Genetic parameter estimates for male and female fertility traits using genomic data to improve fertility in Australian beef cattle
Babatunde S. Olasege A B , Muhammad S. Tahir A B , Gabriela C. Gouveia C , Jagish Kour A , Laercio R. Porto-Neto B , Ben J. Hayes D and Marina R. S. Fortes
A B , Muhammad S. Tahir A B , Gabriela C. Gouveia C , Jagish Kour A , Laercio R. Porto-Neto B , Ben J. Hayes D and Marina R. S. Fortes  A E
A E
A School of Chemistry and Molecular Biosciences, The University of Queensland, Saint Lucia Campus, Brisbane, Qld 4072, Australia.
B CSIRO Agriculture and Food, Saint Lucia, Qld 4067, Australia.
C Animal Science Department, Veterinary School, Federal University of Minas Gerais, Belo Horizonte 31270-901, Brazil.
D Queensland Alliance for Agriculture and Food Innovation (QAAFI), The University of Queensland, Saint Lucia Campus, Brisbane, Qld 4072, Australia.
E Corresponding author. Email: m.fortes@uq.edu.au
Animal Production Science - https://doi.org/10.1071/AN21097
Submitted: 22 February 2021 Accepted: 1 June 2021 Published online: 3 August 2021
Journal Compilation © CSIRO 2021 Open Access CC BY-NC-ND
Abstract
Context: Studies have shown that favourable genetic correlations exist between female and male fertility traits. However, investigations regarding these correlations in Australian tropical beef cattle are limited to either pedigree or single-breed analysis.
Aim: The study aims to use genomic information to estimate genetic parameters of six female and seven male fertility traits measured during the first 2 years of life, in two tropical breeds.
Methods: Single-, bivariate and multi-trait models were used to analyse fertility data from Brahman (BB; 996 cows and 1022 bulls); and Tropical Composite (TC; 1091 cows and 998 bulls) cattle genotyped with high-density single-nucleotide polymorphism chip assay.
Key results: Heritability estimates in BB cows ranged from low (0.07 ± 0.04) for days to calving at the first calving opportunity (DC1, days) to high (0.57 ± 0.08) for age at first corpus luteum (AGECL, days). In BB bulls, estimates varied from low (0.09 ± 0.05) for sperm motility (score 1–5) to high (0.64 ± 0.06) for scrotal circumference (SC) measured at 24 months (SC24, cm). Similarly, heritability estimates in TC cows were low (0.04 ± 0.03) for DC1 and high (0.69 ± 0.02) for AGECL. In TC bulls, the heritability was low (0.09 ± 0.05) for sperm motility and high (0.69 ± 0.07) for SC24. Within-sex for both breeds, blood concentrations of insulin growth-factor 1 (IGF1) measured in cows at 18 months (IGF1c) were negatively correlated with female fertility phenotypes. In BB, across-sex, bulls’ blood concentration of IGF1 measured at 6 months (IGF1b) was a good indicator trait for the following four female traits: AGECL, the first postpartum anoestrus interval, age at first calving and DC1. In TC, IGF1b and percentage normal sperm were good predictors of female fertility phenotypes.
Conclusions: The heritability estimates and genomic correlations from the present study generally support and confirmed the earlier estimates from pedigree analyses. The findings suggest that selection for female fertility traits will benefit male fertility, and vice versa.
Implications: Heritability estimates and genomic correlations suggest that we can select for fertility traits measured early in life, with benefits within and across sex. Using traits available through veterinary assessment of bull fertility as selection indicators will enhance bull and cow fertility, which can lead to better breeding rates in tropical herds.
Keywords: genetic correlation, fertility, heritability, beef cattle, genomics.
Introduction
In cow-calf operations, the number of calves produced per cow in their lifetime is indicative of beef productivity and sustainability (Johnston et al. 2014a; Zhang et al. 2014). Increasing the number of calves to ensure enterprise profitability is a complex challenge that involves both female and male fertility (Hansen 2006). Improving fertility in both sexes will create numerous benefits across the supply chain and contributes to the beef industry as a whole.
Female and male cattle may employ different reproductive strategies, resulting in the evolution of sex dimorphism (Fairbairn et al. 2007). Interestingly, since both sexes share almost identical genomes apart from the sex chromosome, there is a possibility that selection in one sex can result in indirect selection on the other sex, constraining the evolution of sexual dimorphism (Lande 1984; Poissant et al. 2010; Pennell and Morrow 2013). It is then expected that sex-specific genes would allow independent evolution within sexes and genes that are expressed in both males and females would foster similarities in both sexes (Lande 1984). Genetic correlations can be very high across sexes, when studying traits that are homologous in both male and female (Poissant et al. 2010). But there are obviously traits that are not homologous. Reproductive strategies, dimorphism and genetics form an interesting interdisciplinary field, where researchers investigate the intra-locus sexual conflict.
After the study of Land (1973), who first reported the interplay between male and female reproduction in mammals, several studies in beef cattle have shown the existence of favourable genetic correlations between bull and cow fertility (Johnston et al. 2014b; Raidan et al. 2019). The genetic correlation between scrotal circumference (SC) and female fertility traits justifies the commercial use of SC in selection programs across the world (Lunstra and Cundiff 2003; Gargantini et al. 2004). However, investigation about sex interplay in tropical beef cattle in Australia has been limited to either pedigree (Johnston et al. 2014b; Jeyaruban and Johnston 2017; Johnston and Moore 2019) or single-breed analysis (Raidan et al. 2019). Considering the diverse range of breeds, crossbreeds and composites that characterise the Australian beef industry, there is a need to study the relationships between female and male fertility in more than one breed. Therefore, the present study aims to estimate genetic parameters for a range of male and female fertility traits, in two tropical beef breeds of Australia. We also discuss the implication of these results for selective breeding.
Materials and methods
Animals and phenotypes
Institutional Animal Care and Use Committee approval was not needed for the present study because data used were retrieved from existing databases. Animals used in the study were bred by the Cooperative Research Centre for Beef Genetic Technologies (Beef CRC). These animals were reared under a range of extensive environments at four research stations in Queensland. The details on the routine management, feeding regimes and health treatments have been previously described in Johnston et al. (2009), Burns et al. (2013) and Wolcott et al. (2014). Cows were culled only for husbandry reasons (i.e. disease) or if they failed to wean a calf in two consecutive years. Otherwise, they were kept for six mating opportunities. Briefly, 2018 Brahman (BB) cattle (996 cows and 1022 bulls) and 2089 Tropical Composite (TC) cattle (1091 cows and 998 bulls) were used for the study. The same number of traits were studied in the two populations (six cow traits, and seven bull traits), but the exact number of animals recorded for each trait varied (Table 1). BB cattle are of Bos indicus origin, while TC originated from crossing Bos indicus and Bos taurus breeds. The breed composition of these two populations has been previously investigated. TC breed exhibits more genetic variation than BB (Porto-Neto et al. 2013a, 2013b). Since, breed composition can affect genetic parameter estimates, we model the first two principal components (PC1 and PC2) in addition to the genomic relationship matrix (GRM) to account for the, quite varied, breed composition in the TC breed, for all traits studied. The use of PC1 and PC2 was not necessary for the BB herd, which had a more uniform breed composition.

|
Genotypes and imputation
All animals used were either genotyped with the BovineSNP50 (Matukumalli et al. 2009) or the BovineHD (Illumina Inc., San Diego, CA, USA) chips. Animals genotyped with the medium-density single-nucleotide polymorphism (SNP) panel had their genotypes imputed up to 770 000 as described by Bolormaa et al. (2015). Before performing imputation, all the original SNP genotypes were remapped to the new assembly of the bovine reference genome (ARS_UCD1.2, GenBank assembly accession GCA_002263795.2; Rosen et al. 2020) and were phased using Eagle software (Loh et al. 2016). Subsequently, Minimac3 was used to impute the lower-density genotypes for all autosome (Das et al. 2016) and Minimac4 for the X-chromosome. The X-chromosome was imputed after separating the non-pseudoautosomal (nPAR) and pseudoautosomal regions (PAR) following the recent definition of these two regions by Johnson et al. (2019). After imputation, only SNPs with an imputation accuracy higher than 0.8 were retained for subsequent analyses, resulting in a dataset with 722 208 SNPs for both sexes in the two breeds.
Genomic relationship matrix (GRM)
First, we performed quality control by excluding all SNPs with a minor allele frequency smaller than 0.05 within breed for each sex, before constructing the GRM. As a result, 561 080 and 562 974 genotypes remained for BB cows and bulls respectively. In TC, 688 407 and 688 095 genotypes remained for cows and bulls respectively. GRMs were then constructed following Method 1 of VanRanden (VanRaden 2008) implemented in GIBBS2F90 (Misztal et al. 2002). For the across-sex study, we merged the unfiltered genotypes for both sexes in each breed and performed the quality control as described above, resulting in 554 712 and 688 603 SNPs for both BB and TC respectively. Before constructing the GRM, we separated the X-chromosome into nPAR and PAR regions following the boundary described by Johnson et al. (2019). The PAR region is small, consisting of 1977 SNPs, which were removed from the analyses. After removing PAR SNPs, we used the genotypes of 554 712 and 686 626 SNPs for BB and TC respectively, to build the GRM. First, we constructed a GRM using all the SNPs in autosomes, for each breed separately, using GCTA software (Yang et al. 2011). Second, for the X-chromosome nPAR region, we constructed another GRM using the specific function for the nPAR region as specified in GCTA software (Yang et al. 2011). The final GRM for both BB and TC breeds was created by merging both the autosome GRM and the X-chromosome nPAR region GRM. The merged GRM was imported into GIBBS2F90 for further analyses.
Statistical analyses
Within sex, heritability estimates were computed using single- and multi-trait models, while genomic correlations were estimated using bivariate and multi-trait models. Bivariate models were employed to estimate the genomic correlations between male and female traits. All analyses were performed using GIBBS2F90 (Misztal et al. 2002). The general mixed-model equation was as follows:
within-sex: single- and multi-trait models

where y is a vector of observations for the analysed traits within sex for each breed, b is a vector of fixed effects, a is the vector of random additive animal effects, e is the vector of random residual effects, and X and Z are incidence matrices relating records to their respective effects for every individual in the GRM. The assumptions for this analysis were as follows:

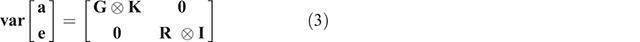
where G is the additive genetic covariance matrix of order n × n, where n is the number of traits analysed, K is the genomic relationship matrix for all animals, R is the residual covariance matrix of order n × n, I is an identity matrix, ⊗ is the Kronecker product operator.
Within-sex bivariate model was as follows:
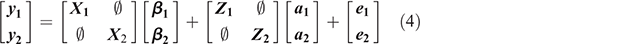
All parameters in Eqn 4 have been described in Eqn 1. Subscripts 1 and 2 here are parameters relative to Traits 1 and 2 within sex for each breed. The assumptions of the structure of (co)variances for bivariate model were as follows:
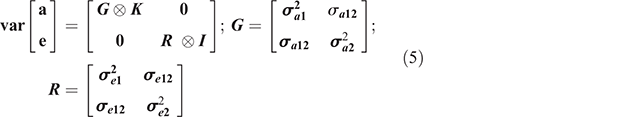
where G and R are the additive and residual covariance matrices of order 2 × 2,  ,
, 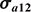 are the additive genetic variances for Trait 1 and Trait 2, σa12 is the covariance for the two traits,
are the additive genetic variances for Trait 1 and Trait 2, σa12 is the covariance for the two traits, 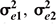 ,
, 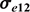 are the residual variances for Trait 1 and Trait 2, and σe12 is the residual (co)variance for the traits.
are the residual variances for Trait 1 and Trait 2, and σe12 is the residual (co)variance for the traits.
Across-sex bivariate model
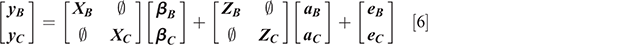
All parameters in Eqn 3 have been described in Eqn 1. Subscripts B and C here are parameters relative to traits measured either in bulls (B) or in cows (C) for each breed. The assumptions of the structure of (co)variance for bivariate model were as follows:

where G and R are the additive and residual covariance matrix of order 2 × 2 (bull and cow), 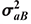 , and
, and  are respectively, the additive genetic variances for the bull and cow, σaBC is the covariance between the two sexes,
are respectively, the additive genetic variances for the bull and cow, σaBC is the covariance between the two sexes,  and
and 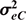 are the residual (co)variances for the bull and cow traits respectively. It is assumed that there is no residual covariance between the two sexes.
are the residual (co)variances for the bull and cow traits respectively. It is assumed that there is no residual covariance between the two sexes.
The fixed effects included in the model were specific for each trait and were deemed significant for the phenotypic variations observed (Table 2). The contemporary group (CG) effect was relevant for most traits. The CG represents cohort of animals born in the same year and raised together under the same management conditions. Information about CG has been described in detail by Barwick et al. (2009), Burns et al. (2013) and Johnston et al. (2014b). In BB, there were 58 levels for males and 78 levels for females in terms of CG. In TC, there were 46 levels for males and 74 levels for females in terms of CG. The laboratory assay batch was an important aspect of the enzyme‐linked immunosorbent assay (ELISA; Moore et al. 1995) used to measure IGF1 (in BB there were 51 batches; in TC, 38 batches). Age of dam and the age of the animal at the time of trait recording were considered and included in the model as linear covariates when significant.

|
Posterior means and standard deviations for (co)variance components, heritabilities and genetic correlations were obtained using the POSTGIBBSF90 (Misztal et al. 2002). The analysis consisted of a single chain of 100 000 cycles discarding the first 20 000 cycles, taking a sample at every 50 iterations to obtain the parameters. The standard error of heritability estimates and genomic correlations were obtained using the OPTION se_covar_function of the POSTGIBBSF90 program (Misztal et al. 2002).
Results
Heritability estimates
The estimates of heritability and their corresponding standard errors for both single- and multi-trait models are presented in Table 3. The estimates for BB cows ranged from low to moderate for both models. Days to calving at the first calving opportunity (DC1) had the lowest heritability for both single-trait (0.07 ± 0.04) and multi-trait (0.17 ± 0.05) models, while AGECL had the highest estimates for both models (single-trait: 0.56 ± 0.08; multi-trait: 0.57 ± 0.08). For TC cows, the estimates of heritability ranged from low to moderate for single-trait models and from low to high for multi-trait models. DC1 had the lowest heritability for both single-trait (0.04 ± 0.03) and multi-trait (0.12 ± 0.03) models, while AGECL had a moderate estimate for the single-trait (0.46 ± 0.08) and a high estimate for the multi-trait (0.69 ± 0.02) model. For BB and TC bulls, the heritability estimates in most cases followed a similar pattern, with the estimates ranging from low to high in both populations. For the single-trait models, sperm motility (MOT) had the lowest heritability estimate in BB (0.09 ± 0.05) and TC (0.09 ± 0.05), while SC24 had the highest estimates in both breeds (0.62 ± 0.07 in BB and 0.62 ± 0.08 in TC). For multi-trait analysis, again SC24 had the highest estimates of heritability in both breeds (0.64 ± 0.06 in BB and 0.69 ± 0.07 in TC), while MOT had the lowest estimate in BB (0.09 ± 0.02) and sperm mass activity (MAS) had the lowest estimate in TC (0.18 ± 0.06). Generally, heritability estimates from the multi-trait analysis were higher than the estimates from a single-trait models and were usually accompanied by lower standard errors.

|
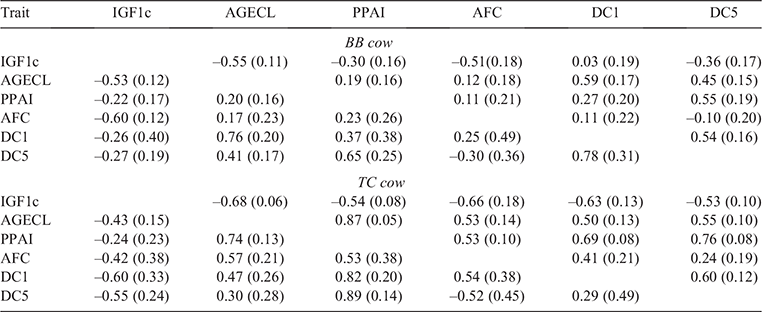
Genomic correlations: cow-fertility traits
Genetic correlation (bivariate and multivariate) among cow-fertility phenotypes for both BB and TC are presented in Table 4. In general, a range of strong positive and negative genetic correlations was recorded between the studied traits for both breeds. Given the low heritabilities of age at first calving (AFC) and DC1 in both breeds, all genetic correlation estimates with these traits had large standard errors. The magnitude of the errors was reduced in the multi-trait model.
For BB cows, the strongest genetic correlation was observed between AGECL and DC1 for both bivariate (BB: 0.76 ± 0.20) and multivariate (BB: 0.59 ± 0.17) model. However, for TC cow, the strongest correlation was observed between PPAI and DC5 (TC: 0.89 ± 0.14) for bivariate and AGECL and PPAI (TC: 0.87 ± 0.05) for multi-trait model.
Genomic correlation: bull fertility
Genetic correlation (bivariate and multivariate) among bull fertility phenotypes for both BB and TC breeds are presented in Table 5. Generally, the estimates of genetic correlation were all positive in both genotypes for both models, indicating a lack of genetic antagonism among the studied traits. A notable exception was the correlation between IGF1b and SC24 (–0.03 ± 0.33) for the bivariate model. However, these estimates were positive for all traits when analysed using a multi-trait model. The strongest genetic correlation was observed between SC18 and SC24 for both bivariate (BB: 0.97 ± 0.02; TC: 0.97 ± 0.01) and multivariate (BB: 0.93 ± 0.02; TC: 0.95 ± 0.01) models.
Across-sex genomic correlations
Estimated genetic correlations (bivariate) between cow and bull traits for BB and TC are presented in Table 6. Some standard errors for these correlations are larger than the genomic correlation itself, implying that the correlations are not significant.
For BB, IGF1b was favourably correlated with all the female fertility phenotypes, with the strongest negative correlation recorded between IGF1b and AGECL (–0.65 ± 0.13) and the lowest observed between IGF1b and DC5 (–0.02 ± 0.20). Semen-quality traits (MAS, MOT and percentage normal sperm (PNS)) had a negative correlation with PPAI, with estimates of –0.81 ± 0.30, –0.75 ± 0.39 and –0.50 ± 0.19 respectively. SC12, SC18 and SC12 had a moderate negative genetic correlation with AGECL (–0.32 ± 0.15) and (–0.36 ± 0.12), (–0.25 ± 0.12) respectively. Moderate negative correlations were also observed between SC12 and DC1(–0.32 ± 0.30) as well as SC12 and DC1 (–0.21 ± 0.31) respectively.
Similar to the result observed for IGF1b in BB, this trait was also favourably correlated with AGECL (–0.55 ± 0.14) and PPAI (–0.17 ± 0.22) in TC, with the strongest estimate being recorded for DC5 (–0.94 ± 0.13). PNS had a strong negative correlation with DC1 (–0.60 ± 0.28) and a moderate negative correlation with AFC (–0.24 ± 0.28). A moderate genetic correlation was also recorded between SC12 and AGECL (–0.26 ± 0.13). SC12 had a negative correlation with DC1 (–0.06 ± 0.58).
Discussion
The objective of most cattle breeding programs is to maximise genetic gain for traits that are of economic relevance to beef production. In part, the success of these breeding programs depends on the accuracies with which the genetic parameters are estimated (Wellmann and Bennewitz 2019). Nowadays, many traits are important to cattle breeders. Producers are often interested in a combined selection objective that uses information from many traits to form a total merit index (Cole and VanRaden 2018). Genetic parameters estimated from a multi-trait model are regarded as more accurate because their estimates utilise additional genomic information and use the complex covariance structure among traits (Analla et al. 1995; Zhang et al. 2018). Moreover, these multi-trait models are expected to produce higher estimates of heritability than single-trait models, due to the additional information. For most traits, we observed higher heritabilities in the multi-trait model. This difference between single- and multi-trait analyses was more evident in TC than in BB. Perhaps, this could be partly explained by the higher genetic variation that composite breeds exhibit than their founders (Rasali et al. 2006). The increase in the proportion of segregating alleles in composite breeds could be advantageous and this information could have been captured in the multi-trait model. In either case, the use of multi-trait models is likely to be advantageous.
Heritability estimates for all phenotypes reported herein, in both BB and TC breeds, are in line with previous estimates for similar traits from single-trait models with either pedigree (Corbet et al. 2009; Corbet et al. 2013; Johnston et al. 2014b; Johnston and Moore 2019) or genomic information (Raidan et al. 2019; Fortes et al. 2020). Estimated heritabilities imply that genetic improvement can be made through selection for these fertility traits. However, some of the studied traits will respond to selection faster than others. The lower heritability estimates for calving traits, such as AFC or DC1, or semen-quality traits, such as MAS and MOT, imply that these traits are not going to respond as fast as AGECL or IGF1 in selective breeding. Probably, the environmental effects are higher for these traits with lower heritabilities and it may be efficient to explore the correlated response via other fertility traits to achieve faster genetic progress (Raidan et al. 2019). Most importantly, MAS and MOT were measured here as visual scores and there is a degree of subjectiveness to these traits. Assessment of semen motility by using computer-assisted semen analysis could remove some of the human error and perhaps provide a more heritable trait for selection.
Several studies have suggested measuring the concentration of IGF1 as an indicator for cow reproductive potential (Taylor et al. 2004; Velazquez et al. 2008). From our results, IGF1c is favourably correlated with most of the female fertility traits studied in both breeds, indicating its multiple roles in reproduction. Therefore, the increased level of IGF1 concentration in the blood could be an indication for cows with the potential to reach puberty early, conceive, calve, and also re-breed early for the next calving season. These favourable correlations between IGF1 and several other fertility traits such as age at first ovulation, conception rate to first service, age at first calving, calving rate, and postpartum anoestrus interval have been reported in several studies (Yilmaz et al. 2004, 2006; Falkenberg et al. 2008; Velazquez et al. 2008). Late onset of puberty is an important component of the reduced lifetime productivity observed in tropically adapted beef (Lesmeister et al. 1973; Johnston et al. 2014a). This explains the estimated positive genetic correlation between AGECL and other female fertility traits in the two breeds. Therefore, genetic selection for increased concentration of IGF1 measured in heifers at ~18 months of age might contribute to early puberty and improve lifetime reproductive rates in Brahman cattle. IGF1 is highly influenced by nutritional status, and so proper nutritional programming will be required to accelerate puberty in heifers (Zulu et al. 2002; Alves et al. 2017).
Similar to our study, Corbet et al. (2013) found no genetic antagonism between bull fertility phenotypes. Regardless, it might be difficult to recommend a single phenotype as a reliable indicator of bull fertility. For instance, if we select bulls on the basis of sperm morphology and other assessments are neglected (i.e. assessment such as physical health that is required for mating in extensive beef farms), then the required genetic progress might not be achieved. Thus, various measurements of bull fertility as obtainable using the bull breeding soundness evaluation (BBSE) are important for sensible breeding decisions (Fordyce et al. 2006). Selecting bulls by using multiple indicator traits provides a better chance of improving the overall fertility of the herd.
Genetic improvement programs for female fertility in beef cattle has been hindered by the difficulty of collecting accurate fertility-related phenotypes needed to improve the predictive ability of genomic selection models (Tiezzi and Maltecca 2011). Some nucleus herds (or research stations) are capable of collecting very detailed, accurate and sometimes expensive fertility phenotypes (Schatz et al. 2010). However, the size from these nucleus herds tends to be relatively small and sample size affects the predictive power of genomic selection. In contrast, commercial herds might be a source of large datasets, with the disadvantage that phenotypes may be less precise (Hayes et al. 2019a). Traits that are directly related to female fertility are not feasible to measure on a large number of cattle in extensively managed herds, which characterise the typical production system of tropically adapted beef cattle in Australia and other countries (Hayes et al. 2019b). This challenge, as well as the complexity of the production systems faced by animal breeders, point to a potential solution, namely, focussing on favourably correlated traits (i.e. traits that are easy to measure early in life) that together act as a fertility index (Brito Lopes et al. 2016). Since bull fertility phenotypes are heritable and less expensive to measure than lifetime female fertility data, identification of traits in male that have a high genetic correlation with female fertility could significantly aid the improvement of reproductive performance of tropically adapted cows (Jeyaruban and Johnston 2017). Moreover, high selection intensity is achievable in males, allowing few genetically superior bulls to contribute to a large percentage of the genetic make-up of the next generation (Martinez-Velazquez et al. 2003; Weigel 2017). From the present study, IGF1 measured in bull favourably correlated with early fertility phenotypes in the BB breed. These favourable across-sex correlations have been reported in previous studies investigating the correlation between male and female reproductive traits such as age at puberty, pregnancy rate, age at first calving, days to calving and calving rate (Yilmaz et al. 2006; Johnston et al. 2014b). This implies that selection for concentration of IGFI as early as at 6 months in bulls might improve reproductive performance in BB cattle. Notwithstanding, it will be important to investigate the cost of measuring IGF1 in bulls on herd profitability before recommending this practice. Even if selection decision is made at this early age, assessment of bulls through BBSE will still be required to ensure that the selected bulls are indeed capable of breeding, because (a) the correlation between IGF1 and BBSE success is far from perfect, and (b) several circumstances and environmental factors, including disease, might affect bull fertility in the timespan between 6 and 24 months of age (i.e. between IGF1 measurement and the first mating season of a bull).
However, in TC breed, IGF1b was favourably correlated only with AGECL, PPAI and DC5, but not with AFC and DC1. These traits (AFC and DC1) were correlated only with PNS in this breed. Nevertheless, the estimates are in tandem with the findings of Johnston et al. (2014b), who reported a favourable correlation between IGF1b and AGECL in TC breed. The authors also found PNS to be genetically correlated with most early female fertility phenotypes. Therefore, IGF1b and PNS could be useful as indirect selection criteria for improving female reproduction in TC breed.
Of note from our study is the difference in the period of measurement of IGF1 across the sexes. In cow, IGF1 was measured at 18 months, whereas the measurement was observed in bull as early as at 6 months of age. There is considerable evidence that IGF1 concentration varies with age (Abribat et al. 1990; Larnkjær et al. 2012; Michaelsen 2013). As we are not certain on the implication of measuring IGF1 early on female fertility, further research is required to firmly establish this trait as a biomarker for pubertal development that might be useful for a selection decision for cattle breeding. The big gain from genomic selection is the ability to estimate genomic breeding values at birth or even at embryo for both male and female calves quite accurately without the need for the animal’s own performance record, as long as there is a sizable and relevant reference population (Hayes and Goddard 2010; Taylor 2014; Hayes et al. 2019b). These genomic estimated breeding values may allow breeders to make an accurate selection decision before the breeding season and assist with stoking decisions.
Findings from Johnston et al. (2014b) have shown that SC is a modest predictor of age at puberty in tropically adapted beef cattle. In the present study, SC18 better reflected puberty in BB, while the estimate could be as short as 12 months in TC. This is not surprising as Brahman are typically late pubertal when compared with Bos taurus breeds (Lunstra and Cundiff 2003; Lopez et al. 2006). Furthermore, SC measured at 12 and 18 months in Brahman was also found to be a moderate predictor for DC1. However, in TC, the estimate of genetic correlation between SC12 and DC1 was negative, although not very large, indicating that selection for SC12 had a favourable influence on DC1. The estimate becomes positive at 18 and 24 months, albeit with a large standard error. These results correspond with the findings of Jeyaruban and Johnston (2017), where moderate genetic correlations were observed between SC measured between 300 and 700 days of age and DC1 for BB breed. The authors also reported a negative estimate for Santa Gertrudis cattle, a stabilised composite with 50% to 30% Bos indicus content (Hayes et al. 2019b). This estimate is consistent with what we observed for TC. Differences in results for SC12, SC18 and SC24 are expected, especially when we consider the breed differences in pubertal development between BB and TC. BB cattle are late pubertal and so it is expected that we observe a lot of variation in SC in the early measurements (Lunstra et al. 1978; Lunstra and Cundiff 2003). As we progress to 24 months of age, most bulls have already passed puberty, which means there is less variation in the trait and SC24 might not be a good indicator of female fertility traits anymore. In TC, at 12 months, many bulls have reached pubertal and are also producing sperm, which is different from the BB (Corbet et al. 2013). Therefore, depending on breed, different ages for SC measurement are required to better capture these favourable correlations with female fertility traits.
Conclusions
The heritability estimates and genomic correlations from the present study generally support and confirmed the earlier estimates from pedigree analyses. Within sex, IGF1c might be used as a predictor of female fertility and BBSE traits might be combined to accurately predict bull fertility. Across sex, IGF1b might be a useful indicator for early-in-life female fertility traits in Brahmans, while both IGF1b and PNS could be indicator traits for female fertility in TC. The findings from this study suggest that selection for fertility traits will have a favourable correlated response to selection within and across sex.
Declaration of funding
BSO was supported by funding from Meat and Livestock Australia (L.GEN.1710; Female Fertility Phenobank and L.GEN.1818; Bull fertility update: historical data, new cohort and advanced genomics) and top-up scholarship from CSIRO.
Conflicts of interests
Marina R. S. Fortes is an Associate Editor of Animal Production Science but was blinded from the peer-review process for this paper. All other authors declare no conflicts of interest.
Acknowledgements
This research was conducted using the legacy database of the Cooperative Centre for Beef Genetics Technologies and their core partners including Meat and Livestock Australia. The authors acknowledge all the participants of the Cooperative Centre for Beef Genetics Technologies for their efforts in conducting the field trials and recording the phenotypes. We also acknowledge the mentorship of Professor Steve Moore to Babatunde Olasege in his PhD.
References
Abribat T, Lapierre H, Dubreuil P, Pelletier G, Gaudreau P, Brazeau P, Petitclerc D (1990) Insulin-like growth factor-I concentration in Holstein female cattle: variations with age, stage of lactation and growth hormone-releasing factor administration. Domestic Animal Endocrinology 7, 93–102.| Insulin-like growth factor-I concentration in Holstein female cattle: variations with age, stage of lactation and growth hormone-releasing factor administration.Crossref | GoogleScholarGoogle Scholar | 2107053PubMed |
Alves BR, Cardoso RC, Doan R, Zhang Y, Dindot SV, Williams GL, Amstalden M (2017) Nutritional programming of accelerated puberty in heifers: alterations in DNA methylation in the arcuate nucleus. Biology of Reproduction 96, 174–184.
Analla M, Munoz‐Serrano A, Cruz J, Serradilla J (1995) Estimation of genetic parameters of growth traits in Segureña lambs. Journal of Animal Breeding and Genetics 112, 183–190.
| Estimation of genetic parameters of growth traits in Segureña lambs.Crossref | GoogleScholarGoogle Scholar |
Barwick S, Johnston D, Burrow H, Holroyd R, Fordyce G, Wolcott ML, Sim WD, Sullivan M (2009) Erratum to: Genetics of heifer performance in ‘wet’ and ‘dry’ seasons and their relationships with steer performance in two tropical beef genotypes. Animal Production Science 49, 727
| Erratum to: Genetics of heifer performance in ‘wet’ and ‘dry’ seasons and their relationships with steer performance in two tropical beef genotypes.Crossref | GoogleScholarGoogle Scholar |
Bolormaa S, Pryce JE, Zhang Y, Reverter A, Barendse W, Hayes BJ, Goddard ME (2015) Non-additive genetic variation in growth, carcass and fertility traits of beef cattle. Genetics, Selection, Evolution 47, 26
| Non-additive genetic variation in growth, carcass and fertility traits of beef cattle.Crossref | GoogleScholarGoogle Scholar | 25880217PubMed |
Brito Lopes F, da Silva MC, Magnabosco CU, Goncalves Narciso M, Sainz RD (2016) Selection indices and multivariate analysis show similar results in the evaluation of growth and carcass traits in beef cattle. PLoS One 11, e0147180
| Selection indices and multivariate analysis show similar results in the evaluation of growth and carcass traits in beef cattle.Crossref | GoogleScholarGoogle Scholar | 26789008PubMed |
Burns B, Corbet N, Corbet D, Crisp J, Venus B, Johnston D, Li Y, McGowan M, Holroyd R (2013) Male traits and herd reproductive capability in tropical beef cattle. 1. Experimental design and animal measures. Animal Production Science 53, 87–100.
| Male traits and herd reproductive capability in tropical beef cattle. 1. Experimental design and animal measures.Crossref | GoogleScholarGoogle Scholar |
Cole J, VanRaden P (2018) Symposium review: possibilities in an age of genomics: the future of selection indices. Journal of Dairy Science 101, 3686–3701.
| Symposium review: possibilities in an age of genomics: the future of selection indices.Crossref | GoogleScholarGoogle Scholar | 29103719PubMed |
Corbet N, Burns B, Corbet D, Johnston D, Crisp J, McGowan M, Prayaga K, Venus B, Holroyd R (2009). Genetic variation in growth, hormonal and seminal traits of young tropically adapted bulls. In ‘Proceedings of the of the 18th Conference of the Association for the Advancement of Animal Breeding and Genetics (AABG)’, Barossa Valley, SA, Australia, 28 September–1 October 2009. pp. 121–124. (Association for the Advancement of Animal Breeding and Genetics)
Corbet N, Burns B, Johnston D, Wolcott ML, Corbet D, Venus B, Li Y, McGowan M, Holroyd R (2013) Male traits and herd reproductive capability in tropical beef cattle. 2. Genetic parameters of bull traits. Animal Production Science 53, 101–113.
| Male traits and herd reproductive capability in tropical beef cattle. 2. Genetic parameters of bull traits.Crossref | GoogleScholarGoogle Scholar |
Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M (2016) Next-generation genotype imputation service and methods. Nature Genetics 48, 1284–1287.
| Next-generation genotype imputation service and methods.Crossref | GoogleScholarGoogle Scholar | 27571263PubMed |
Fairbairn DJ, Blanckenhorn WU, Székely T (2007) ‘Sex, size and gender roles: evolutionary studies of sexual size dimorphism.’ (Oxford University Press)
Falkenberg U, Haertel J, Rotter K, Iwersen M, Arndt G, Heuwieser W (2008) Relationships between the concentration of insulin-like growth factor-1 in serum in dairy cows in early lactation and reproductive performance and milk yield. Journal of Dairy Science 91, 3862–3868.
| Relationships between the concentration of insulin-like growth factor-1 in serum in dairy cows in early lactation and reproductive performance and milk yield.Crossref | GoogleScholarGoogle Scholar | 18832208PubMed |
Fordyce G, Entwistle K, Norman S, Perry V, Gardiner B, Fordyce P (2006) Standardising bull breeding soundness evaluations and reporting in Australia. Theriogenology 66, 1140–1148.
| Standardising bull breeding soundness evaluations and reporting in Australia.Crossref | GoogleScholarGoogle Scholar | 16620941PubMed |
Fortes MR, Porto-Neto LR, Satake N, Nguyen LT, Freitas AC, Melo TP, Scalez DCB, Hayes B, Raidan FS, Reverter A (2020) X chromosome variants are associated with male fertility traits in two bovine populations. Genetics, Selection, Evolution 52, 46
| X chromosome variants are associated with male fertility traits in two bovine populations.Crossref | GoogleScholarGoogle Scholar | 32787790PubMed |
Gargantini G, Cundiff LV, Lunstra DD, Van Vleck LD (2004) Genetic relationships between male and female reproductive traits in beef cattle. Journal of Animal Science 82, 37.
Hansen GR (2006) ‘Managing bull fertility in beef cattle herds.’ (Animal Science Department, Florida Cooperative Extension Service, Institute of Food of Agricultural Sciences, University of Florida: FL, USA)
Hayes B, Goddard M (2010) Genome-wide association and genomic selection in animal breeding. Genome 53, 876–883.
| Genome-wide association and genomic selection in animal breeding.Crossref | GoogleScholarGoogle Scholar | 21076503PubMed |
Hayes B, Fordyce G, Landmark S (2019a) Genomic predictions for fertility traits in tropical beef cattle from a multi-breed, crossbred and composite reference population. In ‘Proceedings of the 23rd Conference of the Association for the Advancement of Animal Breeding and Genetics (AAABG)’, Armidale, NSW, Australia, 27 October – 1 November 2019. pp. 282–285. (Association for the Advancement of Animal Breeding and Genetics)
Hayes BJ, Corbet NJ, Allen JM, Laing AR, Fordyce G, Lyons R, McGowan MR, Burns BM (2019b) Towards multi-breed genomic evaluations for female fertility of tropical beef cattle. Journal of Animal Science 97, 55–62.
| Towards multi-breed genomic evaluations for female fertility of tropical beef cattle.Crossref | GoogleScholarGoogle Scholar | 30371787PubMed |
Jeyaruban M, Johnston D (2017) Genetic Association of Young Male Traits with Female Reproductive Performance in Brahman and Santa Gertrudis Cattle. In ‘Proceedings of the Association for the Advancement of Animal Breeding and Genetics (AABG)’, Townsville, QLD, Australia, 2 July–5 July 2017. pp. 305–308 (Association for the Advancement of Animal Breeding and Genetics.
Johnson T, Keehan M, Harland C, Lopdell T, Spelman R, Davis S, Rosen B, Smith T, Couldrey C (2019) Identification of the pseudoautosomal region in the Hereford bovine reference genome assembly ARS-UCD1. 2. Journal of Dairy Science 102, 3254–3258.
| Identification of the pseudoautosomal region in the Hereford bovine reference genome assembly ARS-UCD1. 2.Crossref | GoogleScholarGoogle Scholar | 30712931PubMed |
Johnston D, Moore K (2019) Genetic Correlations Between Days to Calving and Other Male and Female Reproduction Traits in Brahman Cattle. In ‘Proceedings of the Association for the Advancement of Animal Breeding and Genetics’, pp. 354–357.
Johnston D, Barwick S, Corbet N, Fordyce G, Holroyd R, Williams PJ, Burrow HM (2009) Genetics of heifer puberty in two tropical beef genotypes in northern Australia and associations with heifer-and steer-production traits. Animal Production Science 49, 399–412.
| Genetics of heifer puberty in two tropical beef genotypes in northern Australia and associations with heifer-and steer-production traits.Crossref | GoogleScholarGoogle Scholar |
Johnston D, Barwick S, Fordyce G, Holroyd R, Williams P, Corbet N, Grant T (2014a) Genetics of early and lifetime annual reproductive performance in cows of two tropical beef genotypes in northern Australia. Animal Production Science 54, 1–15.
| Genetics of early and lifetime annual reproductive performance in cows of two tropical beef genotypes in northern Australia.Crossref | GoogleScholarGoogle Scholar |
Johnston D, Corbet N, Barwick S, Wolcott ML, Holroyd R (2014b) Genetic correlations of young bull reproductive traits and heifer puberty traits with female reproductive performance in two tropical beef genotypes in northern Australia. Animal Production Science 54, 74–84.
| Genetic correlations of young bull reproductive traits and heifer puberty traits with female reproductive performance in two tropical beef genotypes in northern Australia.Crossref | GoogleScholarGoogle Scholar |
Land R (1973) The expression of female sex-limited characters in the male. Nature 241, 208–209.
| The expression of female sex-limited characters in the male.Crossref | GoogleScholarGoogle Scholar | 4700891PubMed |
Lande R (1984) The genetic correlation between characters maintained by selection, linkage and inbreeding. Genetical Research 44, 309–320.
| The genetic correlation between characters maintained by selection, linkage and inbreeding.Crossref | GoogleScholarGoogle Scholar | 6530140PubMed |
Larnkjær A, Mølgaard C, Michaelsen KF (2012) Early nutrition impact on the insulin-like growth factor axis and later health consequences. Current Opinion in Clinical Nutrition and Metabolic Care 15, 285–292.
| Early nutrition impact on the insulin-like growth factor axis and later health consequences.Crossref | GoogleScholarGoogle Scholar | 22466924PubMed |
Lesmeister J, Burfening P, Blackwell R (1973) Date of first calving in beef cows and subsequent calf production. Journal of Animal Science 36, 1–6.
| Date of first calving in beef cows and subsequent calf production.Crossref | GoogleScholarGoogle Scholar |
Loh P-R, Danecek P, Palamara PF, Fuchsberger C, Reshef YA, Finucane HK, Schoenherr S, Forer L, McCarthy S, Abecasis GR (2016) Reference-based phasing using the Haplotype Reference Consortium panel. Nature Genetics 48, 1443
| Reference-based phasing using the Haplotype Reference Consortium panel.Crossref | GoogleScholarGoogle Scholar | 27694958PubMed |
Lopez R, Thomas M, Hallford D, Keisler D, Silver G, Obeidat B, Garcia M, Krehbiel C (2006) Metabolic hormone profiles and evaluation of associations of metabolic hormones with body fat and reproductive characteristics of Angus, Brangus, and Brahman heifers. The Professional Animal Scientist 22, 273–282.
| Metabolic hormone profiles and evaluation of associations of metabolic hormones with body fat and reproductive characteristics of Angus, Brangus, and Brahman heifers.Crossref | GoogleScholarGoogle Scholar |
Lunstra D, Cundiff L (2003) Growth and pubertal development in Brahman-, Boran-, Tuli-, Belgian blue-, Hereford- and Angus-sired f1 bulls. Journal of Animal Science 81, 1414–1426.
| Growth and pubertal development in Brahman-, Boran-, Tuli-, Belgian blue-, Hereford- and Angus-sired f1 bulls.Crossref | GoogleScholarGoogle Scholar | 12817488PubMed |
Lunstra D, Ford J, Echternkamp S (1978) Puberty in beef bulls: hormone concentrations, growth, testicular development, sperm production and sexual aggressiveness in bulls of different breeds. Journal of Animal Science 46, 1054–1062.
| Puberty in beef bulls: hormone concentrations, growth, testicular development, sperm production and sexual aggressiveness in bulls of different breeds.Crossref | GoogleScholarGoogle Scholar | 566747PubMed |
Martinez-Velazquez G, Gregory K, Bennett G, Van Vleck LD (2003) Genetic relationships between scrotal circumference and female reproductive traits. Journal of Animal Science 81, 395–401.
| Genetic relationships between scrotal circumference and female reproductive traits.Crossref | GoogleScholarGoogle Scholar | 12643482PubMed |
Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP, O’Connell J, Moore SS, Smith TP, Sonstegard TS (2009) Development and characterization of a high density SNP genotyping assay for cattle. PLoS One 4, e5350
| Development and characterization of a high density SNP genotyping assay for cattle.Crossref | GoogleScholarGoogle Scholar | 19390634PubMed |
Michaelsen KF (2013) Effect of protein intake from 6 to 24 months on insulin-like growth factor 1 (IGF-1) levels, body composition, linear growth velocity, and linear growth acceleration: what are the implications for stunting and wasting? Food and Nutrition Bulletin 34, 268–271.
| Effect of protein intake from 6 to 24 months on insulin-like growth factor 1 (IGF-1) levels, body composition, linear growth velocity, and linear growth acceleration: what are the implications for stunting and wasting?Crossref | GoogleScholarGoogle Scholar | 23964408PubMed |
Misztal I, Tsuruta S, Strabel T, Auvray B, Druet T, Lee D (2002) BLUPF90 and related programs (BGF90). In ‘Proceedings of the 7th world congress on genetics applied to livestock production’, pp. 743–744.
Moore L, Pfeffer A, Chie WN, Miller H, Rogers K, O’Keeffe L (1995) Induction of an acute phase response in lambs causes an increase in plasma levels of GH and IGF-I. The Journal of Endocrinology 144, 243–250.
| Induction of an acute phase response in lambs causes an increase in plasma levels of GH and IGF-I.Crossref | GoogleScholarGoogle Scholar | 7706978PubMed |
Pennell TM, Morrow EH (2013) Two sexes, one genome: the evolutionary dynamics of intralocus sexual conflict. Ecology and Evolution 3, 1819–1834.
| Two sexes, one genome: the evolutionary dynamics of intralocus sexual conflict.Crossref | GoogleScholarGoogle Scholar | 23789088PubMed |
Poissant J, Wilson AJ, Coltman DW (2010) Sex‐specific genetic variance and the evolution of sexual dimorphism: a systematic review of cross‐sex genetic correlations. Evolution 64, 97–107.
| Sex‐specific genetic variance and the evolution of sexual dimorphism: a systematic review of cross‐sex genetic correlations.Crossref | GoogleScholarGoogle Scholar | 19659596PubMed |
Porto-Neto L, Lehnert S, Fortes M, Kelly M, Reverter A (2013a) Population stratification and breed composition of Australian tropically adapted cattle. In ‘Proceedings of the Twentieth Conference of the Association for the Advancement of Animal Breeding and Genetics’, pp. 147–150.
Porto-Neto LR, Sonstegard TS, Liu GE, Bickhart DM, Da Silva MV, Machado MA, Utsunomiya YT, Garcia JF, Gondro C, Van Tassell CP (2013b) Genomic divergence of zebu and taurine cattle identified through high-density SNP genotyping. BMC Genomics 14, 876
| Genomic divergence of zebu and taurine cattle identified through high-density SNP genotyping.Crossref | GoogleScholarGoogle Scholar | 24330634PubMed |
Raidan FS, Porto-Neto LR, Reverter A (2019) Across-sex genomic-assisted genetic correlations for sex-influenced traits in Brahman cattle. Genetics, Selection, Evolution 51, 41
| Across-sex genomic-assisted genetic correlations for sex-influenced traits in Brahman cattle.Crossref | GoogleScholarGoogle Scholar | 31337334PubMed |
Rasali D, Shrestha J, Crow G (2006) Development of composite sheep breeds in the world: a review. Canadian Journal of Animal Science 86, 1–24.
Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E, Rowan TN, Low WY, Zimin A, Couldrey C (2020) De novo assembly of the cattle reference genome with single-molecule sequencing. GigaScience 9, giaa021
| De novo assembly of the cattle reference genome with single-molecule sequencing.Crossref | GoogleScholarGoogle Scholar | 32543654PubMed |
Schatz T, Jayawardhana G, Golding R, Hearnden M (2010) Selection for fertility traits in Brahmans increases heifer pregnancy rates from yearling mating. Animal Production Science 50, 345–348.
| Selection for fertility traits in Brahmans increases heifer pregnancy rates from yearling mating.Crossref | GoogleScholarGoogle Scholar |
Taylor JF (2014) Implementation and accuracy of genomic selection. Aquaculture 420, S8–S14.
| Implementation and accuracy of genomic selection.Crossref | GoogleScholarGoogle Scholar |
Taylor V, Cheng Z, Pushpakumara P, Wathes D, Beever D (2004) Relationships between the plasma concentrations of insulin-like growth factor-I in dairy cows and their fertility and milk yield. The Veterinary Record 155, 583–588.
| Relationships between the plasma concentrations of insulin-like growth factor-I in dairy cows and their fertility and milk yield.Crossref | GoogleScholarGoogle Scholar | 15573950PubMed |
Tiezzi F, Maltecca C (2011) Selecting for female fertility: what can be learned from the dairy experience. Beef Improvement Federation, Research Symposium & Annual Meeting. Montana, USA, pp. 47–60.
VanRaden PM (2008) Efficient methods to compute genomic predictions. Journal of Dairy Science 91, 4414–4423.
| Efficient methods to compute genomic predictions.Crossref | GoogleScholarGoogle Scholar | 18946147PubMed |
Velazquez M, Spicer L, Wathes D (2008) The role of endocrine insulin-like growth factor-I (IGF-I) in female bovine reproduction. Domestic Animal Endocrinology 35, 325–342.
| The role of endocrine insulin-like growth factor-I (IGF-I) in female bovine reproduction.Crossref | GoogleScholarGoogle Scholar | 18703307PubMed |
Weigel KA (2017) Genomic selection of dairy cattle: a review of methods, strategies, and impact. Journal of Animal Breeding and Genetics 1, 1–15.
Wellmann R, Bennewitz J (2019) Key Genetic Parameters for population management. Frontiers in Genetics 10, 667
| Key Genetic Parameters for population management.Crossref | GoogleScholarGoogle Scholar | 31475027PubMed |
Wolcott ML, Johnston D, Barwick S, Corbet N, Burrow HM (2014) Genetic relationships between steer performance and female reproduction and possible impacts on whole herd productivity in two tropical beef genotypes. Animal Production Science 54, 85–96.
| Genetic relationships between steer performance and female reproduction and possible impacts on whole herd productivity in two tropical beef genotypes.Crossref | GoogleScholarGoogle Scholar |
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics 88, 76–82.
| GCTA: a tool for genome-wide complex trait analysis.Crossref | GoogleScholarGoogle Scholar | 21167468PubMed |
Yilmaz A, Davis M, Simmen R (2004) Estimation of (co) variance components for reproductive traits in Angus beef cattle divergently selected for blood serum IGF-I concentration. Journal of Animal Science 82, 2285–2292.
| Estimation of (co) variance components for reproductive traits in Angus beef cattle divergently selected for blood serum IGF-I concentration.Crossref | GoogleScholarGoogle Scholar | 15318726PubMed |
Yilmaz A, Davis M, Simmen R (2006) Analysis of female reproductive traits in Angus beef cattle divergently selected for blood serum insulin-like growth factor I concentration. Theriogenology 65, 1180–1190.
| Analysis of female reproductive traits in Angus beef cattle divergently selected for blood serum insulin-like growth factor I concentration.Crossref | GoogleScholarGoogle Scholar | 16144706PubMed |
Zhang Y, Johnston D, Bolormaa S, Hawken R, Tier B (2014) Genomic selection for female reproduction in Australian tropically adapted beef cattle. Animal Production Science 54, 16–24.
| Genomic selection for female reproduction in Australian tropically adapted beef cattle.Crossref | GoogleScholarGoogle Scholar |
Zhang X, Tsuruta S, Andonov S, Lourenco D, Sapp R, Wang C, Misztal I (2018) Relationships among mortality, performance, and disorder traits in broiler chickens: a genetic and genomic approach. Poultry Science 97, 1511–1518.
| Relationships among mortality, performance, and disorder traits in broiler chickens: a genetic and genomic approach.Crossref | GoogleScholarGoogle Scholar | 29529319PubMed |
Zulu VC, Nakao T, Sawamukai Y (2002) Insulin-like growth factor-I as a possible hormonal mediator of nutritional regulation of reproduction in cattle. The Journal of Veterinary Medical Science 64, 657–665.
| Insulin-like growth factor-I as a possible hormonal mediator of nutritional regulation of reproduction in cattle.Crossref | GoogleScholarGoogle Scholar | 12237508PubMed |