109 Utilising cell-free DNA for detection of gene editing outcomes in rhesus macaque embryos
J. Ryu A , W. Chan A , F. Carvalho B , E. Mishler B , J. Hennebold A and C. Hanna BA Division of Reproductive & Developmental Sciences, Oregon National Primate Research Center, Oregon Health & Science University, Beaverton, OR, USA
B Assisted Reproductive Technologies Core, Oregon National Primate Research Center, Oregon Health & Science University, Beaverton, OR, USA
Reproduction, Fertility and Development 34(2) 291-292 https://doi.org/10.1071/RDv34n2Ab109
Published: 7 December 2021
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing on behalf of the IETS
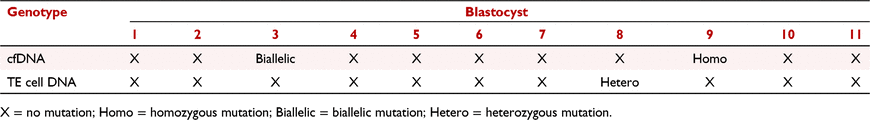
CRISPR/Cas9 injection into zygotes enables the production of genetically modified nonhuman primates (NHPs) essential for modelling specific human diseases. Target gene modification can be confirmed through trophectoderm (TE) cell biopsy and subsequent genotyping to avoid the transfer of non-mutated embryos. However, in addition to being labour-intensive, TE cell biopsies are invasive and require significant disruption to the trophectoderm layer. Recently, isolation of embryonic cell-free DNA (cfDNA) from spent embryo culture medium was proposed for aneuploidy detection in human in vitro-derived embryos. Collecting cfDNA is less labour-intensive than performing TE cell biopsies and is not invasive. Thus, we investigated whether cfDNA can be used to assess CRISPR/Cas9-mediated gene editing in NHP embryos. Cas9 and two guide (g)RNAs targeting exon 3 of the myosin 7A (MYO7A) gene were injected into the pronucleus and cytoplasm of rhesus macaque zygotes 14 h after in vitro insemination. Injected embryos were cultured individually in 20 µL of BO-IVC (IVF Bioscience) under oil at 37°C in 5/5/90 (%CO2, %O2, %N2) mixed gas. Spent embryo culture media and TE cell biopsies were collected from expanded blastocysts (n = 15) on Day 8 to assess MYO7A mutations. The region flanking the two MYO7A gRNA binding sites was PCR amplified from TE cell DNA after whole-genome amplification (WGA) or directly from cfDNA isolated from the culture medium. PCR amplicons were analysed by Sanger DNA sequencing. MYO7A amplicons were obtained from 10 of 15 blastocysts. Biallelic mutations and a homozygous mutation were detected from two cfDNA samples, whereas the corresponding TE cell biopsy of these blastocysts carried no mutations (Table 1). In another blastocyst, the PCR amplified cfDNA possessed the wild-type MYO7A sequence and the amplified DNA from TE cells included a heterozygous MYO7A mutation. For the remaining seven blastocysts, the TE cell DNA and medium cfDNA both possessed the wild-type MYO7A sequence. These results demonstrate that analysis of cfDNA for MYO7A mutations did not consistently identify the same genotype as what was obtained from TE biopsy DNA. Different genotyping outcomes between cfDNA and TE cell biopsy may be caused by maternal DNA contamination such as polar bodies and somatic cells. Moreover, because TE biopsies include a small number of cells (∼10 to 12), it is possible the full complement of mutations are missed. Further optimisation of cfDNA collection and analysis methods is required before it can dependably be used for rhesus macaque embryo genotyping.
|


