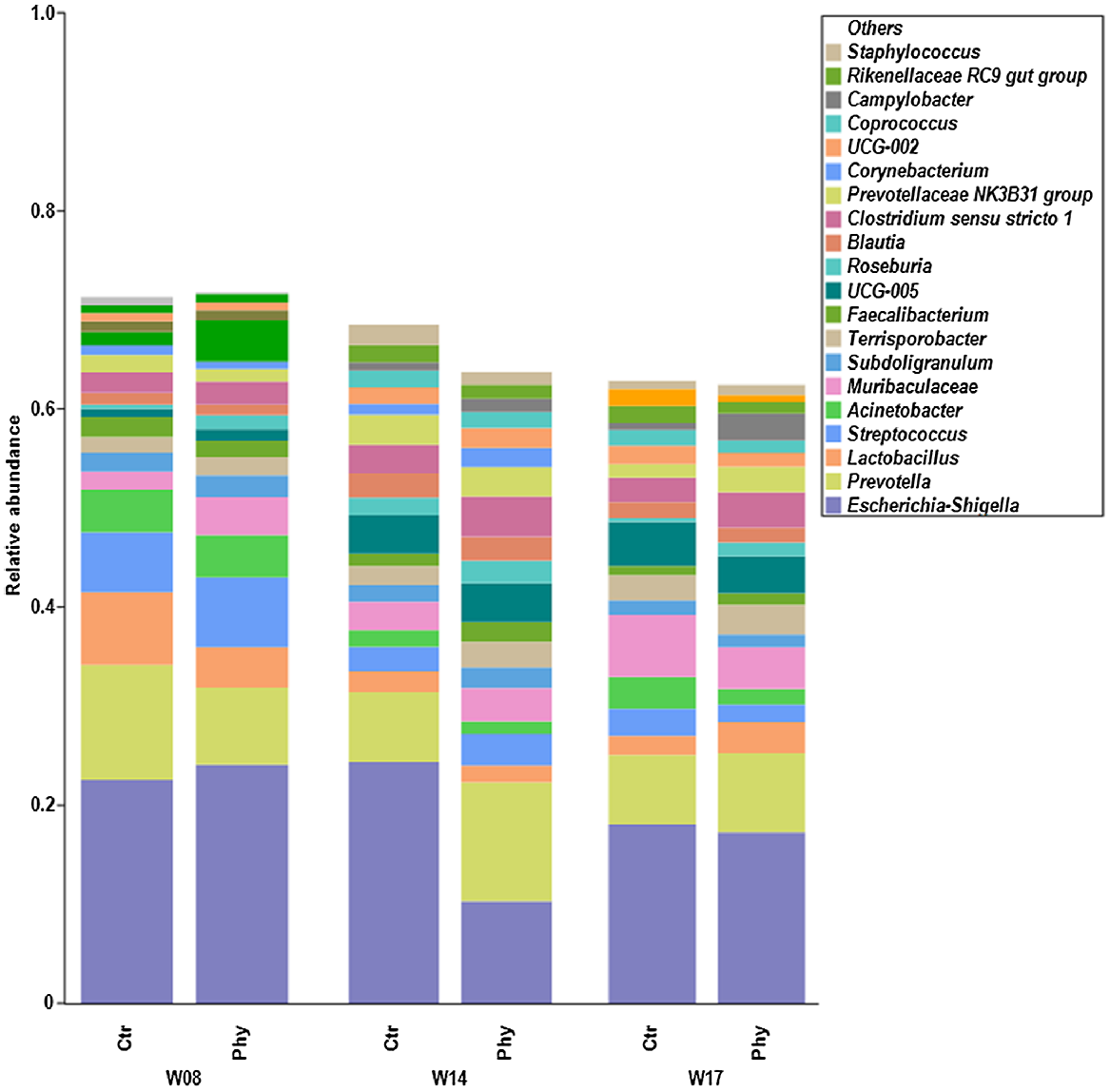
Fig. 3 displays stacked bar charts of the top 20 genera in rectal swabs, sampled at Weeks 8, 14, and 17. The most common genera found were Escherichia–Shigella, Prevotella, Lactobacillus, Streptococcus, Acinetobacter, Muribaculaceae, Subdoligranulum, Terrisporobacter, Faecalibacterium, UCG-005, Roseburia, Blautia, Clostridium sensu stricto 1, Prevotellaceae NK3B31 group, Corynebacterium, UCG-002, Coprococcus, Campylobacter, Rikenellaceae RC9 gut group, and Staphylococcus. At Week 14, there was a significant shift in the microbial community compared with Weeks 8 and 17, with a decreased presence of Escherichia–Shigella and an increased presence of Prevotella in the Phy-treated pigs.
Relative abundance at the genus level, showing the top 20 genera across different time frames (Week 8, W08; Week 14, W14; Week 17, W17) between control (Ctr) and treatment (Phy).
The relative abundance bar chart was generated as in Supplementary Fig. S1 to demonstrate the microbial composition of individual pigs at Week 17. The analysis of rectal swabs from each group displayed that the microbial populations varied among the individual animals.
Alpha and beta diversity
To determine whether the treatment significantly affected the gut microbiota, we used both weighted and unweighted UniFrac distances at ASV level to conduct a PERMANOVA for the main variables and a paired MANOVA for paired comparisons by using Primer-e v7 (Table 2). The analysis used the following three factors: the age of the pigs (measured in weeks), the type of treatment they received (either a control group or the Phy treatment), and the origin of the sample (rectum, colon, caecum, jejunum, ileum, or duodenum). The paired MANOVA and PERMANOVA were performed to assess the impact of these factors on the gut microbiome.
| Age (weeks) | Measure (UniFrac) | P-value | Significance | |
|---|---|---|---|---|
| 8 | Weighted | 0.171 | ||
| Unweighted | 0.02 | * | ||
| 14 | Weighted | 0.001 | *** | |
| Unweighted | 0.001 | *** | ||
| 17 | Weighted | 0.313 | ||
| Unweighted | 0.004 | ** |
*** P < 0.001; ** P < 0.01; * P < 0.05.
The results of the PERMANOVA indicated that both age and origin had a significant (P < 0.001) impact on the gut microbiota, as determined by both weighted and unweighted UniFrac distances (Table S2). The treatment variable was found to significantly influence the microbiota when analysed with the unweighted UniFrac distance (P < 0.01) and Bray–Curtis distance at the genus level (P < 0.05).
The paired-comparison analysis using both weighted and unweighted UniFrac distances between treatment and age showed significant (P < 0.001 and P < 0.01 respectively) differences at Week 14 (Table S3). Furthermore, treatment and origin paired comparisons as shown in Table S4 determined that the differences between the control and Phy groups were most pronounced in the rectal samples when analysed using the unweighted UniFrac distance matrix (P < 0.01).
Microbial community composition in faecal samples was examined using the UniFrac distance metric to assess differences at different ages (Table 2). At Weeks 8 and 17, only the unweighted UniFrac distances showed significant differences, suggesting variations in the presence or absence of specific bacterial species between the two groups. However, weighted distances did not show significant differences at these time points. In contrast, at Week 14, both weighted and unweighted UniFrac distances exhibited significant (P < 0.001) differences, indicating distinct microbial community composition between the control and treatment groups.
The graph that displays the differences in the microbial community structure of different gut regions was created by using the principal coordinate analysis (PCoA) based on both the weighted and unweighted UniFrac distances (Fig. 4). The results from the PCoA plot demonstrated that microbiota from rectal swabs were most distinct relative to other gut regions, such as caecum (Cec), colon (Col), duodenum (Duo), ileum (Ile), and jejunum (Jej). Among intestinal regions, microbiota in the large intestine (caecum and colon) were relatively different from those of the small intestine (duodenum, jejunum and ileum), because samples from large intestine clustered together, whereas samples from the small intestine clustered together away from the large intestine.
The alpha diversity of the microbiota in the rectal-swab samples was evaluated to compare the diversity of microbes in the Ctr and Phy groups during the study period. Shannon and Simpson indices showed significant differences between the Ctr and Phy groups in Weeks 8 and 17. The analysis showed that the microbial community in the Phy-treated pigs was more diverse, as demonstrated by the higher values of the Simpson and Shannon entropy index, than that in the untreated group, as shown in Fig. 5a. However, there was no difference in the diversity in Week 14.
(a) Characterisation of alpha diversity and (b) beta diversity in cloacal swabs before and after treating with Phy in pigs. Mann–Whitney test was used to calculate the significance. (*P < 0.05; **P < 0.01; ns, not significant).
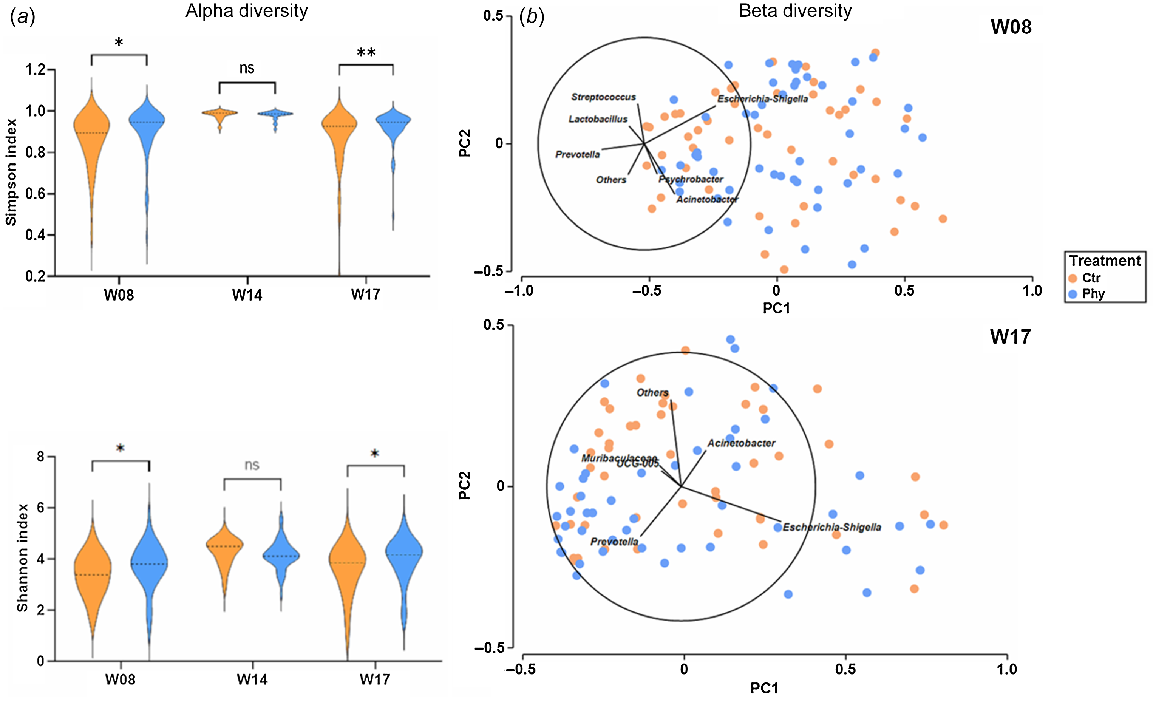
To illustrate the alterations in the composition of the microbial community in the cloacal swabs, a principal-components analysis (PCA) was performed utilising Primer-7e, as presented in Fig. 5b. The Bray–Curtis dissimilarity matrix was employed to quantify the difference between the groups. The loading vectors indicated the direction and magnitude of the contribution of some important variables (microbes) to the variance explained by each principal component.
In Week 8, both the Phy-treated and the untreated groups had a similar distribution, with the highest contribution of Escherichia–Shigella to the variance captured by the principal components in one direction and Acinetobacter in another direction. Other important genera contributing to variance are Psychrobacter, Prevotella, Lactobacillus and Streptococcus. However, at Week 17, Escherichia–Shigella had the highest contribution to the variance in one direction and Prevotella in another direction. Acenetobacter, Muribaculaceae and UCG-005 are other major taxa contributing to the variance defined by the principal components.
Taxonomic alterations
The linear discriminant analysis effect size (LEfSe) analysis, a widely used biomarker discovery tool for genomic studies, was utilised to assess the statistical significance of differences among the microbial communities at Week 17 (Fig. 6). The threshold for linear discriminant analysis (LDA) score was set at 3.3. The LDA score higher than 2.0 is considered to be biologically relevant, indicating that particular feature is significantly enriched in one group compared with the other. Therefore, the higher the LDA score is, the higher is the contribution of specific taxa to the differences between groups (Ctr and Phy). The control group was found to have a higher abundance of the genera Escherichia–Shigella, Fusobacterium, Muribaculaceae, Romboutsia, Acinetobacter, Christensenellaceae R-7 group, and Streptococcus, while the Phy-treated group was enriched with the genera Campylobacter, Clostridium sensu stricto 1, Prevotella, Roseburia, Terrisporobacter, Prevotellaceae NK3B31 group, Lactobacillus, and Alloprevotella.
Discussion
The experimental results indicated that the phytogenic product tested on this scale did not exhibit statistically significant performance differences between the Phy group and the control group in terms of weight gain. Additionally, mortality rates were comparable between both groups, further indicating that the phytogenic product did not influence overall health outcomes in this study.
The findings from the beta diversity analysis indicated that the Phy treatment had a significant effect on the gut microbiota of the pigs, particularly in their rectal swabs at the age of 14 weeks. Furthermore, the PERMANOVA showed that the age of pigs and the origin of samples were also significant factors in the changes to the gut microbiota. It is crucial to consider multiple factors when evaluating the impact of treatment on the gut microbiome, given the influence of age and origin.
It is worth noting that the results of both PERMANOVA and paired MANOVA were consistent with the relative abundance stacked-bar chart, which demonstrated alterations in the microbial community structure in the Phy-treated pigs compared with the control group. These findings further emphasised the potential advantages of using phytogenic supplements to shape the gut microbiota in pigs.
The relative abundance bar chart of the top 20 genera and LEfSe analysis results demonstrated a shift in the microbial community composition in the faeces. The higher values of the Shannon and Simpson entropy indices indicated a more diverse microbial community in the Phy-treated pigs. These changes in the microbial community composition are consistent with previous research on the effect of phytogenic supplements on the gut microbiota (Bajagai et al. 2020, 2022).
LEfSe analysis showed that Escherichia–Shigella, Fusobacterium, Muribaculaceae, Romboutsia, Acinetobacter, Christensenellaceae R-7 group, and Streptococcus were more abundant in the control group, whereas Campylobacter, Clostridium sensu stricto 1, Prevotella, Roseburia, Terrisporobacter, Prevotellaceae NK3B31 group, Lactobacillus, and Alloprevotella were enriched in the treatment group.
Escherichia–Shigella is a bacterial group that contains species from both Escherichia and Shigella genera. In this study, the sequences assigned to this genus predominantly aligned with Escherichia species, which are typical gut bacteria. A significant observation from our dataset was the reduction in the Escherichia–Shigella population within the Phy group. This is particularly relevant because E. coli, a member of this group, is primarily associated with post-weaning diarrhoea (Xu et al. 2023). This condition poses a considerable economic challenge for pig farmers due to its association with reduced weight gain, increased mortality, and higher susceptibility to secondary infections.
The incidence of post-weaning diarrhoea is intricately linked to the gut microbial ecology of piglets and the abundance and diversity of E. coli. Our findings suggest that Phy supplementation might play an important role in modulating this microbial structure. The observed reduction in Escherichia–Shigella in the Phy group underscores the potential of such supplements in mitigating risks associated with post-weaning diarrhoea.
Several other genera exhibited differential abundance in Phy-treated animals. Notably, Roseburia, known for butyrate production, a vital source of energy for colon cells, showed an increased abundance (Tamanai-Shacoori et al. 2017). Likewise, Prevotella and its related groups, which are essential for the breakdown of complex carbohydrates to produce short-chain fatty acids (SCFA), were more abundant (Lecerf 2020). Although some Prevotella species have been implicated in various diseases, their overall role in immune modulation cannot be ignored (Sharma et al. 2022). Beneficial bacteria such as Lactobacillus were also prominent in the Phy group, reinforcing the gut’s defence mechanisms (Hou et al. 2015; Sinclair et al. 2016).
However, certain pathogenic genera such as Clostridium sensu stricto 1 also saw an uptick in the Phy group. It is worth noting that while members of this group, such as C. perfringens, are usually benign residents of the pig gut, under specific conditions, they might become opportunistic pathogens leading to diseases (Posthaus et al. 2020). Moreover, Campylobacter, another genus increased in the Phy group, is linked with gastroenteritis, colitis, and diarrhoea (Denis et al. 2011). Its high prevalence in the Phy-treated group merits further investigation, considering its association with suboptimal growth rates and reduced feed conversion efficiency (Carrique-Mas et al. 2014).
Our data showed that the phytogen reduced the genus Escherichia-Shigella but increased Campylobacter and Clostridium abundance in this trial. It is difficult to directly compare the economic losses caused by these three groups of pathogens in the pig industry, because the severity and frequency of outbreaks can depend on the virulence of the strains involved, the age and health status of the pigs, feed nutritional composition, feed supplements, geographic location, climate (heat stress), management practices and biosecurity measures used in the farm, to name a few.
However, there is an age-related temporal difference in the severity of the two pathogenic genera, namely, Escherichia-Shigella and Campylobacter, which may have a greater economic impact on pig farming in the post-weaning period in younger piglets. C. perfringens can cause a range of diseases mainly in older pigs. However, further investigation is required to study the effects on these pathogens at species and strain levels and their pathogenicity because the methods used in this study can give information only about the changes in microbiota at the genus and higher level of classification.
Conclusions
In conclusion, the findings of this study demonstrated a significant influence of PFA on the abundance of both beneficial and pathogenic bacterial genera in the pig gut. The supplementation led to an increase in beneficial genera such as Roseburia, Prevotella, Alloprevotella, Terrisporobacter and Lactobacillus, which are known to improve gut barrier function, prevent harmful bacterial growth, and helps degrade complex carbohydrates to produce SCFA for improved energy generation. However, there was also an increase in pathogenic genera, including Clostridium sensu stricto 1 and Campylobacter, while abundance of Escherichia-Shigella was decreased. Despite these microbiota changes, no significant improvement in the performance of the animals was observed for the scale of this experiment. However, it is important to recognise that the effect of phytogenic supplements on pig growth are complex and can be influenced by various factors, including environments, age, genetics, and dietary composition. Further studies such as metagenomic sequencing and host-RNA sequencing can give further insights on the role of PFA on gut microbiota and functions at greater resolution and the host microbiota interaction.
Data availability
Raw sequencing data used to make the conclusions of this study are available in the NCBI SRA database under project number PRJNA974158.
Declaration of funding
This work was supported by PhD scholarship funding scheme by AusPac Ingredients Pty Ltd.
Acknowledgements
The data were analysed using the Marie Curie High Performance Computing System at Central Queensland University. We acknowledge and appreciate the help Jason Bell provided in all aspects of high-performance computing. We also thank Nathan Dwyer and Jonothon Doubleday from AusPac Ingredients Pty Ltd for their assistance in organising the animal trial, helping with the sample collection and student support. We extend our gratitude to Riverbend Pork Group for the help provided via consulting and trial logistics.
References
Abdelli N, Sola-Oriol D, Perez JF (2021) Phytogenic feed additives in poultry: achievements, prospective and challenges. Animals 11, 3471.
| Crossref | Google Scholar |
Bajagai YS, Alsemgeest J, Moore RJ, Van TTH, Stanley D (2020) Phytogenic products, used as alternatives to antibiotic growth promoters, modify the intestinal microbiota derived from a range of production systems: an in vitro model. Applied Microbiology and Biotechnology 104, 10631-10640.
| Crossref | Google Scholar | PubMed |
Bajagai YS, Petranyi F, Yu SJ, Lobo E, Batacan R, Jr, Kayal A, Horyanto D, Ren X, Whitton MM, Stanley D (2022) Phytogenic supplement containing menthol, carvacrol and carvone ameliorates gut microbiota and production performance of commercial layers. Scientific Reports 12, 11033.
| Crossref | Google Scholar | PubMed |
Bartos P, Dolan A, Smutny L, Sistkova M, Celjak I, Soch M, Havelka Z (2016) Effects of phytogenic feed additives on growth performance and on ammonia and greenhouse gases emissions in growing-finishing pigs. Animal Feed Science and Technology 212, 143-148.
| Crossref | Google Scholar |
Bauer BW, Gangadoo S, Bajagai YS, Van TTH, Moore RJ, Stanley D (2019) Oregano powder reduces Streptococcus and increases SCFA concentration in a mixed bacterial culture assay. PLoS ONE 14, e0216853.
| Crossref | Google Scholar | PubMed |
Bengtsson B, Wierup M (2006) Antimicrobial resistance in Scandinavia after a ban of antimicrobial growth promoters. Animal Biotechnology 17, 147-156.
| Crossref | Google Scholar | PubMed |
Bhat MA, Mishra AK, Tantray JA, Alatawi HA, Saeed M, Rahman S, Jan AT (2022) Gut microbiota and cardiovascular system: an intricate balance of health and the diseased state. Life 12, 1986.
| Crossref | Google Scholar |
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodriguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, Koester I, Kosciolek T, Kreps J, Langille MGI, Lee J, Ley R, Liu Y-X, Loftfield E, Lozupone C, Maher M, Marotz C, Martin BD, McDonald D, McIver LJ, Melnik AV, Metcalf JL, Morgan SC, Morton JT, Naimey AT, Navas-Molina JA, Nothias LF, Orchanian SB, Pearson T, Peoples SL, Petras D, Preuss ML, Pruesse E, Rasmussen LB, Rivers A, Robeson MS, 2nd, Rosenthal P, Segata N, Shaffer M, Shiffer A, Sinha R, Song SJ, Spear JR, Swafford AD, Thompson LR, Torres PJ, Trinh P, Tripathi A, Turnbaugh PJ, Ul-Hasan S, van der Hooft JJJ, Vargas F, Vazquez-Baeza Y, Vogtmann E, von Hippel M, Walters W, et al. (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology 37, 852-857.
| Crossref | Google Scholar | PubMed |
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nature Methods 13, 581-583.
| Crossref | Google Scholar | PubMed |
Carrique-Mas JJ, Bryant JE, Cuong NV, Hoang NVM, Campbell J, Hoang NV, Dung TTN, Duy DT, Hoa NT, Thompson C, Hien VV, Phat VV, Farrar J, Baker S (2014) An epidemiological investigation of Campylobacter in pig and poultry farms in the Mekong delta of Vietnam. Epidemiology and Infection 142, 1425-1436.
| Crossref | Google Scholar | PubMed |
Chen X, Xu J, Ren E, Su Y, Zhu W (2018) Co-occurrence of early gut colonization in neonatal piglets with microbiota in the maternal and surrounding delivery environments. Anaerobe 49, 30-40.
| Crossref | Google Scholar | PubMed |
Crisol-Martinez E, Stanley D, Geier MS, Hughes RJ, Moore RJ (2017) Understanding the mechanisms of zinc bacitracin and avilamycin on animal production: linking gut microbiota and growth performance in chickens. Applied Microbiology and Biotechnology 101, 4547-4559.
| Crossref | Google Scholar | PubMed |
Dang DX, Kim IH (2020) Effects of dietary supplementation of Quillaja saponin on growth performance, nutrient digestibility, fecal gas emissions, and meat quality in finishing pigs. Journal of Applied Animal Research 48, 397-401.
| Crossref | Google Scholar |
de Aguiar FC, Solarte AL, Tarradas C, Luque I, Maldonado A, Galan-Relano A, Huerta B (2018) Antimicrobial activity of selected essential oils against Streptococcus suis isolated from pigs. MicrobiologyOpen 7, e00613.
| Crossref | Google Scholar | PubMed |
Denis M, Henrique E, Chidaine B, Tircot A, Bougeard S, Fravalo P (2011) Campylobacter from sows in farrow-to-finish pig farms: risk indicators and genetic diversity. Veterinary Microbiology 154, 163-170.
| Crossref | Google Scholar | PubMed |
Dixon P (2003) Vegan, a package of R functions for community ecology. Journal of Vegetation Science 14, 927-930.
| Crossref | Google Scholar |
Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, Ravel J (2014) An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2, 6.
| Crossref | Google Scholar | PubMed |
Ghasemi G, Alirezalu A, Ghosta Y, Jarrahi A, Safavi SA, Abbas-Mohammadi M, Barba FJ, Munekata PES, Dominguez R, Lorenzo JM (2020) Composition, antifungal, phytotoxic, and insecticidal activities of thymus kotschyanus essential oil. Molecules 25, 1152.
| Crossref | Google Scholar |
Hou C, Zeng X, Yang F, Liu H, Qiao S (2015) Study and use of the probiotic Lactobacillus reuteri in pigs: a review. Journal of Animal Science and Biotechnology 6, 14.
| Crossref | Google Scholar | PubMed |
Kabat AM, Srinivasan N, Maloy KJ (2014) Modulation of immune development and function by intestinal microbiota. Trends in Immunology 35, 507-517.
| Crossref | Google Scholar | PubMed |
Karaskova K, Suchy P, Strakova E (2015) Current use of phytogenic feed additives in animal nutrition: a review. Czech Journal of Animal Science 60, 521-530.
| Crossref | Google Scholar |
Kim SW, Holanda DM, Gao X, Park I, Yiannikouris A (2019) Efficacy of a yeast cell wall extract to mitigate the effect of naturally co-occurring mycotoxins contaminating feed ingredients fed to young pigs: impact on gut health, microbiome, and growth. Toxins 11, 633.
| Crossref | Google Scholar |
Konstantinov SR, Awati AA, Williams BA, Miller BG, Jones P, Stokes CR, Akkermans ADL, Smidt H, De Vos WM (2006) Post-natal development of the porcine microbiota composition and activities. Environmental Microbiology 8, 1191-1199.
| Crossref | Google Scholar | PubMed |
Lalles J-P, Boudry G, Favier C, Le Floc’h N, Luron I, Montagne L, Oswald IP, Pie S, Piel C, Seve B (2004) Gut function and dysfunction in young pigs: physiology. Animal Research 53, 301-316.
| Crossref | Google Scholar |
Lecerf JM (2020) Is Prevotella/Bacteroides ratio associated with weight loss. Correspondances En Metabolismes Hormones Diabetes Et Nutrition 24, 182.
| Google Scholar |
Lee YK, Mazmanian SK (2010) Has the microbiota played a critical role in the evolution of the adaptive immune system? Science 330, 1768-1773.
| Crossref | Google Scholar | PubMed |
Liu C, Cui Y, Li X, Yao M (2021) microeco: an R package for data mining in microbial community ecology. FEMS Microbiology Ecology 97, fiaa255.
| Crossref | Google Scholar |
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.Journal 17, 10-12.
| Crossref | Google Scholar |
Mathlouthi N, Bouzaienne T, Oueslati I, Recoquillay F, Hamdi M, Urdaci M, Bergaoui R (2012) Use of rosemary, oregano, and a commercial blend of essential oils in broiler chickens: in vitro antimicrobial activities and effects on growth performance. Journal of Animal Science 90, 813-823.
| Crossref | Google Scholar | PubMed |
Mcmurdie PJ, Holmes S (2013) Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217.
| Crossref | Google Scholar |
Posthaus H, Kittl S, Tarek B, Bruggisser J (2020) Clostridium perfringens type C necrotic enteritis in pigs: diagnosis, pathogenesis, and prevention. Journal of Veterinary Diagnostic Investigation 32, 203-212.
| Crossref | Google Scholar | PubMed |
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41, D590-D596.
| Crossref | Google Scholar | PubMed |
Sharma G, Garg N, Hasan S, Shirodkar S (2022) Prevotella: an insight into its characteristics and associated virulence factors. Microbial Pathogenesis 169, 105673.
| Crossref | Google Scholar |
Shehata AA, Yalcin S, Latorre JD, Basiouni S, Attia YA, Abd El-Wahab A, Visscher C, El-Seedi HR, Huber C, Hafez HM, Eisenreich W, Tellez-Isaias G (2022) Probiotics, prebiotics, and phytogenic substances for optimizing gut health in poultry. Microorganisms 10, 395.
| Crossref | Google Scholar |
Shen YB, Voilque G, Odle J, Kim SW (2012) Dietary L-tryptophan supplementation with reduced large neutral amino acids enhances feed efficiency and decreases stress hormone secretion in nursery pigs under social-mixing stress. The Journal of Nutrition 142, 1540-1546.
| Crossref | Google Scholar | PubMed |
Sidiropoulou E, Marugan-Hernandez V, Skoufos I, Giannenas I, Bonos E, Aguiar-Martins K, Lazari D, Papagrigoriou T, Fotou K, Grigoriadou K, Blake DP, Tzora A (2022) In vitro antioxidant, antimicrobial, anticoccidial, and anti-inflammatory study of essential oils of oregano, thyme, and sage from Epirus, Greece. Life 12, 1783.
| Crossref | Google Scholar |
Sinclair A, Xie X, Saab L, Dendukuri N (2016) Lactobacillus probiotics in the prevention of diarrhea associated with Clostridium difficile: a systematic review and Bayesian hierarchical meta-analysis. CMAJ Open 4, E706-E718.
| Crossref | Google Scholar | PubMed |
Smith S (2019) phylosmith: an R-package for reproducible and efficient microbiome analysis with phyloseq-objects. Journal of Open Source Software 4, 1442.
| Crossref | Google Scholar |
Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, Bonnaure-Mallet M, Jolivet-Gougeon A (2017) Roseburia spp.: a marker of health? Future Microbiology 12, 157-170.
| Crossref | Google Scholar | PubMed |
The Lancet (2022) Antimicrobial resistance: time to repurpose the Global Fund. The Lancet 399, 335.
| Crossref | Google Scholar |
Varel VH (2002) Carvacrol and thymol reduce swine waste odor and pathogens: stability of oils. Current Microbiology 44, 38-43.
| Crossref | Google Scholar | PubMed |
Verschuren LMG, Calus MPL, Jansman AJM, Bergsma R, Knol EF, Gilbert H, Zemb O (2018) Fecal microbial composition associated with variation in feed efficiency in pigs depends on diet and sex. Journal of Animal Science 96, 1405-1418.
| Crossref | Google Scholar | PubMed |
Xu CL, Peng K, She Y, Fu F, Shi Q, Lin Y, Xu CB (2023) Preparation of novel trivalent vaccine against enterotoxigenic Escherichia coli for preventing newborn piglet diarrhea. American Journal of Veterinary Research 84, ajvr.22.10.0183.
| Crossref | Google Scholar |
Yu Z, Morrison M (2004) Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 36, 808-812.
| Crossref | Google Scholar | PubMed |
Yu SJ, Bajagai YS, Petranyi F, de las Heras-Saldana S, Van TTH, Stanley D (2022) Phytogenic blend improves intestinal health and reduces obesity, diabetes, cholesterol and cancers: a path toward customised supplementation. Antibiotics 11, 1428.
| Crossref | Google Scholar |