Transcription profiles of oocytes during maturation and embryos during preimplantation development in vivo in the goat
Yunsheng Li A * , Jiangwen Sun B * , Yinghui Ling A * , Hao Ming C , Zhen Chen A , Fugui Fang A , Ya Liu A , Hongguo Cao A , Jianping Ding A , Zubing Cao
A * , Jiangwen Sun B * , Yinghui Ling A * , Hao Ming C , Zhen Chen A , Fugui Fang A , Ya Liu A , Hongguo Cao A , Jianping Ding A , Zubing Cao  A , Xiaorong Zhang A , Kenneth Bondioli C , Zongliang Jiang
A , Xiaorong Zhang A , Kenneth Bondioli C , Zongliang Jiang  C D and Yunhai Zhang A D
C D and Yunhai Zhang A D
A Anhui Province Key Laboratory of Local Livestock and Poultry Genetical Resource Conservation and Breeding, College of Animal Science and Technology, Anhui Agricultural University, Hefei 230036, China.
B Department of Computer Science, College of Science, Old Dominion University, Norfolk, VA 23529, USA.
C School of Animal Sciences, AgCenter, Louisiana State University, Baton Rouge, LA 70803, USA.
D Corresponding authors. Emails: zjiang@agcenter.lsu.edu; yunhaizhang@ahau.edu.cn
Reproduction, Fertility and Development 32(7) 714-725 https://doi.org/10.1071/RD19391
Submitted: 09 October 2019 Accepted: 30 January 2020 Published: 15 April 2020
Abstract
RNA sequencing performed on goat matured oocytes and preimplantation embryos generated in vivo enabled us to define the transcriptome for goat preimplantation embryo development. The largest proportion of changes in gene expression in goat was found at the 16-cell stage, not as previously defined at the 8-cell stage, and is later than in other mammalian species. In all, 6482 genes were identified to be significantly differentially expressed across all consecutive developmental stage comparisons, and the important signalling pathways involved in each development transition were determined. In addition, we identified genes that appear to be transcribed only at a specific stage of development. Using weighted gene coexpression network analysis, we found nine stage-specific modules of coexpressed genes that represent the corresponding stage of development. Furthermore, we identified conserved key members (or hub genes) of the goat transcriptional networks. Their association with other embryo genes suggests that they may have important regulatory roles in embryo development. Our cross-mammalian species transcriptomic comparisons demonstrate both conserved and goat-specific features of preimplantation development.
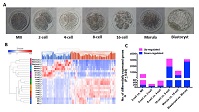
Additional keywords: embryonic genome activation, RNA sequencing.
References
Baguisi, A., Behboodi, E., Melican, D. T., Pollock, J. S., Destrempes, M. M., Cammuso, C., Williams, J. L., Nims, S. D., Porter, C. A., Midura, P., Palacios, M. J., Ayres, S. L., Denniston, R. S., Hayes, M. L., Ziomek, C. A., Meade, H. M., Godke, R. A., Gavin, W. G., Overstrom, E. W., and Echelard, Y. (1999). Production of goats by somatic cell nuclear transfer. Nat. Biotechnol. 17, 456–461.| Production of goats by somatic cell nuclear transfer.Crossref | GoogleScholarGoogle Scholar | 10331804PubMed |
Bao, Z. K., Lin, J., Ye, L. L., Zhang, Q., Chen, J. Q., Yang, Q., and Yu, Q. H. (2016). Modulation of mammary gland development and milk production by growth hormone expression in GH transgenic goats. Front. Physiol. 7, 278.
Boggavarapu, N. R., Lalitkumar, S., Joshua, V., Kasvandik, S., Salumets, A., Lalitkumar, P. G., and Gemzell-Danielsson, K. (2016). Compartmentalized gene expression profiling of receptive endometrium reveals progesterone regulated ENPP3 is differentially expressed and secreted in glycosylated form. Sci. Rep. 6, 33811.
| Compartmentalized gene expression profiling of receptive endometrium reveals progesterone regulated ENPP3 is differentially expressed and secreted in glycosylated form.Crossref | GoogleScholarGoogle Scholar | 27665743PubMed |
Boroviak, T., Stirparo, G. G., Dietmann, S., Hernando-Herraez, I., Mohammed, H., Reik, W., Smith, A., Sasaki, E., Nichols, J., and Bertone, P. (2018). Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development 145, dev167833.
| Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development.Crossref | GoogleScholarGoogle Scholar | 30413530PubMed |
Burton, A., and Torres-Padilla, M. E. (2014). Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 15, 723–735.
| Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis.Crossref | GoogleScholarGoogle Scholar | 25303116PubMed |
Cao, S., Han, J., Wu, J., Li, Q., Liu, S., Zhang, W., Pei, Y., Ruan, X., Liu, Z., Wang, X., Lim, B., and Li, N. (2014). Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing. BMC Genomics 15, 4.
| Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing.Crossref | GoogleScholarGoogle Scholar | 24383959PubMed |
Chuang, C. Y., Lin, K. I., Hsiao, M., Stone, L., Chen, H. F., Huang, Y. H., Lin, S. P., Ho, H. N., and Kuo, H. C. (2012). Meiotic competent human germ cell-like cells derived from human embryonic stem cells induced by BMP4/WNT3A signaling and OCT4/EpCAM (epithelial cell adhesion molecule) selection. J. Biol. Chem. 287, 14389–14401.
| Meiotic competent human germ cell-like cells derived from human embryonic stem cells induced by BMP4/WNT3A signaling and OCT4/EpCAM (epithelial cell adhesion molecule) selection.Crossref | GoogleScholarGoogle Scholar | 22396540PubMed |
Cortazar, D., Kunz, C., Selfridge, J., Lettieri, T., Saito, Y., MacDougall, E., Wirz, A., Schuermann, D., Jacobs, A. L., Siegrist, F., Steinacher, R., Jiricny, J., Bird, A., and Schar, P. (2011). Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470, 419–423.
| Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability.Crossref | GoogleScholarGoogle Scholar | 21278727PubMed |
de Vries, F. A., de Boer, E., van den Bosch, M., Baarends, W. M., Ooms, M., Yuan, L., Liu, J. G., van Zeeland, A. A., Heyting, C., and Pastink, A. (2005). Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 19, 1376–1389.
| Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation.Crossref | GoogleScholarGoogle Scholar | 15937223PubMed |
Deng, M., Liu, Z., Ren, C., Zhang, G., Pang, J., Zhang, Y., Wang, F., and Wan, Y. (2018a). Long noncoding RNAs exchange during zygotic genome activation in goat. Biol. Reprod. 99, 707–717.
| Long noncoding RNAs exchange during zygotic genome activation in goat.Crossref | GoogleScholarGoogle Scholar | 29771291PubMed |
Deng, R., Han, C., Zhao, L., Zhang, Q., Yan, B., Cheng, R., Wei, B., Meng, P., Mao, T., Zhang, Y., and Liu, J. (2018b). Identification and characterization of ERV transcripts in goat embryos. Reproduction 157, 115–126.
| Identification and characterization of ERV transcripts in goat embryos.Crossref | GoogleScholarGoogle Scholar |
Fernandez-Gonzalez, R., Ramirez, M. A., Pericuesta, E., Calle, A., and Gutierrez-Adan, A. (2010). Histone modifications at the blastocyst Axin1(Fu) locus mark the heritability of in vitro culture-induced epigenetic alterations in mice. Biol. Reprod. 83, 720–727.
| Histone modifications at the blastocyst Axin1(Fu) locus mark the heritability of in vitro culture-induced epigenetic alterations in mice.Crossref | GoogleScholarGoogle Scholar | 20650886PubMed |
Graf, A., Krebs, S., Zakhartchenko, V., Schwalb, B., Blum, H., and Wolf, E. (2014). Fine mapping of genome activation in bovine embryos by RNA sequencing. Proc. Natl Acad. Sci. USA 111, 4139–4144.
| Fine mapping of genome activation in bovine embryos by RNA sequencing.Crossref | GoogleScholarGoogle Scholar | 24591639PubMed |
Huan, Y., Wang, H., Wu, Z., Zhang, J., Liu, Z., and He, H. (2015). The expression patterns of DNA methylation reprogramming related genes are associated with the developmental competence of cloned embryos after zygotic genome activation in pigs. Gene Expr. Patterns 18, 1–7.
| The expression patterns of DNA methylation reprogramming related genes are associated with the developmental competence of cloned embryos after zygotic genome activation in pigs.Crossref | GoogleScholarGoogle Scholar | 25917378PubMed |
Jiang, Z., Sun, J., Dong, H., Luo, O., Zheng, X., Obergfell, C., Tang, Y., Bi, J., O’Neill, R., Ruan, Y., Chen, J., and Tian, X. C. (2014). Transcriptional profiles of bovine in vivo pre-implantation development. BMC Genomics 15, 756.
| Transcriptional profiles of bovine in vivo pre-implantation development.Crossref | GoogleScholarGoogle Scholar | 25185836PubMed |
Kawamura, Y., Uchijima, Y., Horike, N., Tonami, K., Nishiyama, K., Amano, T., Asano, T., Kurihara, Y., and Kurihara, H. (2010). Sirt3 protects in vitro-fertilized mouse preimplantation embryos against oxidative stress-induced p53-mediated developmental arrest. J. Clin. Invest. 120, 2817–2828.
| Sirt3 protects in vitro-fertilized mouse preimplantation embryos against oxidative stress-induced p53-mediated developmental arrest.Crossref | GoogleScholarGoogle Scholar | 20644252PubMed |
Kepkova, K. V., Vodicka, P., Toralova, T., Lopatarova, M., Cech, S., Dolezel, R., Havlicek, V., Besenfelder, U., Kuzmany, A., Sirard, M. A., Laurincik, J., and Kanka, J. (2011). Transcriptomic analysis of in vivo and in vitro produced bovine embryos revealed a developmental change in cullin 1 expression during maternal-to-embryonic transition. Theriogenology 75, 1582–1595.
| Transcriptomic analysis of in vivo and in vitro produced bovine embryos revealed a developmental change in cullin 1 expression during maternal-to-embryonic transition.Crossref | GoogleScholarGoogle Scholar | 21411133PubMed |
Knijn, H. M., Wrenzycki, C., Hendriksen, P. J., Vos, P. L., Zeinstra, E. C., van der Weijden, G. C., Niemann, H., and Dieleman, S. J. (2005). In vitro and in vivo culture effects on mRNA expression of genes involved in metabolism and apoptosis in bovine embryos. Reprod. Fertil. Dev. 17, 775–784.
| In vitro and in vivo culture effects on mRNA expression of genes involved in metabolism and apoptosis in bovine embryos.Crossref | GoogleScholarGoogle Scholar | 16476204PubMed |
Kouznetsova, A., Benavente, R., Pastink, A., and Hoog, C. (2011). Meiosis in mice without a synaptonemal complex. PLoS One 6, e28255.
| Meiosis in mice without a synaptonemal complex.Crossref | GoogleScholarGoogle Scholar | 22164254PubMed |
Kues, W. A., Sudheer, S., Herrmann, D., Carnwath, J. W., Havlicek, V., Besenfelder, U., Lehrach, H., Adjaye, J., and Niemann, H. (2008). Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc. Natl Acad. Sci. USA 105, 19768–19773.
| Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo.Crossref | GoogleScholarGoogle Scholar | 19064908PubMed |
Langfelder, P., and Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559.
| WGCNA: an R package for weighted correlation network analysis.Crossref | GoogleScholarGoogle Scholar | 19114008PubMed |
Langfelder, P., Luo, R., Oldham, M. C., and Horvath, S. (2011). Is my network module preserved and reproducible? PLOS Comput. Biol. 7, e1001057.
| Is my network module preserved and reproducible?Crossref | GoogleScholarGoogle Scholar | 21283776PubMed |
Laplante, M., and Sabatini, D. M. (2009). mTOR signaling at a glance. J. Cell Sci. 122, 3589–3594.
| mTOR signaling at a glance.Crossref | GoogleScholarGoogle Scholar | 19812304PubMed |
Lee, M. T., Bonneau, A. R., and Giraldez, A. J. (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol. 30, 581–613.
| Zygotic genome activation during the maternal-to-zygotic transition.Crossref | GoogleScholarGoogle Scholar | 25150012PubMed |
Li, T., Vu, T. H., Ulaner, G. A., Littman, E., Ling, J. Q., Chen, H. L., Hu, J. F., Behr, B., Giudice, L., and Hoffman, A. R. (2005). IVF results in de novo DNA methylation and histone methylation at an Igf2–H19 imprinting epigenetic switch. Mol. Hum. Reprod. 11, 631–640.
| IVF results in de novo DNA methylation and histone methylation at an Igf2–H19 imprinting epigenetic switch.Crossref | GoogleScholarGoogle Scholar | 16219628PubMed |
Lonergan, P., Rizos, D., Gutierrez-Adan, A., Fair, T., and Boland, M. P. (2003). Effect of culture environment on embryo quality and gene expression – experience from animal studies. Reprod. Biomed. Online 7, 657–663.
| Effect of culture environment on embryo quality and gene expression – experience from animal studies.Crossref | GoogleScholarGoogle Scholar | 14748964PubMed |
Menchaca, A., Anegon, I., Whitelaw, C. B. A., Baldassarre, H., and Crispo, M. (2016). New insights and current tools for genetically engineered (GE) sheep and goats. Theriogenology 86, 160–169.
| New insights and current tools for genetically engineered (GE) sheep and goats.Crossref | GoogleScholarGoogle Scholar | 27155732PubMed |
Misirlioglu, M., Page, G. P., Sagirkaya, H., Kaya, A., Parrish, J. J., First, N. L., and Memili, E. (2006). Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc. Natl Acad. Sci. USA 103, 18905–18910.
| Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos.Crossref | GoogleScholarGoogle Scholar | 17142320PubMed |
Murakami, M., Ichisaka, T., Maeda, M., Oshiro, N., Hara, K., Edenhofer, F., Kiyama, H., Yonezawa, K., and Yamanaka, S. (2004). mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 24, 6710–6718.
| mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells.Crossref | GoogleScholarGoogle Scholar | 15254238PubMed |
Noguchi, T., Aizawa, T., Munakata, Y., and Iwata, H. (2019). Comparison of gene expression and mitochondria number between bovine blastocysts obtained in vitro and in vivo. J. Reprod. Dev. 66, 35–39.
| Comparison of gene expression and mitochondria number between bovine blastocysts obtained in vitro and in vivo.Crossref | GoogleScholarGoogle Scholar | 31748448PubMed |
Pan, H., and Schultz, R. M. (2011). Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol. Reprod. 85, 409–416.
| Sox2 modulates reprogramming of gene expression in two-cell mouse embryos.Crossref | GoogleScholarGoogle Scholar | 21543769PubMed |
Paramio, M. T., and Izquierdo, D. (2014). Current status of in vitro embryo production in sheep and goats. Reprod. Domest. Anim. 49, 37–48.
| Current status of in vitro embryo production in sheep and goats.Crossref | GoogleScholarGoogle Scholar | 25277431PubMed |
Paramio, M. T., and Izquierdo, D. (2016). Recent advances in in vitro embryo production in small ruminants. Theriogenology 86, 152–159.
| Recent advances in in vitro embryo production in small ruminants.Crossref | GoogleScholarGoogle Scholar | 27157391PubMed |
Pfeifer, N., Baston-Bust, D. M., Hirchenhain, J., Friebe-Hoffmann, U., Rein, D. T., Krussel, J. S., and Hess, A. P. (2012). Selection of the in vitro culture media influences mRNA expression of Hedgehog genes, Il-6, and important genes regarding reactive oxygen species in single murine preimplantation embryos. ScientificWorldJournal 2012, 479315.
| Selection of the in vitro culture media influences mRNA expression of Hedgehog genes, Il-6, and important genes regarding reactive oxygen species in single murine preimplantation embryos.Crossref | GoogleScholarGoogle Scholar | 22919324PubMed |
Picelli, S., Björklund, Å. K., Faridani, O. R., Sagasser, S., Winberg, G., and Sandberg, R. (2013). Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098.
| Smart-seq2 for sensitive full-length transcriptome profiling in single cells.Crossref | GoogleScholarGoogle Scholar | 24056875PubMed |
Pivko, J., Grafenau, P., and Kopecny, V. (1995). Nuclear fine structure and transcription in early goat embryos. Theriogenology 44, 661–671.
| Nuclear fine structure and transcription in early goat embryos.Crossref | GoogleScholarGoogle Scholar | 16727764PubMed |
Ramsköld, D., Luo, S., Wang, Y. C., Li, R., Deng, Q., Faridani, O. R., Daniels, G. A., Khrebtukova, I., Loring, J. F., Laurent, L. C., Schroth, G. P., and Sandberg, R. (2012). Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 30, 777–782.
| Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells.Crossref | GoogleScholarGoogle Scholar | 22820318PubMed |
Saenz-de-Juano, M. D., Naturil-Alfonso, C., Vicente, J. S., and Marco-Jimenez, F. (2013). Effect of different culture systems on mRNA expression in developing rabbit embryos. Zygote 21, 103–109.
| Effect of different culture systems on mRNA expression in developing rabbit embryos.Crossref | GoogleScholarGoogle Scholar | 21838964PubMed |
Saito, Y., Ono, T., Takeda, N., Nohmi, T., Seki, M., Enomoto, T., Noda, T., and Uehara, Y. (2012). Embryonic lethality in mice lacking mismatch-specific thymine DNA glycosylase is partially prevented by DOPS, a precursor of noradrenaline. Tohoku J. Exp. Med. 226, 75–83.
| Embryonic lethality in mice lacking mismatch-specific thymine DNA glycosylase is partially prevented by DOPS, a precursor of noradrenaline.Crossref | GoogleScholarGoogle Scholar | 22200605PubMed |
Wang, X., Yu, H., Lei, A., Zhou, J., Zeng, W., Zhu, H., Dong, Z., Niu, Y., Shi, B., Cai, B., Liu, J., Huang, S., Yan, H., Zhao, X., Zhou, G., He, X., Chen, X., Yang, Y., Jiang, Y., Shi, L., Tian, X., Wang, Y., Ma, B., Huang, X., Qu, L., and Chen, Y. (2015). Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci. Rep. 5, 13878.
| Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system.Crossref | GoogleScholarGoogle Scholar | 26354037PubMed |
Wang, T. R., Zhang, M., Jiang, Z. L., and Seli, E. (2017). Mitochondrial dysfunction and ovarian aging. Am. J. Reprod. Immunol. 77, e12651.
Xu, X., Watt, D. S., and Liu, C. (2016). Multifaceted roles for thymine DNA glycosylase in embryonic development and human carcinogenesis. Acta Biochim. Biophys. Sin. (Shanghai) 48, 82–89.
| 26370152PubMed |
Xue, Z., Huang, K., Cai, C., Cai, L., Jiang, C. Y., Feng, Y., Liu, Z., Zeng, Q., Cheng, L., Sun, Y. E., Liu, J. Y., Horvath, S., and Fan, G. (2013). Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 500, 593–597.
| Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing.Crossref | GoogleScholarGoogle Scholar | 23892778PubMed |
Yu, D. W., Wang, J., Zou, H. Y., Feng, T., Chen, L., Li, J., Qi, X. L., Li, Z. F., Duan, X. Y., Xu, C. L., Zhang, L., Long, X., Lan, J., Chen, C., Wang, C., Xu, X. Y., Ren, J. L., Zhao, Y. Q., Hu, X. X., Lian, Z. X., Men, H. S., Pan, D. K., Li, N., Capecchi, M. R., Du, X. G., Zhao, Y. F., and Wu, S. (2018). Silencing of retrotransposon-derived imprinted gene RTL1 is the main cause for postimplantational failures in mammalian cloning. Proc. Natl Acad. Sci. USA 115, E11071–E11080.
| Silencing of retrotransposon-derived imprinted gene RTL1 is the main cause for postimplantational failures in mammalian cloning.Crossref | GoogleScholarGoogle Scholar |
Yurttas, P., Vitale, A. M., Fitzhenry, R. J., Cohen-Gould, L., Wu, W., Gossen, J. A., and Coonrod, S. A. (2008). Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo. Development 135, 2627–2636.
| Role for PADI6 and the cytoplasmic lattices in ribosomal storage in oocytes and translational control in the early mouse embryo.Crossref | GoogleScholarGoogle Scholar | 18599511PubMed |
Zhang, B., and Horvath, S. (2005). A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4, e17.
| A general framework for weighted gene co-expression network analysis.Crossref | GoogleScholarGoogle Scholar | 16646834PubMed |
Zhang, K., and Smith, G. W. (2015). Maternal control of early embryogenesis in mammals. Reprod. Fertil. Dev. 27, 880–896.
| Maternal control of early embryogenesis in mammals.Crossref | GoogleScholarGoogle Scholar | 25695370PubMed |
Zhang, M., Bener, M. B., Jiang, Z. L., Wang, T. R., Esencan, E., Scott, R., Horvath, T., and Seli, E. (2019). Mitofusin 1 is required for female fertility and to maintain ovarian follicular reserve. Cell Death Dis. 10, 560.
| Mitofusin 1 is required for female fertility and to maintain ovarian follicular reserve.Crossref | GoogleScholarGoogle Scholar | 31332167PubMed |
Zhu, H. M., Hu, L. Y., Liu, J., Chen, H. T., Cui, C. C., Song, Y. J., Jin, Y. P., and Zhang, Y. (2016). Generation of beta-lactoglobulin-modified transgenic goats by homologous recombination. FEBS J. 283, 4600–4613.
| Generation of beta-lactoglobulin-modified transgenic goats by homologous recombination.Crossref | GoogleScholarGoogle Scholar |


