Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring
Nicolas Virlet A , Kasra Sabermanesh A , Pouria Sadeghi-Tehran A and Malcolm J. Hawkesford A BA Department of Plant Biology and Crop Science, Rothamsted Research, Harpenden, Herts AL5 2JQ, UK.
B Corresponding author. Email: malcolm.hawkesford@rothamsted.ac.uk
Functional Plant Biology 44(1) 143-153 https://doi.org/10.1071/FP16163
Submitted: 28 April 2016 Accepted: 2 September 2016 Published: 2 November 2016
Journal Compilation © Published Open Access CC BY 2017 Open Access CC BY-NC-ND
Abstract
Current approaches to field phenotyping are laborious or permit the use of only a few sensors at a time. In an effort to overcome this, a fully automated robotic field phenotyping platform with a dedicated sensor array that may be accurately positioned in three dimensions and mounted on fixed rails has been established, to facilitate continual and high-throughput monitoring of crop performance. Employed sensors comprise of high-resolution visible, chlorophyll fluorescence and thermal infrared cameras, two hyperspectral imagers and dual 3D laser scanners. The sensor array facilitates specific growth measurements and identification of key growth stages with dense temporal and spectral resolution. Together, this platform produces a detailed description of canopy development across the crops entire lifecycle, with a high-degree of accuracy and reproducibility.
Additional keywords: data processing, computer vision, field scanalyzer, nitrogen, phenomics, scanalyzer.
Introduction
The requirement for crop production is projected to double by 2050 to meet the demand represented by the rapidly growing human population and environmental changes (Tilman et al. 2011). Furthermore, the increase in crop production must be achieved sustainably, whereby reducing agricultural inputs, especially nitrogenous fertilisers, if we are to reduce environmental degradation caused by our agricultural footprint (Tester and Langridge 2010). In order to meet crop production demands, yields need to increase by 2.4% per annum. However, although breeding and agronomic efforts over the past 50 years have been responsible for tripling cereal yields (Pingali 2012), annual yield increase targets for all three major cereal crops (rice, maize and wheat) are no longer being achieved through traditional breeding programs (Tester and Langridge 2010). This highlights a need to exploit advances in genotyping and phenotyping methods to identify novel traits, and broaden the available genetic diversity of existing traits, in order to accelerate increases in genetic improvement and yield. There has been a rapid progression in functional genomics and genetic technologies over the past 20 years; however, the ability to exploit available genomic tools (sequencing technology, molecular markers) to their full potential are now limited by the ability to phenotype (Araus and Cairns 2014).
Several phenotyping platforms have been developed to increase the precision, resolution, and throughput of phenotyping, including controlled environment-based systems, aerial solutions and various ground-based platforms, each with their own advantages and limitations (Deery et al. 2014). Indeed, high-throughput phenotyping platforms (HTPPs) in controlled environments have enabled detailed non-invasive observations of individual plants in potted soil. However, the resulting quantitative trait loci and candidate genes identified have generally not translated into gains in grain yield in the field, as potted plants experience far different environments to those in the field (Passioura and Angus 2010; Passioura 2012). The current generation of aerial HTPPs, such as unmanned aerial vehicles (UAVs) (Sankaran et al. 2015), and blimps (Losos et al. 2013) enable rapid data acquisition of numerous plots within a field, simultaneously. Although blimps have a payload of several kilograms, UAVs, such as polycopters, generally only carry up to 2 kg, limiting the number of sensors that can be used at any given time (Sankaran et al. 2015). Spatial resolution is also usually limited in such aerial vehicles.
Ground-based HTPPs for the field (often referred to as ‘phenomobiles’) hold an advantage over controlled environment- based platforms, as they operate directly in the field, can be used across multiple sites and have a potential for high spatial resolution. The attachment of cameras and sensors onto a tall-erecting vehicle, such as a cherry-picker (often referred to as ‘phenotowers’), increase the throughput of image acquisition, relative to phenomobiles, however achieve with a lower spatial resolution (Rascher et al. 2011). Several different GPS-navigated vehicles, manned or semi-autonomous (Deery et al. 2014), have been developed in recent years and are equipped to simultaneously collect detailed phenotypic data of row crops using spectral sensors, as well as thermal and 3D imagers, at plot-level. However, a key caveat with the use of such vehicles is that they still require some level of supervision, and the mass of the vehicles may detrimentally influence the underlying soil structure, particularly over repeated-measurements, which is critical for the support of plant life (Bronick and Lal 2005). Here, we present the field-based HTPP installed at Rothamsted Research by LemnaTec GmbH, the ‘Field Scanalyzer’, which is being used for crop phenotyping.
The Field Scanalyzer
Rothamsted’s Field Scanalyzer is a fully-automated, high-throughput, fixed-site phenotyping platform, carrying multiple sensors for non-invasive monitoring of plant growth, morphology, physiology and health. Automation enables the Field Scanalyzer to be exploited for detailed large scale screening of germplasm with high temporal resolution. Detailed monitoring of crops in the field will help to (i) quantify traits contributing to crop performance and may serve as yield predictors, (ii) identify novel traits, and (iii) dissect genetic control of complex traits. The information obtained are outputs that may be utilised directly by breeders to generate new elite germplasm.
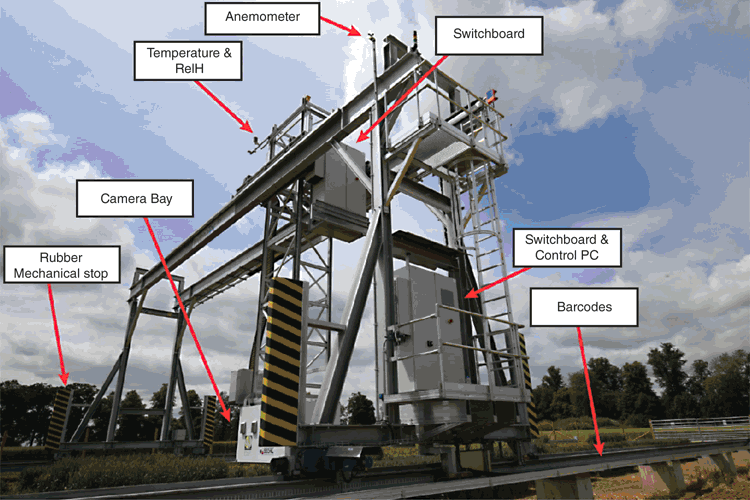
The platform incorporates an overhead gantry that carries a camera box, which houses eight sensors and moves in three main axes; along rails (x-axis); perpendicular to rails with trolley (y-axis); lifter arm which moves the camera box (z-axis). All three axes have end position safety limiters in three hierarchies, including software and hardware end position, as well as a mechanical rubber end stop (Fig. 1). The Field Scanalyzer is robust enough to cope with harsh environmental conditions, with high sampling frequency and greenhouse accuracy. However, the system has been programmed to secure itself in wind-lock mode (safe position), in the middle of the field, when threshold wind speeds are reached (10 m s–1).
The platform is 125 m × 15 m × 6 m in size and crops within a specified area of xmax = 115.8 m; ymax = 11.3 m; zmax = 4.1 m can be monitored throughout the season with a high degree of resolution (spatial and temporal) and reproducibility. The 6 m height of the platform was chosen to provide sufficient vertical clearance to operate any farm machinery if required. The concept is not restricted to these dimensions. LemnaTec control software is used on a master computer to run the gantry in automatic mode and transfer data to a dedicated database that is accessible remotely via the network. The platform operates through scripts that are written in C# programming language. The scripts select and initialise different cameras and illumination settings for the appropriate application. Once the platform is programmed, the system can operate 24 h day–1, throughout the year, with minimal human supervision. The platform may also be remotely controlled in manual mode.
For any moving system, it is essential that parts work together seamlessly, with high level of accuracy. It is the combination of high-resolution sensors and accurate positioning of these sensors that ensures reproducible measurements, which can then be used to accurately monitor growth and development. In order to achieve precise gantry positioning, barcodes are installed on the rails of the x- and y-axes (Fig. 1), which determines the position and speed relative to a barcode tape. In order to evaluate the repositioning accuracy of the Field Scanalyzer, a test was carried out sending the gantry to the test position, returning to a random position on the field over 100 iterations. At each iteration, the on-board RGB camera imaged a checkerboard, resulting in a series of images that were used to calculate a displacement of the sensor position and presented a high accuracy of 25 pixels as the mean x-axis repositioning error, and 5 pixels for the y-axis. In this case, where the red, green, blue (RGB) camera is 2.5 m above the canopy, a resolution of 1 pixel is equivalent to ~0.15 mm.
Sensor array
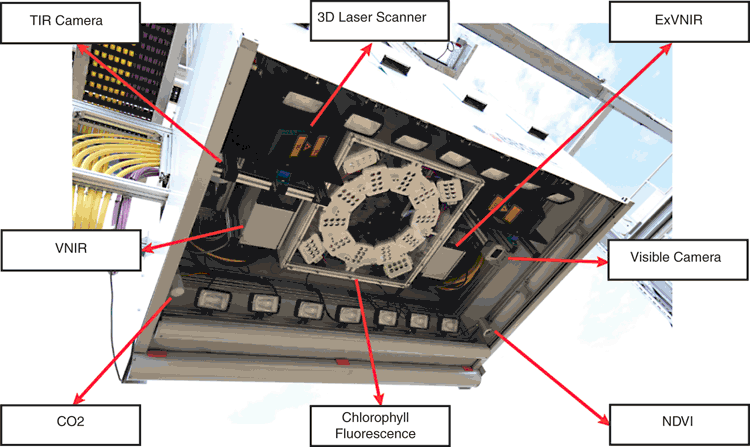
The camera box is equipped with state of the art cameras, sensors and illumination, as well as a temperature sensors to regulate temperature. All cameras are mounted in separate weatherproof housings, and are moved over the field by the supporting gantry (Fig. 2). The maximum total payload of the camera box is 500 kg, which enables the capacity to carry less portable camera technologies, such as the Field Scanalyzer’s chlorophyll fluorescence imager (CFI; 120 kg) and two hyperspectral mirror scanners (30 kg each). The Field Scanalyzer can be programmed to capture images using one or multiple cameras and sensors during a scan, sequentially. However, no two sensors can simultaneously capture images. Fig. 3 illustrates examples taken from five sensors within the Field Scanalyzer’s sensor array.
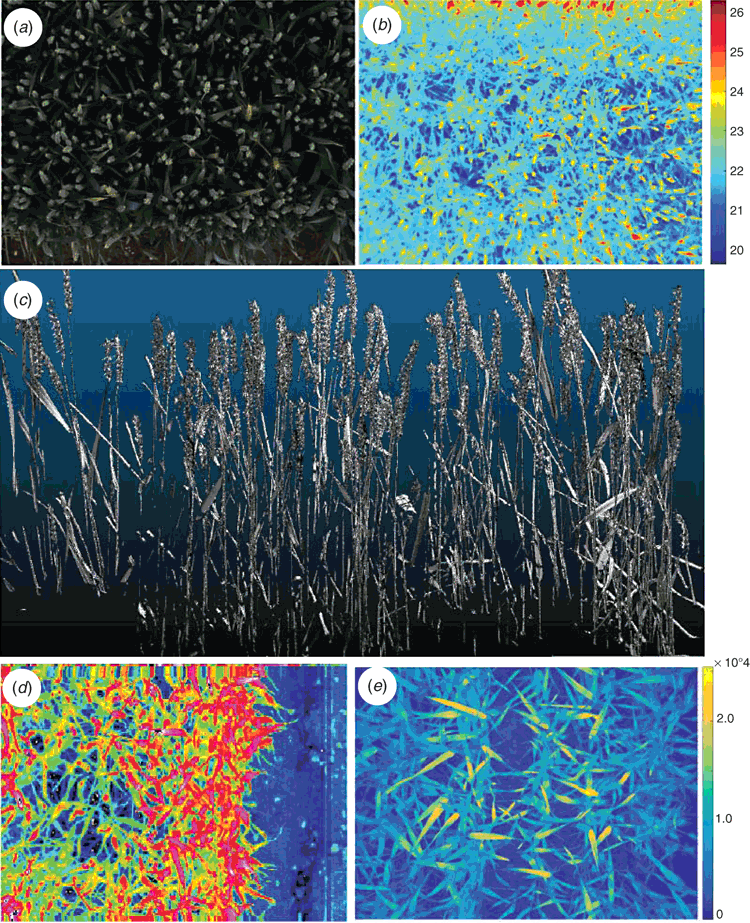
A high-resolution (3296 × 2472 pixels) 12 colour bit Prosilica GT3300 (Allied Vision), with a maximum frame rate of 14.7 frames s–1 is employed as the visible camera. The lens of camera is directed vertically downward to the ground, and set up in auto-exposure mode to compensate outdoor light effects. Images of multiple plots can be quickly acquired (~180 images h–1, inclusive of crane positioning time), to monitor canopy closure, by removing the soil background from an RGB image and quantifying the percentage of green pixels (vegetation) within an image (Fig. 4a). Given the autonomous operation of the Field Scanalyzer, images can be acquired with high-temporal resolution, to resolve canopy development in detail. Fig. 4b provides an example of how an RGB image series from the Field Scanalyzer can be used to monitor canopy development of two UK wheat varieties during the 2015–16 winter period. RGB images can also be utilised to automatically detect and quantify plant organs, such as the ears of wheat (Fig. 4c), providing a high-throughput approach to quantifying this important yield component, compared with current labour-intensive methods (P. Sadeghi-Tehran; K Sabermanesh, N Virlet, MJ Hawkesford, unpubl. data).

|
A FLIR A645SC (FLIR Systems Inc.) is employed as a thermal infrared camera (TIR; 640 × 480 pixel matrix) and covers the spectral range of 7.5–13 µm. The TIR images acquired have a radiometric resolution of 0.05°C and an absolute precision of 2°C. The TIR camera is directed vertically downward to the ground and can collect 330 images h–1, inclusive of crane positioning time. Data is recorded in raw 16-bit format and the digital number intensities are later converted to radiometric temperature using the RBF equation provided by FLIR Systems.
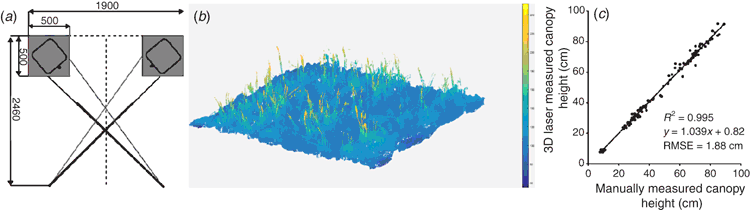
Twin 3D laser scanners (Fraunhofer Institute) are mounted in the camera bay, opposing each other (Fig. 5a), and are capable of scanning plant canopies with high resolution (0.25 mm) in all three axes, using an NIR laser (840 nm) to ensure high reflectance by plant tissue and minimal physiological interaction. The laser scanners have a throughput of 30 plots h–1 and a field of view of ~0.5 m width and 0.5 m depth. Through the analysis of point cloud images, morphological traits of crop plots, such as plant height may be quantified (Fig. 5b). Fig. 5c represents the comparison of plant height measurements of seedlings of six wheat varieties commonly grown in the UK (Triticum aestivum vars. Avalon, Cadenza, Crusoe, Gatsby, Soissons and Maris Widgeon), obtained manually and from the on-board 3D lasers at 90, 196 and 226 days after sowing (DAS). Height measurements obtained from the 3D lasers are comparable to manual measurements (R2 = 0.99; Root Mean Square Error (RMSE) = 1.88 cm), evidencing the reliability of quantification of a simple morphological trait. The manual height measurements appear, on average, to be slightly lower than those measured from the 3D lasers, especially for the highest values. However, this could likely due to human-error, given that manual measurements are acquired by taking the mean height of only 12 individual plants within an entire 1.5 m2 plot.
The hyperspectral system comprises two mirror-scanning Hyperspec Inspector (Headwall Photonic) visible and near-infrared (VNIR) and extended VNIR (ExVNIR) cameras, together covering the 400–1700 nm range. The VNIR and ExVNIR cameras have 1600/923 (0.7 nm step) and 320/229 (4.6 nm step) spatial/spectral resolution, with an f/2 optical aperture. Both VNIR and ExVNIR cameras are directed vertically downward to the ground and scan 46 and 80 plots h–1, respectively. During acquisition, the crane remains stationary and the resulting hypercube is collected from the motion of an internal concave mirror. Data is recorded in their original 16-bit format.
A four-channel amplified radiometer normalised difference vegetation index (NDVI) light sensor (Skye Instruments Ltd) is fitted on the Field Scanalyzer, with two channels calibrated for red (633 ± 19 nm) wavebands and the others for NIR (800 ± 17 nm). One combination of waveband channels is positioned on top of the Field Scanalyzer, pointing at the sky to measure incident solar radiation, whilst the other pair points directly at the ground, to simultaneously measure radiation reflected upwards. A test was performed to compare NDVI values computed from the VNIR sensor (raw data, without radiometric correction) and those from the Skye Instruments sensor (accounts for incoming solar radiation) (Fig. 6). Comparison of NDVI values between the corrected and uncorrected sensors highlighted that, despite the strong linear relationship between NDVI values of the two sensors (R2 = 0.89–0.90 at 150 and 174 DAS respectively), values generated from the VNIR were consistently lower than the corresponding values of the Skye Instruments sensor, which accounts for incoming solar radiation. The difference in NDVI values may be due to comparing narrowband (VNIR) with broadband (Skye Instruments) measurements. Additionally, the constant within the linear equation differed between 150 and 174 DAS, reinforcing the necessity to apply post-acquisition radiometric correction, through the use of on-field reflectance standards (Comar et al. 2012).

|
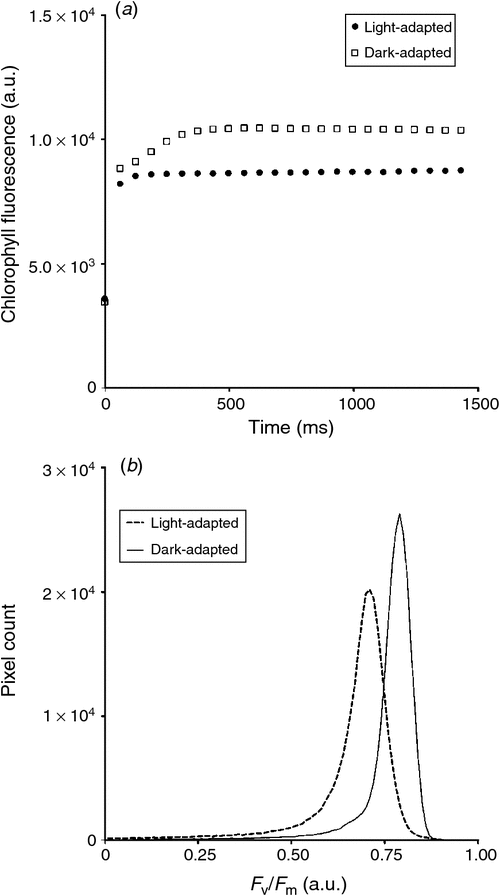
The on-board CFI (CropReporter) is provided by PhenoVation, and enables fluorescence measurements emitted mainly by PSII. The CropReporter is equipped with prime lens, having a fixed wide focal length (18 mm) and a large aperture (f/2), with a CCD matrix of 1.4 Mp (1388 × 1038 pixels). The CFI is a LED (light emitting diode) induced chlorophyll fluorescent transient imager (Jalink and van der Schoor 2011) and the chlorophyll fluorescence (CF) is induced by a flash of red light (620 nm) for 1400 ms, saturating the electron transfer between the two photosystems (PSII and PSI), and records 24 images within that time. At the rising edge of LED illumination, the trigger is being used to start the exposure time of the first image and is called ground fluorescence. The intensity of the flash of light (0–4000 µmol m–2 s–1) is set up according to the distance between the camera and the canopy, and sunlight intensity (for light-adapted measurement). The optimal distance between the sensor and canopy is defined at 70 cm, permitting a final field of view of ~40 cm × 30 cm. The CropReporter generates 14-bit data and can acquire CF measurements at a rate of 90 plots h–1 (inclusive of crane repositioning time). Fig. 7a provides an example of in-situ CF measurements, which can be used to obtain ground and maximum fluorescence (F0 and Fm respectively), values of light- and dark-adapted wheat, to subsequently calculate various photosynthetic parameters, such as Fv/Fm, of individual pixels (Fig. 7b).
|
Data storage and management
Any successful large-scale non-destructive plant analysis requires integrating cultivation systems, automated precise environment monitoring and robust data management system (Araus and Cairns 2014). In order to evaluate and analyse large-scale phenotyping data, it is essential to standardise data storage, as an efficient data management system can improve data accessibility and speed up data evaluation.
HTPPs such as the Field Scanalyzer produce big datasets, generating gigabytes of data in short amounts of time. For instance, it takes only 7 min to acquire data from all sensors in the camera bay from one plot, which generates 800 MB data. Rothamsted and LemnaTec GmbH developed a comprehensive data solution for the raw data acquired by the Field Scanalyzer to be handled and stored automatically. A high quality data management and data analytic system was created to securely access the data instantly at any time from various workstations and different locations.
Data and meta-information for each sampled plot is acquired and automatically transferred from the Field Scanalyzer to the dedicated database, through optical cables. The meta-information comprises a plot ID tag, timestamp, position of plots, and corresponding images (i.e. N1-Rep1–22-Crusoe_2015–07–13_10–34–46), and facilitates seamless navigation of datasets for quality control, evaluation and analysis. Software provided by LemnaTec GmbH, such as LemnaControl and LemnaBase, allow users to graphically check the status of the phenotyping equipment and also preview any acquired data.
Raw data itself has little value and needs to be combined and analysed before we can gain knowledge from it. We used a combination of common image processing toolbox from Matlab, Python and R to develop a direct interface for image analysis from the database. Pipelines are developed according to the sensor technology, in order to automatically extract relevant quantitative information concerning the trait of interest. Extracted results (i.e. spread-sheets, figures) are then uploaded to the database and can be used in subsequent years by other partners and researchers. An overview of the data management and data processing workflow is illustrated in Fig. 8.
Advantages and limitations
As the Field Scanalyzer operates on rails, it preserves the below soil structure by avoiding compaction as a result of repeated-measurements (Bronick and Lal 2005). Although the platform was designed to be compatible with farming operations, the use of farming equipment is restricted to sowing and harvest operations, in order to minimise soil compaction. As the experimental plots can be quite condensed, given the area covered by the platform is relatively small compared with standard field experiments, herbicides, pesticides and nutrients are applied manually.
The framework of the platform is also a limiting factor. Unlike the Zürich Field Phenotyping Platform (ETH-Zurich), which covers ~1 ha and is potentially transferrable to another site, the Field Scanalyzer covers only a 0.12 ha area and does not allow multi-site experiments without investing in duplicate platforms. Moreover, as the main crop of interest is wheat, a three-year rotation is implemented (wheat, oilseed rape and oat), to maintain soil-nutrient balance, as well as to avoid root diseases, such as take-all. In consequence, the effective area for the main experiment is further restricted to one-third of the platform area in any one year.
A key advantage of the Field Scanalyzer comes directly from its framework, which enables the capacity to carry a 500 kg payload within the camera bay. This high payload provides the opportunity to carry together, all the main imagers currently used in field phenotyping, as well as less portable imagers normally used in controlled environment facilities. The volume and the high payload also offers room for the expansion of the sensor array to test and validate new sensor technologies that are not portable, before possible miniaturisation for UAV platforms. Currently, only ~300 kg of the payload is in use.
The Field Scanalyzer was designed for high spatial resolution phenotyping. As screening is generally performed 2–3 m above the canopy, according to the cameras focal-point, with exception of the fluorescence imager (70 cm above canopy), full advantage of the resolution of each camera is exploited to collect high quality images. High resolution images, such as those obtained from the RGB camera and the 3D laser scanner, potentially allow for plant organ recognition and quantification, as shown previously (Fig. 4).
As a consequence of close range phenotyping, the area covered by each camera is limited by their field of view. Unlike UAVs, which captures multiple plots in a single snapshot, plots are imaged individually, reducing the throughput. Nevertheless, scanning technologies, such as the 3D lasers and hyperspectral imagers require 1–2 min to collect data at each plot. This makes their outputs particularly susceptible to wind and, thus, affecting the image quality. Another limiting factor induced by the close range, common to every ground platform, is the effect of varying ambient illumination, which makes comparison within and/or between days challenging. For example, hyperspectral imagers equipped on the Field Scanalyzer have to be set-up for exposure time before each run, according the ambient illumination. Whilst clear skies, or homogenous overcast are the best conditions for collecting spectral data (Rundquist et al. 2014), variations could likely occur during a 2–3 h scan of a field. This runs the risk of generating under- or overexposed images that cannot be corrected for, particularly when it also occurs on the reflectance standard panel.
The key advantage of the Field Scanalyzer is the implementation as an autonomous system, similar to the Zürich Field Phenotyping Platform (ETH-Zurich). Unlike UAVs and most mobile platforms, which require the supervision of a technician or pilot to ensure safe operation, the Field Scanalyzer requires human interaction only for the initial programming and loading of the script for image acquisition. To prevent any script programming errors, LemnaControl (LemnaTec GmbH), the software controlling the platform, provides the opportunity to simulate the loaded script and track any errors. The only sensor requiring human intervention are the hyperspectral imagers. An initial scan has to be performed before each run on a reflectance standard panel in order to set up an optimal exposure time to avoid saturation of the images when slight changes in illumination occur.
The system was designed for 24 h day–1, 7 days week–1 operation. In practice, sequential sensor runs are performed according to the data needed for the experiment and the forecast weather conditions. Moreover, 3D laser scanning is performed during the night, due to (i) the low throughput of the sensor, freeing up daylight hours for other cameras, and (ii) less windy conditions at night, promoting better image quality. The CFI is also used at night, as it allows collection of fluorescence parameters in the field, simplifying the task of quantifying photosynthetic capacity in dark-adapted conditions, which remains laborious with handheld fluorimeters. Additionally, acquisition of fluorescence, thermal infrared and hyperspectral data throughout 24 h periods can also facilitate a better understanding of crops and plant diurnal rhythm kinetics.
Key challenges
The objective of collecting field data in high throughput is close to being achieved: now the phenotyping community is needed to validate the relevance of the developed approaches for the trait being measured. Although the ability to collect data is now established, challenges remain in terms of dealing with changing ambient illumination, data processing, analysis and the interpretation of the image variables and their potential value as a descriptor of a plant trait.
Illumination conditions
Changes in ambient illumination are inevitable when collecting a data series in the field throughout the growing season. Screening should be performed in the most optimal weather conditions (e.g. minimal wind, even/no cloud cover, no rain), although the temperate climate in some regions remains uncertain, notably in the UK. Acquiring data at the same time interval between days is not always possible because of weather conditions. For RGB cameras, Casadesús et al. (2007) suggests that in order to capture well-exposed images, the camera should be set to automatic exposure mode to adapt the aperture, shutter speed and gain, according to the intensity measured by the sensor.
Acquiring data with hyperspectral imagers at close range adds difficulty to the interpretation of data collected from a series of plots. This is because only one plot is captured in a single image and ambient illumination may vary during the acquisition time of multiple plots. To minimise the effect of its variation, exposure time must be manually configured before each run and ensure the run does not exceed 3 h. Additionally, an on-field reflectance standard is systematically added within the data acquisition sequence (every six or 12 plots), allowing radiometric correction to minimise illumination effects between images.
As described by Murchie and Lawson (2013), active CFIs require large LED arrays to ensure even actinic illumination and fully saturating pulses over the entire imaging area, making the instruments relatively large in size and not very portable as the imager equipped on the Field Scanalyzer. Due to its high payload capacity, using this heavy device is not a problem. However, being able to fully saturate a complex canopy in the field remains challenging, especially for the deeper layer of the vegetation as well as the application during the day (Li et al. 2014).
Data processing
The ability to efficiently process big datasets collected remains a challenge, as well as the ability to develop robust and fast algorithms according to the sensor used. Recent developments in data processing of RGB images allow the separation of plants from the background in various ambient illumination conditions. Some approaches to achieve this are based on colour spaced transformation, where colour thresholds are applied to differentiate between green and senescent leaves (Casadesús et al. 2007; Casadesús and Villegas 2014). Machine learning offers another solution to differentiate vegetation from background (Guo et al. 2013), where the model can be trained using plant in various illuminating conditions.
For TIR imaging, different methods have been investigated to extract plant temperature from the image as reviewed in Jones and Sirault (2014). The task is not simple for a heterogeneous canopy, where the presence of mixed pixels (pixel containing signals from both soil and vegetation) is a recurring problem (Jones et al. 2009; Hackl et al. 2012). It is generally considered that directly using the surface temperature of vegetation is risky, because the weight of mixed or soil pixels in porous plant cover can shift towards the soil surface temperature (Jackson et al. 1981). To overcome the problem of mixed pixels, various image pre-processing methods based on both histogram analysis on the raw TIR image or data fusion with RGB image can be applied to exclude mixed pixels (Jones and Sirault 2014). These methods are time-consuming and can also be subjective, as they depend on the threshold chosen. However, automated segmentation techniques have recently been developed to select pure vegetation pixels in TIR images (Jerbi et al. 2015).
Processing images generated from hyperspectral imagers and laser scanners likely remain the most challenging, as they easily generate up to a gigabyte of data from a single scan. Thus, the efficiency of processing tools must be considered when repeated-measurements may generate hundreds of gigabytes to terabytes of data throughout the season.
For hyperspectral imaging, Römer et al. (2012) proposed a new method based on unsupervised classification to accelerate the computation of large datasets. This method, which allows computation of how similar spectrum is to observed typical spectra, was suggested to be better than spectral reflectance indices (SRI) for the early detection of drought or nitrogen stress in cereal crops, even in field conditions. Another factor to consider is the pipeline of hyperspectral imaging for field phenotyping, as it should integrate robust and fast algorithms for the various data processing steps before extracting the features of interest: (i) pre-processing of the raw hypercube, (ii) exploration, (iii) segmentation, and (iv) image processing (Amigo 2010).
Processing data generated from 3D laser scanner technology is mainly performed by converting point clouds into (i) distances and angles to generate 2D images or (ii) a voxel image, where a voxel (volume element) is the 3D equivalent to a pixel (Deery et al. 2014), which can both be subsequently processed using standard image processing software. However, point clouds converted into the image equivalent adds a few additional steps to data processing and could induce a loss of information. Recently, a new approach based on surface feature classification was developed for high throughput data analysis of 3D data (Paulus et al. 2013). In the mentioned study, the authors used the laser scanning outputs directly to establish a reliable and fast technique to differentiate individual plant organs, avoiding any transformation of point clouds.
Data analysis and interpretation
Interpreting a TIR signal is not simple, on account of the high sensitivity of stomatal response to environmental micro-variations. Numerous indices have been developed to assess crop water status from canopy surface temperature (Ts) acquired from aerial or ground platforms (Maes and Steppe 2012). Subtracting air temperature (Ta) from Ts generates a raw variable that is easy to extract from images, but remains sensitive to solar radiation, wind speed and vapour pressure deficit (Maes and Steppe 2012). Temporal comparison of plant transpiration based on this variable (Ts – Ta) requires controlled ambient conditions (Li et al. 2014), or environmental conditions to be similar on measurement days (Idso et al. 1981). The crop water stress index (CWSI) was developed to take into account ambient meteorological conditions, including the vapour pressure deficit, which also influences the canopy temperature (Idso et al. 1981). In recent decades, semi-analytical and empirical approaches to facilitate the computation and the use of CWSI have been developed (Maes and Steppe 2012). However, the application of CWSI and its derivatives is limited to full-cover vegetation and requires calibration from natural (leaf or canopy) or artificial reference surfaces to define the lower and upper baseline necessary to compute those indices. Moreover, the lower baseline (corresponding to a non-water-stressed state) may be genotype dependent, therefore limiting its applications for high-throughput phenotyping where a panel of genotypes is used (Bellvert et al. 2015).
Different approaches have been developed to predict crop performance using multi- or hyperspectral data. Statistical methods are most commonly used and are based on univariate and multivariate regression models (Capolupo et al. 2015). Although both regression models aim to predict crop performance traits, the univariate approach only uses a limited set of SRI (Gonzalez-Dugo et al. 2015), whereas the multivariate approach utilises the entire spectrum to model plant response (Kipp et al. 2014). Some authors show that, in comparison to univariate approaches, multivariate approaches were able to provide better results in detection of early stages of biotic stress (Römer et al. 2011), or in the predict nitrogen and water content (Kusnierek and Korsaeth 2015). Another approach, based on radiative transfer models, seems promising for field phenotyping, as this takes into account biochemical and structural properties of the leaf and canopy (Thorp et al. 2015).
Given that the platform’s camera housing is fixed above the canopy, the majority of cameras and sensors are directed vertically downward to the ground. Consequently, sensors relying on quantifying reflected or emitted radiation off the canopy are sensitive to leaf and canopy structure, with areas of the crop canopy directly parallel to the sensor lens reflecting wavelengths at a higher intensity, compared with those not parallel (see Fig. 3e). Moreover, the leaf optical properties have to be taken into account as they affect the directional reflectance. This is because it may become a source of bias in the (i) quantified signal measured, (ii) prediction of leaves biochemical components, and (iii) estimation of canopy cover or greenness using the bidirectional reflectance distribution function (BRDF) (Comar et al. 2012). In addition, BRDF varies at different observation angles and also at different solar elevation (Deery et al. 2014). This last point must be taken into account when using hyperspectral imaging in field phenotyping through the season. A solution could be to test radiative transfer models, such as PROSAIL (Jacquemoud et al. 2009), to allow the simulation of leaf and canopy reflectance from 400–2500 nm (in 1 nm steps), as well as the BRDF. By using the BRDF output, the effects of canopy structure, composition and geometry, as well as illumination on the spectra collected, can be corrected for. Moreover, model inversion techniques and reflectance measurements can be used to obtain input parameters of the PROSAIL model, such as chlorophyll concentration, dry matter content or leaf water thickness (Thorp et al. 2015). A challenging approach to overcoming issues caused by canopy geometry may be through the fusion of 3D point cloud images with 2D spectral images, whereby each point cloud within a 3D image is assigned a value corresponding to the same point in an image from a spectral image, however this is an avenue requiring further exploration.
Regardless of the quality and robustness of the phenotyping vector used to acquire data, the data itself is only as valuable as the analyses that can be performed on them. Biological noise is present even in controlled conditions and must be accounted for in statistical models to assess phenotypic data. Variations in environmental conditions will add further noise and these caveats present a challenge to develop robust statistical models that can be applied to analyse phenotypic data (Cobb et al. 2013). However, White et al. (2012) indicated that the analysis of data from a temporal series can be difficult because many of the observed traits are auto-correlated and integrate multiple effects from the underlying physiological mechanisms, which each operate on different time scales. Analysis by linear mixed models can be used to detect auto-correlation effects (Piepho et al. 2004); however, the collection of data at regular time intervals is required.
Acknowledgements
Rothamsted Research receives support from the Biotechnology and Biological Sciences Research Council (BBSRC) of the UK as part of the 20 : 20 Wheat project. The authors declare that no conflicts of interests exist.
References
Amigo J (2010) Practical issues of hyperspectral imaging analysis of solid dosage forms. Analytical and Bioanalytical Chemistry 398, 93–109.| Practical issues of hyperspectral imaging analysis of solid dosage forms.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXmsVeqtLs%3D&md5=62475d030a577f711674e36cea9ee6acCAS | 20496027PubMed |
Araus JL, Cairns JE (2014) Field high-throughput phenotyping: the new crop breeding frontier. Trends in Plant Science 19, 52–61.
| Field high-throughput phenotyping: the new crop breeding frontier.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhs1CrtbjL&md5=1b60756cded27f694ab74a260c975182CAS | 24139902PubMed |
Bellvert J, Marsal J, Girona J, Zarco-Tejada PJ (2015) Seasonal evolution of crop water stress index in grapevine varieties determined with high-resolution remote sensing thermal imagery. Irrigation Science 33, 81–93.
| Seasonal evolution of crop water stress index in grapevine varieties determined with high-resolution remote sensing thermal imagery.Crossref | GoogleScholarGoogle Scholar |
Bronick CJ, Lal R (2005) Soil structure and management: a review. Geoderma 124, 3–22.
| Soil structure and management: a review.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhtVOru7jP&md5=b828d03799da7c4a58df6083362baf98CAS |
Capolupo A, Kooistra L, Berendonk C, Boccia L, Suomalainen J (2015) Estimating plant traits of grasslands from UAV-acquired hyperspectral images: a comparison of statistical approaches. ISPRS International Journal of Geo-Information 4, 2792–2820.
| Estimating plant traits of grasslands from UAV-acquired hyperspectral images: a comparison of statistical approaches.Crossref | GoogleScholarGoogle Scholar |
Casadesús J, Villegas D (2014) Conventional digital cameras as a tool for assessing leaf area index and biomass for cereal breeding. Journal of Integrative Plant Biology 56, 7–14.
| Conventional digital cameras as a tool for assessing leaf area index and biomass for cereal breeding.Crossref | GoogleScholarGoogle Scholar | 24330531PubMed |
Casadesús J, Kaya Y, Bort J, Nachit MM, Araus JL, Amor S, Ferrazzano G, Maalouf F, Maccaferri M, Martos V, Ouabbou H, Villegas D (2007) Using vegetation indices derived from conventional digital cameras as selection criteria for wheat breeding in water-limited environments. Annals of Applied Biology 150, 227–236.
| Using vegetation indices derived from conventional digital cameras as selection criteria for wheat breeding in water-limited environments.Crossref | GoogleScholarGoogle Scholar |
Cobb JN, DeClerck G, Greenberg A, Clark R, McCouch S (2013) Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theoretical and Applied Genetics 126, 867–887.
| Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement.Crossref | GoogleScholarGoogle Scholar | 23471459PubMed |
Comar A, Burger P, de Solan B, Baret F, Daumard F, Hanocq J-F (2012) A semi-automatic system for high throughput phenotyping wheat cultivars in-field conditions: description and first results. Functional Plant Biology 39, 914–924.
| A semi-automatic system for high throughput phenotyping wheat cultivars in-field conditions: description and first results.Crossref | GoogleScholarGoogle Scholar |
Deery D, Jimenez-Berni J, Jones H, Sirault X, Furbank R (2014) Proximal remote sensing buggies and potential applications for field-based phenotyping. Agronomy 4, 349–379.
| Proximal remote sensing buggies and potential applications for field-based phenotyping.Crossref | GoogleScholarGoogle Scholar |
ETH-Zurich ‘ETH – crop science – field phenotyping platform (FIP).’ Available at http://www.kp.ethz.ch/infrastructure/FIP.html [Verified 10 September 2016].
Gonzalez-Dugo V, Hernandez P, Solis I, Zarco-Tejada P (2015) Using high-resolution hyperspectral and thermal airborne imagery to assess physiological condition in the context of wheat phenotyping. Remote Sensing 7, 13586–13605.
| Using high-resolution hyperspectral and thermal airborne imagery to assess physiological condition in the context of wheat phenotyping.Crossref | GoogleScholarGoogle Scholar |
Guo W, Rage UK, Ninomiya S (2013) Illumination invariant segmentation of vegetation for time series wheat images based on decision tree model. Computers and Electronics in Agriculture 96, 58–66.
| Illumination invariant segmentation of vegetation for time series wheat images based on decision tree model.Crossref | GoogleScholarGoogle Scholar |
Hackl H, Baresel JP, Mistele B, Hu Y, Schmidhalter U (2012) A comparison of plant temperatures as measured by thermal imaging and infrared thermometry. Journal Agronomy & Crop Science 198, 415–429.
| A comparison of plant temperatures as measured by thermal imaging and infrared thermometry.Crossref | GoogleScholarGoogle Scholar |
Idso SB, Jackson RD, Pinter PJ, Reginato RJ, Hatfield JL (1981) Normalizing the stress-degree-day parameter for environmental variability. Agricultural Meteorology 24, 45–55.
| Normalizing the stress-degree-day parameter for environmental variability.Crossref | GoogleScholarGoogle Scholar |
Jackson RD, Idso SB, Reginato RJ, Pinter PJ (1981) Canopy temperature as a crop water-stress indicator. Water Resources Research 17, 1133–1138.
| Canopy temperature as a crop water-stress indicator.Crossref | GoogleScholarGoogle Scholar |
Jacquemoud S, Verhoef W, Baret F, Bacour C, Zarco-Tejada PJ, Asner GP, Francois C, Ustin SL (2009) PROSPECT plus SAIL models: a review of use for vegetation characterization. Remote Sensing of Environment 113, S56–S66.
| PROSPECT plus SAIL models: a review of use for vegetation characterization.Crossref | GoogleScholarGoogle Scholar |
Jalink H, van der Schoor R (2011) LED induced chlorophyll fluorescence transient imager for measurements of health and stress status of whole plants. In ‘Proceedings of the International Symposium on High Technology for Greenhouse Systems: GreenSys2009, Quebec, Canada, 14–19 June, 2009’. Available at http://edepot.wur.nl/169025 [Verified 10 September 2016].
Jerbi T, Wuyts N, Cane MA, Faux PF, Draye X (2015) High resolution imaging of maize (Zea mays) leaf temperature in the field: the key role of the regions of interest. Functional Plant Biology 42, 858–864.
| High resolution imaging of maize (Zea mays) leaf temperature in the field: the key role of the regions of interest.Crossref | GoogleScholarGoogle Scholar |
Jones H, Sirault X (2014) Scaling of thermal images at different spatial resolution: the mixed pixel problem. Agronomy 4, 380
| Scaling of thermal images at different spatial resolution: the mixed pixel problem.Crossref | GoogleScholarGoogle Scholar |
Jones HG, Serraj R, Loveys BR, Xiong LZ, Wheaton A, Price AH (2009) Thermal infrared imaging of crop canopies for the remote diagnosis and quantification of plant responses to water stress in the field. Functional Plant Biology 36, 978–989.
| Thermal infrared imaging of crop canopies for the remote diagnosis and quantification of plant responses to water stress in the field.Crossref | GoogleScholarGoogle Scholar |
Kipp S, Mistele B, Schmidhalter U (2014) Identification of stay-green and early senescence phenotypes in high-yielding winter wheat, and their relationship to grain yield and grain protein concentration using high-throughput phenotyping techniques. Functional Plant Biology 41, 227–235.
| Identification of stay-green and early senescence phenotypes in high-yielding winter wheat, and their relationship to grain yield and grain protein concentration using high-throughput phenotyping techniques.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXisF2rs7g%3D&md5=4e9a0f450ad52374d346776af8389adeCAS |
Kusnierek K, Korsaeth A (2015) Simultaneous identification of spring wheat nitrogen and water status using visible and near infrared spectra and powered partial least squares regression. Computers and Electronics in Agriculture 117, 200–213.
| Simultaneous identification of spring wheat nitrogen and water status using visible and near infrared spectra and powered partial least squares regression.Crossref | GoogleScholarGoogle Scholar |
Li L, Zhang Q, Huang D (2014) A review of imaging techniques for plant phenotyping. Sensors 14, 20078
| A review of imaging techniques for plant phenotyping.Crossref | GoogleScholarGoogle Scholar | 25347588PubMed |
Losos JB, Arnold SJ, Bejerano G, Brodie E, Hibbett D, Hoekstra HE, Mindell DP, Monteiro A, Moritz C, Orr HA (2013) Evolutionary biology for the 21st century. PLoS Biology 11, e1001466
| Evolutionary biology for the 21st century.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXis1aktr4%3D&md5=8408453d75b30c1a58716e664109f659CAS | 23319892PubMed |
Maes WH, Steppe K (2012) Estimating evapotranspiration and drought stress with ground-based thermal remote sensing in agriculture: a review. Journal of Experimental Botany 63, 4671–4712.
| Estimating evapotranspiration and drought stress with ground-based thermal remote sensing in agriculture: a review.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xht1Kjsb7O&md5=a5b4ac8ce868b2a904630c8f9862df9fCAS | 22922637PubMed |
Murchie EH, Lawson T (2013) Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications. Journal of Experimental Botany 64, 3983–3998.
| Chlorophyll fluorescence analysis: a guide to good practice and understanding some new applications.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhs1SmtLbJ&md5=8b721e3e9d9bbcbce496be3070f4c2b1CAS | 23913954PubMed |
Passioura JB (2012) Phenotyping for drought tolerance in grain crops: when is it useful to breeders? Functional Plant Biology 39, 851–859.
| Phenotyping for drought tolerance in grain crops: when is it useful to breeders?Crossref | GoogleScholarGoogle Scholar |
Passioura JB, Angus JF (2010) Improving productivity of crops in water-limited environments. In ‘Advances in agronomy. Vol. 106’. (Ed. DL Sparks) pp. 37–75. (Elsevier Academic Press Inc.: San Diego, CA, USA)
Paulus S, Dupuis J, Mahlein AK, Kuhlmann H (2013) Surface feature based classification of plant organs from 3D laserscanned point clouds for plant phenotyping. BMC Bioinformatics 14, 238
| Surface feature based classification of plant organs from 3D laserscanned point clouds for plant phenotyping.Crossref | GoogleScholarGoogle Scholar | 23890277PubMed |
Piepho HP, Büchse A, Richter C (2004) A mixed modelling approach for randomized experiments with repeated measures. Journal Agronomy & Crop Science 190, 230–247.
| A mixed modelling approach for randomized experiments with repeated measures.Crossref | GoogleScholarGoogle Scholar |
Pingali PL (2012) Green revolution: impacts, limits, and the path ahead. Proceedings of the National Academy of Sciences of the United States of America 109, 12302–12308.
| Green revolution: impacts, limits, and the path ahead.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtleqsrvJ&md5=d060d381551681eefa22ed93fd23955aCAS | 22826253PubMed |
Rascher U, Blossfeld S, Fiorani F, Jahnke S, Jansen M, Kuhn AJ, Matsubara S, Märtin LL, Merchant A, Metzner R (2011) Non-invasive approaches for phenotyping of enhanced performance traits in bean. Functional Plant Biology 38, 968–983.
| Non-invasive approaches for phenotyping of enhanced performance traits in bean.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhsFKktLvP&md5=c2f321d5c90b65e2b2b8f10d5cfb6f23CAS |
Römer C, Bürling K, Hunsche M, Rumpf T, Noga G, Plümer L (2011) Robust fitting of fluorescence spectra for pre-symptomatic wheat leaf rust detection with support vector machines. Computers and Electronics in Agriculture 79, 180–188.
| Robust fitting of fluorescence spectra for pre-symptomatic wheat leaf rust detection with support vector machines.Crossref | GoogleScholarGoogle Scholar |
Römer C, Wahabzada M, Ballvora A, Pinto F, Rossini M, Panigada C, Behmann J, Leon J, Thurau C, Bauckhage C, Kersting K, Rascher U, Plumer L (2012) Early drought stress detection in cereals: simplex volume maximisation for hyperspectral image analysis. Functional Plant Biology 39, 878–890.
| Early drought stress detection in cereals: simplex volume maximisation for hyperspectral image analysis.Crossref | GoogleScholarGoogle Scholar |
Rundquist D, Gitelson A, Leavitt B, Zygielbaum A, Perk R, Keydan G (2014) Elements of an integrated phenotyping system for monitoring crop status at canopy level. Agronomy 4, 108
| Elements of an integrated phenotyping system for monitoring crop status at canopy level.Crossref | GoogleScholarGoogle Scholar |
Sankaran S, Khot LR, Espinoza CZ, Jarolmasjed S, Sathuvalli VR, Vandemark GJ, Miklas PN, Carter AH, Pumphrey MO, Knowles NR, Payek MJ (2015) Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: a review. European Journal of Agronomy 70, 112–123.
| Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: a review.Crossref | GoogleScholarGoogle Scholar |
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327, 818–822.
| Breeding technologies to increase crop production in a changing world.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhslWisLg%3D&md5=b2caf8a1006ace209e819db8aed0f155CAS | 20150489PubMed |
Thorp KR, Gore MA, Andrade-Sanchez P, Carmo-Silva AE, Welch SM, White JW, French AN (2015) Proximal hyperspectral sensing and data analysis approaches for field-based plant phenomics. Computers and Electronics in Agriculture 118, 225–236.
| Proximal hyperspectral sensing and data analysis approaches for field-based plant phenomics.Crossref | GoogleScholarGoogle Scholar |
Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. Proceedings of the National Academy of Sciences of the United States of America 108, 20260–20264.
| Global food demand and the sustainable intensification of agriculture.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhs1yqsbnM&md5=cb16304f14cb2df9b11230252f1e0616CAS | 22106295PubMed |
White JW, Andrade-Sanchez P, Gore MA, Bronson KF, Coffelt TA, Conley MM, Feldmann KA, French AN, Heun JT, Hunsaker DJ, Jenks MA, Kimball BA, Roth RL, Strand RJ, Thorp KR, Wall GW, Wang GY (2012) Field-based phenomics for plant genetics research. Field Crops Research 133, 101–112.
| Field-based phenomics for plant genetics research.Crossref | GoogleScholarGoogle Scholar |