Plants versus pathogens: an evolutionary arms race
Jonathan P. Anderson A , Cynthia A. Gleason A , Rhonda C. Foley A , Peter H. Thrall B , Jeremy B. Burdon B and Karam B. Singh A C DA CSIRO Plant Industry, Centre for Environment and Life Sciences, Private Bag #5, Wembley, WA 6913, Australia.
B CSIRO Plant Industry, GPO Box 1600, Canberra, ACT 2601, Australia.
C The Institute of Agriculture, The University of Western Australia, 35 Stirling Highway, Crawley, Perth, WA 6009, Australia.
D Corresponding author. Email: karam.singh@csiro.au
This paper is part of an ongoing series: ‘The Evolution of Plant Functions’.
Functional Plant Biology 37(6) 499-512 https://doi.org/10.1071/FP09304
Submitted: 21 December 2009 Accepted: 7 April 2010 Published: 20 May 2010
Abstract
The analysis of plant–pathogen interactions is a rapidly moving research field and one that is very important for productive agricultural systems. The focus of this review is on the evolution of plant defence responses and the coevolution of their pathogens, primarily from a molecular-genetic perspective. It explores the evolution of the major types of plant defence responses including pathogen associated molecular patterns and effector triggered immunity as well as the forces driving pathogen evolution, such as the mechanisms by which pathogen lineages and species evolve. Advances in our understanding of plant defence signalling, stomatal regulation, R gene–effector interactions and host specific toxins are used to highlight recent insights into the coevolutionary arms race between pathogens and plants. Finally, the review considers the intriguing question of how plants have evolved the ability to distinguish friends such as rhizobia and mycorrhiza from their many foes.
Additional keywords: bacteria, defence, disease, fungus, insect, symbiosis.
Introduction
Pathogens and insect pests cause widespread losses to agriculture and damage to natural plant ecosystems on an annual basis. However, the occurrence of disease on an individual plant is relatively infrequent, even though plants are commonly in contact with numerous potential pathogens. The ecological and epidemiological interactions in plant–enemy systems represent a dynamic situation that provides the basis for a full-blown coevolutionary arms race that varies in both space and time. The selective pressures driving these processes are very strong with an advance by one partner, for example, the increased virulence of a pathogen, placing strong selective pressure on the plant host to increase or modify specific aspects of its defence response. If this is done too successfully, selective pressures increase on the pathogen for compensatory changes in its ability to overcome these defences.
The challenges posed for plants by the microbial world are considerable. Plants face a wide variety of organisms that range from being extremely pathogenic to being highly beneficial. Major pathogenic organisms include viruses, bacteria, fungi, nematodes and insect pests that have evolved quite distinct and specialised strategies for attacking plants. For example, some fungi have a biotrophic lifestyle that involves feeding on living plant tissue, whereas others have a necrotrophic lifestyle that involves killing plant tissue and feeding on dead or dying cells (Glazebrook 2005). Others have evolved to be hemibiotrophs, with an initial biotrophic phase followed by a necrotrophic phase. Insect pests also display a range of feeding strategies and include chewing, cell content feeding and sucking sap. Overall, the low frequency of disease is testament to the evolution of plant defence systems that, although effective, still permit beneficial associations to occur (e.g. N2-fixing bacteria). However, there is a cost associated with strong defences and plant investment in defence strategies can result in decreased fitness (e.g. yield penalties in crops). For example, in the absence of infection by Stagonospora nodorum (Berk.) E. Castell & Germano resistant varieties of wheat (Triticum aestivum L.) have lower yield potential (5–20%) than susceptible plants (Oliver et al. 2008). To help deal with this issue, plants have evolved tightly regulated inducible defence systems that are less costly to maintain and include mechanisms to restrict defence deployment unless completely necessary. For example, heat stress limits the ability of Alternaria brassicicola (Schwein.) Wiltshire to infect hosts efficiently; consequently, plant defence genes are negatively regulated by heat stress transcription factors in order to avoid the occurrence of ‘unnecessary’ defence responses (Kumar et al. 2009).
Major plant defence responses
The plant defence process consists of both preformed and induced defences that can either prevent the pathogen from entering the plant or from obtaining nutrient for growth and reproduction (Thatcher et al. 2005; Jones and Dangl 2006). In recent times, the term ‘basal resistance’ has been adopted to refer to two distinct aspects of the plant-pathogen interaction. The first meaning refers to constitutive defences that discourage or provide a physical barrier to pathogen and pest ingress, and these are generally classified as ‘pre-invasive’ defences. The vast majority of potential pathogens coming into contact with a plant’s surface are kept at bay by the plant, with most microbes unable to penetrate the outer epidermal wall (Hardham et al. 2007). The plant cytoskeleton is also an important obstacle encountered by pathogens (Thordal-Christensen 2003). An elegant example of the importance of the cytoskeleton was shown by the disruption of the actin cytoskeleton of barley (Hordeum vulgare L.), wheat, cucumber (Cucumis sativus L.) and tobacco (Nicotiana tabacum L.) that resulted in cellular penetration by several non-host fungi (Kobayashi et al. 1997).
Preformed defences also include chemical barriers, which are referred to as phytoanticipins (Morrissey and Osbourn 1999). Many of these compounds had or have roles in plant growth and development and, through selection pressures, may have been recruited or modified for roles in defence. One of the better characterised examples of a phytoanticipin is the saponin avenacin of oats (Avena sativa L.), which is produced in roots. Mutant lines with reduced production of avenacin are more susceptible to the non-host pathogens Gaeumannomyces graminis var. tritici J. Walker and Fusarium culmorum (W.G. Sm.) McAlpine (Papadopoulou et al. 1999; Bednarek and Osbourn 2009). Other antimicrobial chemicals produced by plants include steroids, glycoalkaloids and glucosinalates, the latter being stored during normal growth and converted by myrosinases to active defence compounds.
Another aspect of basal resistance refers to non-specific defences that are induced following the perception of more or less generic microbe or pathogen associated molecular patterns (MAMPs or PAMPs). Such non-specific defence mechanisms are more accurately referred to as PAMP triggered immunity (PTI). Because PTI has been described in several recent review papers (Bent and Mackey 2007; de Wit 2007; Boller and He 2009), we will primarily address this from an evolutionary point of view in this paper.
PAMP triggered immunity
PAMP triggered immunity (PTI) is initiated upon plant recognition of PAMPs through pattern recognition receptors (PRRs) (Chisholm et al. 2006). These PAMPs are molecules associated with a range of pathogens; plants possessing the appropriate PRR are able to detect the presence of the pathogen at very low concentrations (Boller and He 2009). Perhaps the most well known PAMP–PRR system involves the perception of a stretch of the bacterial flagellin through a 22 amino acid epitope known as flg22 by the concomitant PRR, FLS2 (Gomez-Gomez and Boller 2000; Chinchilla et al. 2006). The 22 amino acid flg22 is a highly conserved region and is functionally important for the bacterial flagellin (Boller and He 2009), suggesting that plants have evolved a system to detect the broadest array of potential pathogens while giving the bacteria the lowest possibility of evading detection through mutation of the PAMP. Other conserved PAMPs and MAMPs recognised by plants include chitin, a basic building block of fungal cell walls, a quorum sensing molecule from the pathogen Xanthomonas oryzae (Ishiyama) Dowson that attacks rice (Oryza sativa L.), a glucan from Phytophthora megasperma Drechsler and the bacterial elongation factor Tu (EF-Tu) (see de Wit 2007 and references therein).
A large number of induced defence responses occur during PTI; these include molecular, morphological and physiological changes (reviewed in Altenbach and Robatzek 2007). Early changes occurring within seconds to minutes include ion-flux across the plasma membrane, an oxidative burst, mitogen activated protein (MAP) kinase activation and protein phosphorylation (Schwessinger and Zipfel 2008). This is followed by substantial transcriptional reprogramming within the first hour of PTI involving up to 3% of the transcriptome in Arabidopsis thaliana (L.) Heynh. There is strong evidence for significant overlap in the response to different PAMPs and the defence signalling molecule salicyclic acid (SA) plays an important role (Sato et al. 2007; Tsuda et al. 2008). Later changes include callose deposition, which serves as a physical barrier at infection sites, and stomatal closure. Stomata provide a major entry point for many plant pathogens and A. thaliana stomata have been shown to close within 1 h in response to PAMPs as part of PTI (Melotto et al. 2006). The PAMP triggered stomatal response involves K+ channel regulation and a heterotrimeric G-protein (Zhang et al. 2008).
The evolution of PTI appears to have occurred early, as FLS2 homologues exist in all sequenced higher plants (Boller and He 2009). Moreover, functional conservation of FLS2 has been demonstrated by expression of the rice FLS2 gene, OsFLS2, in an A. thaliana fls2 mutant (Takai et al. 2008), suggesting that the associated signalling pathways are also functionally conserved. However continued evolution of PTI is evident from the perception of the EF-Tu by the receptor-like kinase (RLK) protein EFR, which only occurs in the Brassicaceae (Kunze et al. 2004). Nonetheless, transfer of EFR from A. thaliana into tobacco, which normally lacks a response to EF-Tu, resulted in responsiveness to the PAMP (Zipfel et al. 2006) suggesting conservation of downstream PTI signalling pathways.
In fact, many bacterial effectors including flg22, HrpZ and EF-Tu may use the same downstream signalling pathway involving a MAP kinase cascade and the RLK BAK1 (Lee et al. 2001; Asai et al. 2002; He et al. 2006; Zipfel et al. 2006). Although PTI triggered by the fungal PAMP chitin appears to use at least some different downstream components as it is independent of BAK1, MAP kinase activity is still observed 10 min after chitin treatment. Interestingly, Gimenez-Ibanez et al. (2009) found that PTI against the bacterial pathogen Pseudomonas syringae van Hall also involved the BAK1-independent pathway and was dependent on CERK1 of the chitin PTI pathway. Together, these results suggest that plants have evolved the ability to detect a diverse array of pathogen associated signals with a degree of redundancy such that an individual pathogen may trigger several independent or linked PTI pathways. The situation whereby a single pathogen may trigger the activation of several PTI pathways, each activating an array of defences, may be hypothesised to contribute to the broad spectrum effectiveness of PTI. In some cases, each PTI signalling pathway may converge to activate a largely conserved defence response.
Effector triggered or induced susceptibility
Given that PTI appears to be widespread and effective against the majority of potential pathogens (Shan et al. 2007), how do successful pathogens cause disease? In recent years, an emerging paradigm suggests that successful pathogens are able to (a) suppress the induction of PTI through the production of effectors, (b) circumnavigate the activity of PTI through the production of toxin type effectors or (c) degrade bioactive products of PTI through sophisticated detoxification mechanisms. Considerable research effort has focussed on investigating the mechanisms by which pathogens are able to suppress host defence responses through the secretion of effectors. In the case of gram negative bacterial pathogens, these effectors are typically introduced into the plant cytoplasm through a type III secretion system (TTSS). In the case of fungi, effectors may be secreted into the apoplast or delivered into the host cytoplasm by an as yet unknown mechanism(s). Effectors from oomycete pathogens that are delivered into the host cytoplasm possess a conserved RXLR domain that may gain entry to host cells by exploiting the plant’s endocytic pathway (Rehmany et al. 2005; Birch et al. 2008; Dou et al. 2008).
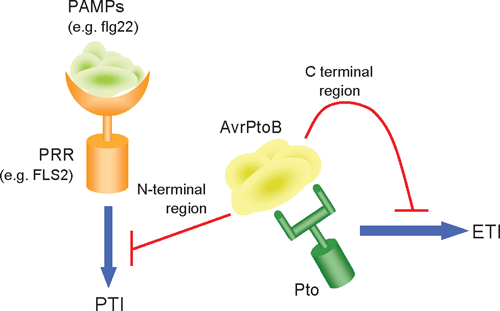
Several examples of pathogen effectors suppressing specific aspects of the plant’s defence response have been found. For example, in A. thaliana, activation of PTI following perception of flg22 induces a range of defences through signalling along a MAP kinase pathway. The P. syringae TTSS effectors AvrPto, AvrPtoB and HOPAI1 have been shown to suppress PTI by blocking the activation of this MAP kinase pathway in A. thaliana (de Torres et al. 2006; He et al. 2006; Zhang et al. 2007). Furthermore, AvrPtoB has been shown to be a potent inhibitor of both BAK1-dependent and BAK1-independent PTI triggered by a range of PAMPs including flg22, HrpZ, NPP1 and chitin (Heese et al. 2007; Gimenez-Ibanez et al. 2009). AvrPtoB contains an N-terminal domain capable of inhibiting the kinase domain of several PTI proteins including FLS2, BAK1 and CERK1 (Shan et al. 2008; Gimenez-Ibanez et al. 2009). AvrPtoB also contains a C-terminal domain that mimics a host E3 ubiqiutin ligase that ubiquitinates host defence proteins for subsequent degradation by the host’s proteosome pathway, thereby preventing immunity associated programmed cell death (Janjusevic et al. 2006; Gimenez-Ibanez et al. 2009).
Effector triggered immunity (ETI) in turn triggers an array of antimicrobial defences aimed at limiting the pathogen’s ability to cause disease. One of the defences plants have evolved is the closure of stomata as innate immunity gates to prevent bacteria from entering the leaf. This active defence response was found to be elicited by MAMPs associated with both plant and human pathogenic bacteria, and was dependent on the activity of FLS2 and the production of nitric oxide (NO) (Melotto et al. 2006). The P. syringae virulence effector coronatine was found to specifically inhibit the stomatal closure response independently of NO but dependent on the plant defence signalling components COI1 and MPK3, while Xanthomonas campestris pv campestris (Pammel) Dowson was found to produce an unknown diffusible factor that also modulates stomatal aperture through MPK3 (Melotto et al. 2006, 2008; Gudesblat et al. 2009). The suppression of stomatal defences by bacterial effectors is hypothesised to be a key adaptation enabling the transition from an epiphytic lifestyle to endophytic parasitism (Melotto et al. 2008).
In addition to suppression of host defences, some effectors may also assist the pathogen in evading detection by PRRs. One such effector is Avr4, a chitin-binding protein from the fungus Cladosporium fulvum Cooke. As mentioned previously, chitin is a major component of fungal cell walls and a PAMP that is recognised by plants. Avr4 is thought to shield the fungal cell wall from plant chitinases, thereby inhibiting the release of PTI triggering polymers (van den Burg et al. 2003). Avr4 is required for virulence by C. fulvum (van Esse et al. 2007) and as such can be considered a counter-defensive effector. The diversity of action of effectors, along with the observation that P. syringae secretes more than 40 effectors (Chang et al. 2005) and oomycete genomes contain hundreds of potential effectors containing the RXLR sequence (Birch et al. 2008), clearly shows that in coevolved pathosystems, the interaction between pathogen effectors and host defence responses can be complex.
Effector triggered immunity
Even though pathogens appear to have suites of effectors to induce susceptibility in the host, successful infection (and disease) is still a relatively rare condition. Continued coevolution of plant–pathogen systems has led to the ability of the host to detect pathogen effectors and mount a more rapid, targeted defence response. Often this response involves programmed cell death (PCD) during a hypersensitive response (HR) but the response is also often tailored to the challenging pathogen. The strong response following perception of pathogen effectors is mediated through plant resistance (R) proteins that either directly recognise pathogen effectors or guard and detect any modification of key plant proteins (Dangl and Jones 2001).
For an elegant example of the role of PTI and ETI and differing layers of plant defence, we can again turn to the extensively studied pathosystem involving A. thalinana and P. syringae pv. tomato. As mentioned previously, perception of P. syringae pv. tomato PAMPs through PRRs induces PTI characterised by closure of stomata, deposition of callose and reduced bacterial growth. A. thaliana lines harbouring a knockout of RIN4 showed enhanced callose deposition and restricted pathogen growth, and P. syringae pv. tomato virulence factors could not re-open stomata, suggesting that RIN4 is a negative regulator of PTI (Kim et al. 2005; Liu et al. 2009). RIN4 also interacts with several P. syringae pv. tomato effectors. Either AvrRpm1 or AvrB can cause the hyperphosphorylation of RIN4, and AvrRpt2 is a protease that degrades RIN4 (Mackey et al. 2002, 2003; Axtell and Staskawicz 2003). Targeting of RIN4 by AvrRPM1 or AvrB in turn leads to activation of RPM1, whereas degradation of RIN4 by AvrRpt2 activates RPS2. RPM1 and RPS2 are resistance proteins that guard RIN4 and activate ETI following modulation by bacterial effectors, thus demonstrating that RIN4 is a point of convergence for both PTI and ETI.
The P. syringae effector protein AvrPtoB provides a good example of the evolutionary arms race occurring between pathogen and host (Fig. 1). As mentioned previously, AvrPtoB contains an N-terminal domain between residues 1 and 307 that is involved in inhibiting several components of PTI, including FLS2, BAK1 and CERK1, which are involved in the perception and response to the PAMPs flg22 and chitin among others. Plants containing the PTO and Fen resistance proteins are able to recognise AvrPtoB via residues 307 and 387. Recognition of a truncated version of AvrPtoB containing residues 1–400 leads to induction of ETI and host resistance. However, the full length AvrPtoB protein also contains a C-terminal E3 ligase domain (residues 400–550) that has been shown to ubiquitinate Fen for degradation, thereby removing the ability of the plant to detect the N-terminal region of the protein and induce PTI. This results in the plant once again being susceptible to P. syrinage containing the full length AvrPtoB (Janjusevic et al. 2006; Gimenez-Ibanez et al. 2009) and demonstrates the complex interplay between coevolved hosts and pathogens.
|
An important question that the field has been grappling with is under what conditions evolution has favoured direct or indirect interactions between R genes and their corresponding effectors. An example of indirect recognition of effectors is given by the interaction between A. thaliana and P. syringae, which is governed by the plant resistance genes RPS2 and RPM1, and the corresponding pathogen effector genes AvrRpt2, AvrB and AvrRpm1, which target the host protein RIN4. The activity of the effectors leads to the degradation or inactivation of RIN4 and thereby the activation of RPT2 or RPS2 and resistance to P. syringae (Kim et al. 2005). In this case, the R proteins have evolved to detect the activity of the pathogen effectors and therefore mutation of pathogen effectors that leads to loss of recognition by the host R proteins would also mean loss of effector activity.
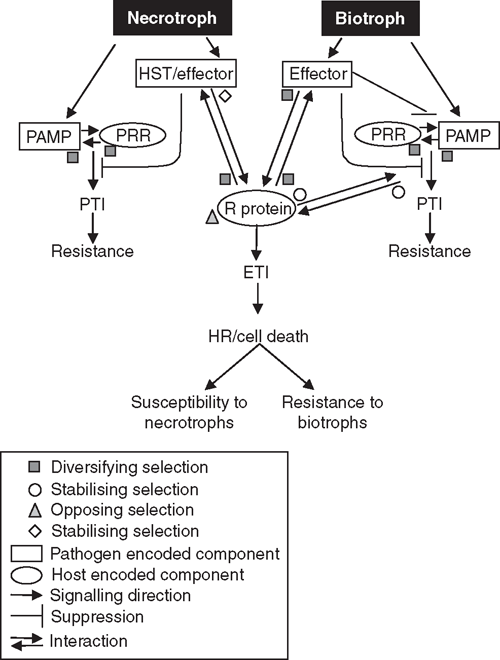
An example of a direct interaction between host R protein and a pathogen effector has been demonstrated for the interaction between flax (Linum usitatissimum L.) and flax rust (Melampsora lini (Ehrenb.)) (Ellis et al. 2007). Yeast-two-hybrid studies have found specific direct interaction of rust avirulence products and flax R genes in a manner consistent with the specificities of the respective resistance and avirulence proteins (Dodds et al. 2006). In keeping with a direct interaction, the resistance proteins appear to be under diversifying selection to introduce additional variation in the LRR domain that is involved in the avirulence protein interaction. If the R proteins were detecting or guarding a host protein, the protein–protein interaction domains may be expected to be under stabilising selection pressure to maintain specificity for or coevolve with, their own guarded protein. Ellis et al. go on to postulate that a potential selective penalty on pathogens recognised indirectly is that escaping recognition requires loss of effector function and thus loss of pathogenicity (Ellis et al. 2007). For obligate pathogens without a saprophytic phase, this loss of pathogenicity would be more detrimental and thus for obligate pathogens, such as flax rust, the direct recognition of effectors by plant R proteins is hypothesised to be more likely and the impact of effectors on host proteins more subtle (Ellis et al. 2007). A diagrammatic summary of the molecular dialogue between plant and pathogen and possible selection pressures exerted on each component is provided in Fig. 2. The presence of several corresponding R and Avr genes in host–pathogen systems, such as the flax–flax rust pathosystem, supports an ongoing coevolution driven by pressure on the host to detect new strains of the pathogen and pressure on the pathogen to evade detection by the host (Dodds and Thrall 2009).
|
Recent progress on the cloning of R genes is shedding light on their diversity and evolutionary origins. R genes with the capacity to provide resistance against viral, bacterial or fungal pathogens, as well as against nematodes or insect pests, have now been isolated from a wide range of plant species. Interestingly, despite the wide range of taxa in which R genes have been described, to date, only five main classes of proteins have been identified (Martin et al. 2003), with the majority being classified as nucleotide binding site leucine rich repeat (NBS-LRR) proteins. The NBS-LRR class can be further divided into those that have N-terminal homology to the Toll and Interleukin-1 receptor (TIR) genes (TIR-NBS-LRR), a leucine-zipper (LZ-NBS-LRR) or a coiled-coil motif (CC) (CC-NBS-LRR).
The conservation of the defence response is also evident in the identification of similar pathogenesis related proteins from a broad range of host species. For example, analysis of the protein sequences of members of the PR-12 (plant defensin) class from a range of monocot and dicot plants demonstrates that diverse plant species contain defence response proteins with substantial homology (Thomma et al. 2002). The majority of plant defensins isolated from a range of plant species exhibit direct antifungal activity against a broad spectrum of fungi, and some inhibit insect gut α-amylase activity and protein synthesis (Thomma et al. 2002). The sequence and functional similarity between plant defensins from a broad range of plants suggests that although these plants may be widely divergent, they have maintained similar mechanisms to counter pathogen challenges. Interestingly, plant defensins have substantial similarity to defensins from insects, molluscs and mammals. However, the specificity of antimicrobial activity of defensins appears to differ according to the relative importance of infection pressure from different microbes on the host organisms (Thomma et al. 2002).
The overlap in antimicrobial compounds is just one area where there appears to be striking conservation in plant and animal defence responses (reviewed in Staskawicz et al. 2001; Nurnberger et al. 2004). For example, in some cases, there is clear conservation of plant R genes with genes in animals that serve similar roles in animal immune responses, suggesting an ancient evolutionary origin for these proteins. Thus several plant R proteins contain the TIR and LRR domains that are also found in animal defence receptors such as the drosophila TOLL protein and TOLL-like receptors in mammals. There is also conservation of downstream signalling components such as mitogen activated protein kinase cascades. These striking similarities suggest that some elements of the pathogen defence response, such as defensins, are either ancient defence mechanisms that have been conserved through the evolution of a broad range of taxa or else convergent evolution led to the development of similar defence mechanisms.
Toxins as effectors
The suppression of plant defence responses is central to the success of many plant pathogens, particularly those that gain energy and nutrients from living plant tissues, the biotrophs. In this case, a plant defence response that involves PCD is able to restrict pathogen growth by cutting off the nutrient supply. However, another class of pathogens, the necrotrophs, gain nutrients from dead or dying tissue. For these pathogens, PCD does not restrict nutrient supply and may even enhance the success of the pathogen (Thomma et al. 2001). For example, Govrin and Levine (2000) found that infection by the necrotroph Botrytis cinerea Pers. was enhanced by first challenging the plant with a HR inducing strain of the bacterial pathogen P. syringae. There are several examples of necrotrophic pathogens producing toxins that specifically induce PCD and are essential for successful infection (Liu et al. 2006; Friesen et al. 2008, 2009).
A compatible host–toxin interaction often relies on the specific recognition of the toxin by a dominant host protein, which leads to toxin sensitivity and enhanced disease susceptibility. An absence of either the toxin or the related host gene produces a resistant response. In some cases, it appears that host specific toxins (HSTs) target host R proteins to induce the HR, conferring resistance to biotrophs but enhancing infection by the necrotroph (Lorang et al. 2007). This results in toxins being under a different selection pressure compared to biotrophic pathogen effectors. Effectors of biotrophs are under pressure to evade detection by the host while maintaining the defence suppressive function, whereas HSTs are under pressure to maintain detection by the host, as this is required for successful infection (Stukenbrock and McDonald 2009). This suggests that selection pressures on plants are likely to be complex, given that, on the one hand, proliferation of R genes is advantageous for detection and defence against biotrophs, whereas on the other hand, such proliferation also provides additional potential targets for HSTs from necrotrophs. The evolution of R gene diversity may therefore partly depend on the relative frequency of interactions with biotrophic and necrotrophic pathogens (Stukenbrock and McDonald 2009); this will vary geographically within particular plant–pathogen interactions, as well as between pathosystems.
Pathogen evolution
So far, this review has focussed mainly on the plant side of the plant–microbe interaction. However, the evolutionary processes occurring in the pathogen and how pathogen populations adapt to changes in host resistance or susceptibility are equally important in the evolutionary arms race. At the most immediate level, evolutionary responses in pathogen populations to changes in the frequency of host resistance are achieved through shifts in the frequency of pre-existing pathogen strains without any underlying mutational or structural change in the genome. Such microevolutionary changes are regularly recorded in both cultivated and wild plant–pathogen interactions, and are the day-to-day embodiment of the interplay of host resistance frequencies and numbers affecting the relative survival and epidemiology of different pathogen genotypes. As an aside, just as pathogen race frequencies change in response to the frequency of host resistance genes, so too does the frequency of host resistance genes in response to selection imposed by the pathogen.
In natural populations, changes in the frequency of avirulence genes, driven by host population resistance structure, are complemented by genetic drift, which can generate marked differences between small neighbouring demes as a consequence of chance survival or extinction of individual pathogen strains when population sizes crash. On the other hand, gene flow or migration from other pathogen populations may lead to the founding of new pathogen populations or the introduction of novel virulence combinations into existing populations.
It is worth highlighting that pathogen life history features will play a direct role in determining the relative selective impact of migration and gene flow (e.g. dispersal ability), and recolonisation–extinction dynamics (host range, presence of resistant spore stages). For example, many bacterial pathogens, such as P. syringae, are highly efficient saprophytes – theoretical studies of the dynamics of soil-borne pathogens have shown that saprophytic ability can have major impacts on disease epidemiology and persistence (Thrall et al. 1997). Such genotypic and phenotypic interactions are central to the within- and among-population epidemiological and evolutionary processes that determine patterns of disease occurrence and prevalence. However, below the surface of these processes lies a range of mechanisms whereby individual pathogen lineages or species may gain variation and evolve. In the following sections, we will explore some of these processes, mainly using examples from fungal and bacterial pathogens. The first and most obvious of these processes is mutation.
Mutation
Solid epidemiological evidence for mutation leading to changes in the virulence profile of individual members of a pathogen lineage is most apparent in systems in which the pathogen is trapped in a neverending cycle of asexual reproduction. This mechanism is undoubtedly the origin of the majority of new pathotypes of the fungus Puccinia graminis f. sp. tritici Erikss. & Henning, Puccinia graminis f. avenae Erikss. & Henning and Puccinia striiformis Westend that have arisen in various clonal lineages in Australia (Watson 1980; Wellings and McIntosh 1990; Haque et al. 2008), and of P. g. tritici (Burdon and Roelfs 1985) in the USA. In these situations, the appearance of novel pathotypes differs from pre-existing ones by just one or two avirulence genes. This has been parsimoniously interpreted as arising through point or small deletions, or the insertion of transposable elements. To date, though, relatively little is known about the precise molecular basis of the sequence differences responsible for the generation of new pathogen strain variation.
In a landmark paper assessing variation in the M. lini AvrL567 genes, Dodds et al. (2006) demonstrated amino acid sequence differences with several polymorphic sites being associated with specificity variation. Careful analysis of the impact of these changes on recognition proteins indicated that changes in recognition were most probably generated by differences in surface-exposed residues rather than any major structural changes. Analysis of sequence variation at two effector loci, including AvrL567, in populations of M. lini associated with native Australian flax (Linum marginale A.Cunn) provided strong support for the role of host associated selection in maintaining adaptive polymorphisms at these loci (Barrett et al. 2009). Similar evidence for the importance of host resistance and coevolutionary processes in the diversification and maintenance of mutation driven variation in bacterial type III effector proteins has been obtained from recent studies of P. syringae (Ma et al. 2006; Kunkeaw et al. 2010).
Sexual recombination
In recent years, our understanding of the extent of the occurrence of sexual recombination in fungal pathogens has been under revision. Molecular studies have frequently failed to find evidence of linkage disequilibrium – a classic sign of a lack of recombination – and, spurred by this, searches for evidence of ‘cryptic sex’ have often been successful (although it should be noted that many recent studies have also found strong support for clonality: e.g. Høvmoller et al. 2002; Enjalbert et al. 2005; Barrett et al. 2008). Sexual recombination creates a broad range of new virulence combinations from existing pathogenic variation. In doing so, it gives pathogens a highly effective means of countering the generation of resistance gene pyramids in hosts. Indeed, detailed analysis has demonstrated that the risk of effective evolutionary response by pathogens to changing resistance patterns in host populations is greatest among pathogens that possess mixed mating systems. Such a mating strategy provides both the benefits of new allelic combinations and often effective off-season survival mechanisms (sexual reproduction) with the advantages of rapid increase of individual clonal lines (asexual reproduction) that are particularly suited to exploiting specific host environments (McDonald and Linde 2002). Furthermore, theoretical models suggest that the level of recombination can impact on the level of selection against ‘unnecessary’ virulence (Brown 1995), indicating that understanding pathogen mating systems is critical to predicting the evolutionary potential of populations.
Lateral gene transfer
Bacteria are remarkably flexible in their ability to interact with one another and to exchange genetic material across broad ranges of genetic relatedness (Ochman and Moran 2001). Fungi also demonstrate considerable capacity for gene transfer, although perhaps not to the same extent as bacteria. Thus, in many fungal species, there is strong evidence that within host tissue, hyphae may undergo anastomosis, providing the opportunity for recombination of cytoplasmic or nuclear factors. For example, cytoplasmic exchange has been demonstrated in M. lini with novel combinations of dsRNAs being recovered from new isolates generated when parental lines with distinct dsRNA profiles were grown together (Lawrence et al. 1988). While dsRNAs have no apparent impact on performance in M. lini, dsRNA viruses are the mechanism behind switches from hyper- to hypo-virulence seen in several fungal pathogens including Cryphonectria parasitica (Murrill) M.E. Barr (Van Alfen et al. 1975), Ophiostoma novo-ulmi Brasier (Brasier 1990) and Sclerotinia sclerotiorum (Lib.) de Bary (Boland 1992).
With the advent of molecular technologies that have allowed more precise analysis of the genetic basis of unusual pathogenic changes in fungi, it has become apparent that this same mechanism of hyphal fusion is also associated with significant nuclear exchanges. These events may range in magnitude from those that are relatively discrete and involve the lateral transfer of single genes through to situations where whole nuclei are exchanged in dikaryotic or coenocytic fungi.
Lateral gene transfer is a well recognised process in bacterial pathogens (typified by the rapid movement of antibiotic resistance genes or even large chromosomal segments across genera and families) and, in fact, this may well be the primary mechanism behind the emergence of novel pathogenic types in bacteria (e.g. Araki et al. 2006; Lovell et al. 2009; Naum et al. 2009). Such processes may also be an important source of variation in fungal pathogens, although once again, detailed molecular evidence for such events is limited. However, recently this mechanism has been identified as the basis behind the emergence of Pyrenophora tritici-repentis (Died.) Drechsler (tan or yellow spot) as a new disease responsible for significant yield losses in wheat (Friesen et al. 2006). In this instance the sequence coding for the production of a toxin (ToxA) was transferred from a different pathogen, S. nodorum, where a genomic sequence with 99.7% similarity to the P. tritici-repentis ToxA and possessing the same three exons and two introns has been identified. Indeed, the ToxA sequences from the two pathogen species differed at four fixed nucleotide sites only. Comparisons of genes elsewhere in the genome of the two pathogen species, for example, glyceraldehyde 3-phosphate dehydrogenase and the ITS region showed only 80% and 83% similarity respectively, suggesting that the ToxA gene was introduced by some form of lateral gene transfer rather than by fusion and recombination of the two fungal genomes. These findings open up the possibility that lateral gene transfer may be a contributing factor to other fungal diseases of crop plants (Oliver and Solomon 2008). Indeed, a recent molecular analysis of members of the Fusarium oxysporum Schltdl. complex has demonstrated the occurrence of lineage specific genomic regions that include four entire chromosomes. These regions are rich in genes related to pathogenicity, and the transfer of two of these chromosomes between pathogenic and non-pathogenic strains of F. oxysporum resulted in conversion of the non-pathogenic strain into a pathogen (Ma et al. 2010). Furthermore, transfer of these chromosomes between otherwise genetically isolated strains explains the polyphyletic origin of host specificity and the emergence of new pathogenic lineages in the F. oxysporum species complex.
Whole genome exchange
Within existing pathogen–host associations, totally new lineages with markedly different virulence spectra on existing hosts may arise from integration of two, albeit very different, strains of the same pathogen (e.g. the evolutionary process involved in the Australian origin of P. graminis tritici lineage 34 as a consequence of somatic hybridisation and whole nuclear exchange between lineages 126 and 21 (Burdon et al. 1982)). Alternatively, genomic transfer events may lead to the formation of a new pathogen species with an extended host range within its parents’ current host genus (e.g. a combination of Melampsora medusae Thüm. and Melampsora larici populina Kleb. giving a novel Melampsora species (Melampsora medusae-populina Spiers) with a wide host range in Populus (Spiers and Hopcroft 1994), or to a pathogen species that is pathogenic on hosts well beyond the range observed in either parent. Thus in Europe, hybridisation between Phytophthora cambivora (Petri) Buisman (an introduced hardwood pathogen) and Phytophthora fragariae Hickman (pathogenic on strawberries and raspberries) has given rise to a group of heteroploid hybrid taxa, causing significant destruction to Alnus spp. – a totally new host (Brasier et al. 2004). A clear example of the importance of recombination and hybridisation in bacterial pathogens can be seen in the gastroenteric pathogen Campylobacter jejuni (Jones et al. 1931) Veron and Chatelain, where ongoing evolution and adaptation is at least partly driven by interactions with its sister species C. coli (Wilson et al. 2009). More subtle effects of hybridisation have also been documented. For example, molecular studies of M. lini on L. marginale have documented the existence of a widespread clonal pathogen lineage of hybrid origin. This lineage differs considerably in its environmental requirements from a second sexual lineage (Barrett et al. 2007, 2008).
Chromosomal instability
On a larger genomic scale, chromosome instability is a further source of variation. In the sexual fungi, recombination during meiosis may generate significant karyotype variation due to the random assortment of parental homologues of different size (Plummer and Howlett 1995). However, chromosome polymorphism is particularly widespread in imperfect fungi where chromosome aberrations may accumulate without the purging effects of meiosis. Indeed, in Nectria haematococca Berk. & Broome, a sudden increase in pathogenicity against previously immune host species has been associated with the loss of a chromosome (VanEtten et al. 1994). Just how important are these different mechanisms in contributing to the evolutionary dynamics of pathogens and their hosts? While the frequency of detection of such events has increased considerably in recent years (due undoubtedly to the availability of a range of molecular markers and sequence information), there appears to be a generally inverse relationship between the likelihood of occurrence of the event and its potential to totally redirect the nature of any given pathogen–host association. Broadly speaking, simple changes in the frequency of specific virulence occur on a more or less continuous basis as a consequence of the cumulative effect of differential selective interactions between pathogen isolates and different host resistance genes, genetic drift at the end of the epidemic season and migration from other pathogen demes. These represent the fundamental interactions that lead to pathogen population evolution through time within existing host–pathogen interactions. At the other end of the scale, the frequency of occurrence of large scale genomic transfer events is far smaller but when this does occur, it may be hugely disruptive, as seen in examples of host shifts or hybridisation events that may result in the emergence of novel pathogens.
Plant–pathogen coevolution at the defence signalling interface
In this section, we will use plant defence signalling as a case study to explore the coevolutionary arms race between pathogens and plants in more detail. Plant hormones are key players for regulating many aspects of plant development and signalling responses to various stresses. Some of the better understood signalling molecules for biotic responses are SA, jasmonic acid (JA) and ethylene. However, work over the last few years has illustrated the importance of other plant hormones including auxins, cytokinin, ABA, giberellin and brassinosteroids in plant defence responses. These hormones are not only involved in helping to restrict pathogen colonisation, but also to redistribute resources within the plant. These developments have been covered in recent reviews (see Robert-Seilaniantz et al. 2007; Bari and Jones 2009), thus the links between these hormones and defence signalling will not be discussed in depth here. Rather, we will focus on how pathogens have targeted various signalling pathways to increase their virulence and the steps plants have taken to counter pathogen evolution.
Pathogens have also evolved mechanisms to produce plant hormones themselves or to manipulate host hormone biosynthesis to suppress plant defence responses and cause disease. Interestingly, the biosynthetic pathways used by pathogens to synthesise plant hormones are quite distinct from those used by plants, demonstrating independent evolution of these pathways. Different strategies are employed by phytopathogens to alter the endogenous hormones of their host (Jameson 2000). For example, galls incited by A. tumefaciens possess pathogen derived biosynthetic genes for cytokinin and auxin under the regulation of plant derived promoters integrated into the plant genome (Zambryski 1992). As with the gall-forming bacteria other pathogens can produce their own hormones (Spaepen et al. 2007) or significantly interfere with endogenous plant hormone levels (Fraser and Whenham 1982; Clarke et al. 1999). For example, the P. syringae type III effector AvrRpt2 has been shown to alter auxin physiology in the host, and the elevated levels of auxin suppress plant defences and promote disease (Chen et al. 2007). Several pathogenic microbes express oxylipins that are similar to the phytohormone JA (see Robert-Seilaniantz et al. 2007; López et al. 2008; Walling 2009) For example, P. syringae contains a phytotoxin, coronatine, which mimics JA-Ile and promotes virulence by overcoming SA-dependendent defences (Zhao et al. 2003; Brooks et al. 2005). Pathogens synthesise other hormones to interfere with plant defence mechanisms, with the classic example being the fungus Gibberella fujikuroi (Sawada) Wollenw., which produces gibberellins in order to cause disease in rice.
Interactions between plants and insect pests contain a great deal of overlap with plant–microbe interactions, with many of the same signalling pathways induced in response to both types of challenge (Edwards and Singh 2006; Howe and Jander 2008). Like pathogens, some insect pests have also been shown to modulate plant defence signalling. The silver whitefly enhances nymph development by manipulating SA–JA crosstalk, thereby repressing JA mediated defence responses (Zarate et al. 2007; Walling 2009). Plants may also interfere with growth and development of insects and nematodes through the production of hormone mimics (phytoecdysteroids) that interfere with moulting and metamorphosis (Dinan 2001; Soriano et al. 2004).
Other forms of plant defence
In addition to the forms of plant defence described thus far, many more highly specialised forms of plant defence exist and there are a few in particular that are worth noting briefly. Plants have evolved mechanisms to interfere with pest biology in addition to the hormone mimics mentioned earlier. For example, plants produce various protein and secondary metabolite inhibitors of insect processes including food ingestion, assimilation and digestive enzymes such as proteases and amylases (Chen 2008). Moreover, in addition to direct interactions between plant and pest, a tripartite relationship has been described for plant–insect interactions where parasitic insects, typically parasitic wasps or predatory mites, are co-opted by plants to control the infesting pest. Specific volatile organic compounds (VOCs) produced by the plants enable predators to not only identify potential food sources but also to discriminate among plants infested by different herbivore species and among different plants infested by the same herbivore (Heil 2008). An intriguing question remains as to how these relationships might have evolved. For example, did parasitoids develop the ability to associate pre-existing plant volatiles with a food source or did plants evolve volatiles to mimic existing compounds recognised by parasitoids?
A similar situation may exist with microbial endophytes (non-pathogenic bacteria and fungi that reside within plant tissues). Endophytes have been reported to promote plant growth and yield, and have a role in suppressing plant pathogens (Rosenblueth and Martinez-Romero 2006). For example, while an endophytic fungus may obtain protein and nutrients from a plant, the plant may gain protection from herbivores by accumulating toxic alkaloids produced by the endophyte (Schardl et al. 2004). This relationship is, however, often dynamic and, under certain circumstances, the endophyte may become pathogenic (Schulz and Boyle 2005). The fossils of the early Devonian Rhynie chert contain fungal endophytes (Krings et al. 2007), and analysis of these fossil specimens show ancient endophytes apparently inducing a plant host reaction; evidence of cell wall thickening, encasement of the intercellular hyphae and tissue degradation in response to the endophyte suggests that pathogen induced responses in extant plants were in position over 400 million years ago (Krings et al. 2007). The presence of non-pathogenic – even beneficial – microbes within a plant poses important questions relating to how plants distinguish between friend and foe, how they modulate their defence responses to keep these organisms in check and how they simultaneously balance interactions with beneficial and pathogenic microbes.
Evolving the ability to distinguish friend from foe
From an evolutionary perspective, one of the oldest and most widespread forms of symbiosis is between land plants and arbuscular mycorrhiza (AM). In this interaction, the fungus helps the plant acquire water and nutrients such as phosphate and nitrogen, while carbohydrates from the plant are supplied to the fungus. AM fungi have been in existence for over 400 million years (Remy et al. 1994) and because AM fungi are widely distributed and occur in 70–90% of all land plants, this mutualism was probably present in ancestral plants. In fact, it has been theorised that the presence of fungal symbionts in early Devonian plants was pivotal for plant colonisation of the land (Remy et al. 1994).
AM fungi can infect a wide range of plant taxa, and the consistency of several traits of the AM infection processes suggests that the molecular mechanisms of AM symbiosis was present in the ancestral lineage (Bonfante and Genre 2008). These molecular mechanisms may have been hijacked by the fungus from pre-existing plant cellular mechanisms and subsequently transformed into a genetic program required for successful colonisation and symbiotic interaction. For example, the AM pre-penetration apparatus (PPA), which guides the penetrating hypha through plant cells (Genre et al. 2005), has overlap with cell division processes. Both cell division and PPA assembly consist of cytoskeletal, endoplasmic reticulum and secretory elements, and both require the production of a cell wall within the lumen rather than along the existing cell wall surface (Bonfante and Genre 2008). How the AM fungus is able to evade plant host immunity is still a matter of investigation, but recent work comparing pathogenic and symbiotic fungal colonisation of Medicago trunculata Gaertn. showed that the plant had similar pre-infection nuclear repositioning responses to both the AM symbiont and the fungal pathogens, suggesting that there is a common primary plant response to these organisms that is further modified depending on the nature of the interacting organism (Genre et al. 2009).
The ancient symbiotic interaction between plant and AM fungi is believed to be the ancestor of bacterial root endosymbioses based on the hypothesis that nodulation co-opted genes from a pre-existing genetic program involving AM fungal symbiosis. Mutagenesis studies defined a common symbiosis pathway between symbiotic rhizobia and AM fungi, and infection processes for these symbionts share a similar requirement for nuclear re-positioning and cytoskeletal rearrangements to guide the microbe through the plant cells to the inner cortex (Genre et al. 2005). Plants evolved the means to form symbiotic relationships with nitrogen-fixing bacteria ~58 Ma (Sprent and James 2007) enabling legumes to overcome one of the main restrictions of plant growth, limited availability of soil nitrogen. With the exception of Gunnera, all nodulating, flowering plants are restricted to Rosid I clade in Eurosid I, which suggests a common ancestor with a pre-disposition for nodulation (Soltis et al. 1995). Subsequent parallel evolutionary events in the genetic pathway may be responsible in root nodule symbiosis in a diversity of plants in this clade (Soltis et al. 1995; Markmann and Parniske 2009).
How rhizobia are able to establish themselves in host plants without triggering plant defence responses is still largely unknown. Infection threads are formed by the plant and encase the bacteria in components derived from the cell wall, and may represent a form of plant defence response to the bacteria. If the bacteria are perceived as possible pathogens, they remain sequestered in infection threads, but if perceived as beneficial, they are released into symbiosomes (Sprent 2007). Treatment of legume roots with purified Nod factor causes influxes of calcium, changes in pH and a transient induction of reactive oxygen species (ROS) in a response highly reminiscent of plant responses to PAMPs; however, the rhizobia fail to trigger a strong plant defence response (Felle et al. 2000; Ramu et al. 2002; Cárdenas et al. 2008).
Many of the weapons that plants use to fight bacterial pathogens are being initiated during nodulation; however, in many instances, the rhizobia manipulate these responses to drive nodulation positively (Soto et al. 2009). There are several lines of evidence to show that responses often associated with defence can have both a positive and negative role in rhizobial symbiosis. The presence of ROS around aborted infection threads could be an indication that in these instances, the plant recognises the bacteria as pathogenic and mounts a defence against invasion (Vasse et al. 1993). However, a positive role for the involvement of ROS in nodulation is supported by the regulation of root hair curling and infection thread formation by ROS production (Peleg-Grossman et al. 2007). This ROS accumulation is transient and it may be this precise control of ROS levels that provides a mechanism for rhizobia to enter the plant without triggering a defence response (Cárdenas et al. 2008). NO is also required for both defence and the legume–rhizobia interaction. NO plays a role in HR and activating defence genes; however, it positively mediates indeterminate nodule formation and is found at high levels within nodules (Baudouin et al. 2006; Pii et al. 2007; Soto et al. 2009).
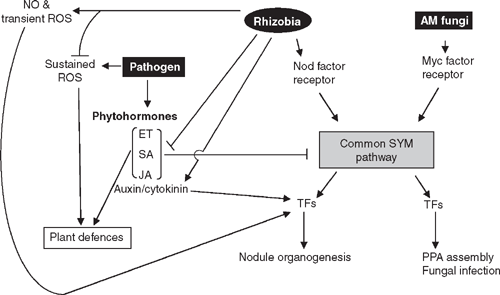
Further evidence for rhizobial manipulation of host defences is provided by the defence suppressing activity of bacterially derived Nod factors on their symbiotic hosts. On non-host plants, Nod factors can trigger defence reactions, which are absent in hosts, suggesting that the defence suppression ability of Nod factors has evolved recently. Nod factors possess a backbone with high similarity to chitin, the building block of fungal cell walls and a potent inducer of PTI. Both chitin and Nod factors are thought to be perceived in plants by LysM-RLKs (Madsen et al. 2003; Radutoiu et al. 2003), suggesting that chitin and Nod factor perception share an evolutionary relationship (Wan et al. 2008). Further similarity between pathogenesis and symbiosis mechanisms is highlighted in a system that is highly reminiscent of the effectors described for the bacterial pathogen, P. syringae. Type III and type IV secretion systems (T3SS and T4SS respectively) facilitate the delivery of nodulation outer proteins (Nops) that encode homologues of effector proteins from pathogenic bacteria and contribute to host specificity (Soto et al. 2006; Deakin and Broughton 2009). Some legumes recognise specific Nops, leading to a blocked infection process, whereas in other legumes, these same Nops enhance the symbiosis (Kambara et al. 2009). The evolution of root nodule symbiosis indicates that the bacteria have hijacked existing genetic pathways from another symbiont during the evolutionary process and have evolved sophisticated methods of evading detection and suppressing defence responses in their cognate legumes. A summary of the interactions between symbiosis and defence pathways is presented in Fig. 3.
|
Conclusions
Plant–pathogen interactions will continue to drive relatively rapid evolutionary change on both sides of the interaction, particularly in terms of the specificities involved in pathogen recognition. Moreover, the impact of human interference through agriculture and forestry is increasingly a major driving force that is heavily influencing the evolutionary arms race between many plant pathogens and their plant hosts. However, it appears that regardless of the microbe encountered, the plant is likely to mount distinct but overlapping defence reactions to all potential evaders and the onus is placed on the microbe to suppress the plant’s defences in order to either cause disease or enter into a symbiotic interaction. The field is in a dynamic phase that will be further enhanced by the rapid development of pathogenomics and the development of technologies such as next generation sequencing to study both sides of plant–pathogen interactions simultaneously. Moreover, the advent of metagenomics opens up opportunities to study complex multiple microbe–host interactions that more closely reflect real world situations and these studies are likely to provide new insight into plant–pathogen coevolution.
Acknowledgements
We thank members of the Singh laboratory for helpful comments. The work in the authors’ laboratories on biotic stress is supported by CSIRO, The Grains Research and Development Corporation and the National Institute of Health (NIH grant 5RO1 GM074265–01A2). We apologise to those colleagues we could not cite due to the broad scope of this review and space limitations.
Altenbach D, Robatzek S
(2007) Pattern recognition receptors: from the cell surface to intracellular dynamics. Molecular Plant–Microbe Interactions 20, 1031–1039.
| Crossref | GoogleScholarGoogle Scholar |

Araki H,
Tian D,
Goss EM,
Jakob K,
Halldorsdottir SS,
Kreitman M, Bergelson J
(2006) Presence/absence polymorphism for alternative pathogenicity islands in Pseudomonas viridiflava, a pathogen of Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 103, 5887–5892.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Asai T,
Tena G,
Plotnikova J,
Willmann MR,
Chiu WL,
Gomez-Gomez L,
Boller T,
Ausubel FM, Sheen J
(2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Axtell MJ, Staskawicz BJ
(2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112, 369–377.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Bari R, Jones J
(2009) Role of plant hormones in plant defence responses. Plant Molecular Biology 69, 473–488.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Barrett LG,
Thrall PH, Burdon JJ
(2007) Evolutionary diversification through hybridization in a wild host–pathogen interaction. Evolution 61, 1613–1621.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Barrett LG,
Thrall PH,
Burdon JJ,
Nicotra AB, Linde CC
(2008) Population structure and diversity in sexual and asexual populations of the pathogenic fungus Melampsora lini. Molecular Ecology 17, 3401–3415.
| Crossref |
PubMed |

Barrett LG,
Thrall PH,
Dodds PN,
van der Merwe M,
Linde CC,
Lawrence GH, Burdon JJ
(2009) Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini. Molecular Biology and Evolution 26, 2499–2513.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Baudouin E,
Pieuchot L,
Engler G,
Pauly N, Puppo A
(2006) Nitric oxide is formed in Medicago truncatula–Sinorhizobium meliloti functional nodules. Molecular Plant–Microbe Interactions 19, 970–975.
| Crossref | GoogleScholarGoogle Scholar |

Bednarek P, Osbourn A
(2009) Plant–microbe interactions: chemical diversity in plant defense. Science 324, 746–748.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Bent AF, Mackey D
(2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annual Review of Phytopathology 45, 399–436.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Birch PRJ,
Boevink PC,
Gilroy EM,
Hein I,
Pritchard L, Whisson SC
(2008) Oomycete RXLR effectors: delivery, functional redundancy and durable disease resistance. Current Opinion in Plant Biology 11, 373–379.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Boland G
(1992) Hypovirulence and double-stranded RNA in Sclerotinia sclerotiorum. Canadian Journal of Plant Pathology 14, 10–17.

Boller T, He SY
(2009) Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Bonfante P, Genre A
(2008) Plants and arbuscular mycorrhizal fungi: an evolutionary-developmental perspective. Trends in Plant Science 13, 492–498.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Brasier CM,
Kirk SA,
Delcan J,
Cooke DEL,
Jung T, Man In’t Veld WA
(2004)
Phytophthora alni sp. nov. and its variants: designation of emerging heteroploid hybrid pathogens spreading on Alnus trees. Mycological Research 108, 1172–1184.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Brooks DM,
Bender CL, Kunkel BN
(2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Molecular Plant Pathology 6, 629–639.
| Crossref | GoogleScholarGoogle Scholar |

Brown JKM
(1995) Recombination and selection in populations of plant pathogens. Plant Pathology 44, 279–293.
| Crossref | GoogleScholarGoogle Scholar |

Burdon JJ, Roelfs AP
(1985) Isozyme and virulence variation in asexually reproducing populations of Puccinia graminis and P. recondita on wheat. Phytopathology 75, 907–913.
| Crossref | GoogleScholarGoogle Scholar |

Burdon JJ,
Marshall DR,
Luig NH, Gow DJS
(1982) Isozyme studies on the origin and evolution of Puccinia graminis f. sp. tritici in Australia. Australian Journal of Biological Sciences 35, 231–238.

Cárdenas L,
Martínez A,
Sánchez F, Quinto C
(2008) Fast, transient and specific intracellular ROS changes in living root hair cells responding to Nod factors (NFs). The Plant Journal 56, 802–813.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chang JH,
Urbach JM,
Law TF,
Arnold LW,
Hu A,
Gombar S,
Grant SR,
Ausubel FM, Dangl JL
(2005) A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proceedings of the National Academy of Sciences of the United States of America 102, 2549–2554.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chen MS
(2008) Inducible direct plant defense against insect herbivores: a review. Insect Science 15, 101–114.
| Crossref | GoogleScholarGoogle Scholar |

Chen Z,
Agnew JL,
Cohen JD,
He P,
Shan L,
Sheen J, Kunkel BN
(2007)
Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proceedings of the National Academy of Sciences of the United States of America 104, 20 131–20 136.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chinchilla D,
Bauer Z,
Regenass M,
Boller T, Felix G
(2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. The Plant Cell 18, 465–476.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chisholm ST,
Coaker G,
Day B, Staskawicz BJ
(2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Clarke SF,
McKenzie MJ,
Burritt DJ,
Guy PL, Jameson PE
(1999) Influence of white clover mosaic potexvirus infection on the endogenous cytokinin content of bean. Plant Physiology 120, 547–552.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Dangl JL, Jones JDG
(2001) Plant pathogens and integrated defence responses to infection. Nature 411, 826–833.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

de Torres M,
Mansfield JW,
Grabov N,
Brown IR,
Ammouneh H,
Tsiamis G,
Forsyth A,
Robatzek S,
Grant M, Boch J
(2006)
Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. The Plant Journal 47, 368–382.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

de Wit PJGM
(2007) How plants recognize pathogens and defend themselves. Cellular and Molecular Life Sciences 64, 2726–2732.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Deakin WJ, Broughton WJ
(2009) Symbiotic use of pathogenic strategies: rhizobial protein secretion systems. Nature Reviews Microbiology 7, 312–320.
| PubMed |

Dinan L
(2001) Phytoecdysteroids: biological aspects. Phytochemistry 57, 325–339.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ding Y, Oldroyd GE
(2009) Positioning the nodule, the hormone dictum. Plant Signaling & Behavior 4, 89–93.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Dodds PN, Thrall P
(2009) Recognition events and host-pathogen co-evolution in gene-for-gene resistance to flax rust. Functional Plant Biology 36, 395–408.
| Crossref | GoogleScholarGoogle Scholar |

Dodds PN,
Lawrence GJ,
Catanzariti AM,
Teh T,
Wang CIA,
Ayliffe MA,
Kobe B, Ellis JG
(2006) Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proceedings of the National Academy of Sciences of the United States of America 103, 8888–8893.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Dou DL,
Kale SD,
Wang X,
Jiang RHY,
Bruce NA,
Arredondo FD,
Zhang XM, Tyler BM
(2008) RXLR-mediated entry of Phytophthora sojae effector Avr1b into soybean cells does not require pathogen-encoded machinery. The Plant Cell 20, 1930–1947.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Edwards O, Singh KB
(2006) Resistance to insect pests: what do legumes have to offer? Euphytica 147, 273–285.
| Crossref | GoogleScholarGoogle Scholar |

Ellis JG,
Dodds PN, Lawrence GJ
(2007) Flax rust resistance gene specificity is based on direct resistance–avirulence protein interactions. Annual Review of Phytopathology 45, 289–306.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Enjalbert J,
Duan M,
Leconte M,
Hovmøller MS, de Vallavieille-Pope C
(2005) Genetic evidence of local adaptation of wheat yellow rust (Puccinia striiformis f. sp. tritici) within France. Molecular Ecology 14, 2065–2073.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Felle HH,
Kondorosi E,
Kondorosi A, Schultze M
(2000) How alfalfa root hairs discriminate between Nod factors and oligochitin elicitors. Plant Physiology 124, 1373–1380.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Fraser RSS, Whenham RJ
(1982) Plant-growth regulators and virus-infection – a critical review. Plant Growth Regulation 1, 37–59.
| Crossref | GoogleScholarGoogle Scholar |

Friesen T,
Stukenbrock EH,
Lui Z,
Meinhardt S,
Ling H,
Faris JD,
Rasmussen JB,
Solomon PS,
McDonald BA, Oliver RP
(2006) Emergence of a new disease as a result of interspecific virulence gene transfer. Nature Genetics 38, 953–956.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Friesen TL,
Faris JD,
Solomon PS, Oliver RP
(2008) Host-specific toxins: effectors of necrotrophic pathogenicity. Cellular Microbiology 10, 1421–1428.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Friesen TL,
Chu CG,
Liu ZH,
Xu SS,
Halley S, Faris JD
(2009) Host-selective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theoretical and Applied Genetics 118, 1489–1497.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Frugier F,
Kosuta S,
Murray JD,
Crespi M, Szczyglowski K
(2008) Cytokinin: secret agent of symbiosis. Trends in Plant Science 13, 115–120.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Genre A,
Chabaud M,
Timmers T,
Bonfante P, Barker DG
(2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. The Plant Cell 17, 3489–3499.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Genre A,
Ortu G,
Bertoldo C,
Martino E, Bonfante P
(2009) Biotic and abiotic stimulation of root epidermal cells reveals common and specific responses to arbuscular mycorrhizal fungi. Plant Physiology 149, 1424–1434.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gimenez-Ibanez S,
Hann DR,
Ntoukakls V,
Petutschnig E,
Lipka V, Rathjen JP
(2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Current Biology 19, 423–429.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Glazebrook J
(2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43, 205–227.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gomez-Gomez L, Boller T
(2000) FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Molecular Cell 5, 1003–1011.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Govrin EM, Levine A
(2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Current Biology 10, 751–757.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Grunewald W,
van Noorden G,
Van Isterdael G,
Beeckman T,
Gheysen G, Mathesius U
(2009) Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. The Plant Cell 21, 2553–2562.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gudesblat GE,
Torres PS, Vojnov AA
(2009)
Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiology 149, 1017–1027.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Haque S,
Park RF,
Keiper FJ,
Bariana HS, Wellings CR
(2008) Pathogenic and molecular variation support the presence of genetically distinct clonal lineages in Australian populations of Puccinia graminis f. sp. avenae. Mycological Research 112, 663–673.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hardham AR,
Jones DA, Takemoto D
(2007) Cytoskeleton and cell wall function in penetration resistance. Current Opinion in Plant Biology 10, 342–348.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

He P,
Shan L,
Lin NC,
Martin GB,
Kemmerling B,
Nurnberger T, Sheen J
(2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125, 563–575.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Heese A,
Hann DR,
Gimenez-Ibanez S,
Jones AME,
He K,
Li J,
Schroeder JI,
Peck SC, Rathjen JP
(2007) The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proceedings of the National Academy of Sciences of the United States of America 104, 12 217–12 222.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Heil M
(2008) Indirect defence via tritrophic interactions. New Phytologist 178, 41–61.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hovmøller MS,
Justesen AF, Brown JKM
(2002) Clonality and long-distance migration of Puccinia striiformis f. sp. tritici in north-west Europe. Plant Pathology 51, 24–32.
| Crossref | GoogleScholarGoogle Scholar |

Howe GA, Jander G
(2008) Plant immunity to insect herbivores. Annual Review of Plant Biology 59, 41–66.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Jameson PE
(2000) Cytokinins and auxins in plant–pathogen interactions – an overview. Plant Growth Regulation 32, 369–380.
| Crossref | GoogleScholarGoogle Scholar |

Janjusevic R,
Abramovitch RB,
Martin GB, Stebbins CE
(2006) A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311, 222–226.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Jones JDG, Dangl JL
(2006) The plant immune system. Nature 444, 323–329.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kambara K,
Ardissone S,
Kobayashi H,
Saad MM,
Schumpp O,
Broughton WJ, Deakin WJ
(2009) Rhizobia utilize pathogen-like effector proteins during symbiosis. Molecular Microbiology 71, 92–106.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kim MG,
da Cunha L,
McFall AJ,
Belkhadir Y,
DebRoy S,
Dangl JL, Mackey D
(2005) Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121, 749–759.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kobayashi Y,
Kobayashi I,
Funaki Y,
Fujimoto S,
Takemoto T, Kunoh H
(1997) Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non-host resistance in barley coleoptile cells. The Plant Journal 11, 525–537.
| Crossref | GoogleScholarGoogle Scholar |

Krings M,
Taylor TN,
Hass H,
Kerp H,
Dotzler N, Hermsen EJ
(2007) Fungal endophytes in a 400-million-yr-old land plant: infection pathways, spatial distribution, and host responses. New Phytologist 174, 648–657.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kumar M,
Busch W,
Birke H,
Kemmerling B,
Nurnberger T, Schoffl F
(2009) Heat shock factors HsfB1 and HsfB2b are involved in the regulation of Pdf1.2 expression and pathogen resistance in Arabidopsis. Molecular Plant 2, 152–165.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kunkeaw S,
Tan S, Coaker G
(2010) Molecular and evolutionary analyses of Pseudomonas syringae pv. tomato race 1. Molecular Plant–Microbe Interactions 23, 415–424.
| Crossref | GoogleScholarGoogle Scholar |

Kunze G,
Zipfel C,
Robatzek S,
Niehaus K,
Boller T, Felix G
(2004) The N-terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. The Plant Cell 16, 3496–3507.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Lawrence GJ,
Boelen MG, Pryor A
(1988) Transmission of double-stranded RNAs in flax rust, Melampsora lini. Canadian Journal of Botany 66, 61–66.

Lee J,
Klessig DF, Nurnberger T
(2001) A harpin binding site in tobacco plasma membranes mediates activation of the pathogenesis-related gene HIN1 independent of extracellular calcium but dependent on mitogen-activated protein kinase activity. The Plant Cell 13, 1079–1093.
| Crossref |
PubMed |

Liu ZH,
Friesen TL,
Ling H,
Meinhardt SW,
Oliver RP,
Rasmussen JB, Faris JD
(2006) The Tsn1-ToxA interaction in the wheat–Stagonospora nodorum pathosystem parallels that of the wheat–tan spot system. Genome 49, 1265–1273.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Liu J,
Elmore JM,
Fuglsang AT,
Palmgren MG,
Staskawicz BJ, Coaker G
(2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. Plos Biology 7, e1000139.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

López MA,
Bannenberg G, Castresana C
(2008) Controlling hormone signaling is a plant and pathogen challenge for growth and survival. Current Opinion in Plant Biology 11, 420–427.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Lorang JM,
Sweat TA, Wolpert TJ
(2007) Plant disease susceptibility conferred by a “resistance” gene. Proceedings of the National Academy of Sciences of the United States of America 104, 14 861–14 866.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Lovell HC,
Mansfield JW,
Godfrey SAC,
Jackson RW,
Hancock JT, Arnold DL
(2009) Bacterial evolution by genomic island transfer occurs via DNA transformation in planta. Current Biology 19, 1586–1590.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ma WB,
Dong FFT,
Stavrinides J, Guttman DS
(2006) Type III effector diversification via both pathoadaptation and horizontal transfer in response to a coevolutionary arms race. PLOS Genetics 2, 2131–2142.
| Crossref | GoogleScholarGoogle Scholar |

Ma L-J,
van der Does HC,
Borkovich KA,
Coleman JJ, Daboussi M-J ,
et al
.
(2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium oxysporum. Nature 464, 367–373.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Mackey D,
Holt BF,
Wiig A, Dangl JL
(2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Mackey D,
Belkhadir Y,
Alonso JM,
Ecker JR, Dangl JL
(2003)
Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112, 379–389.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Madsen EB,
Madsen LH,
Madsen EB,
Madsen LH, Radutoiu S ,
et al
.
(2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Markmann K, Parniske M
(2009) Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends in Plant Science 14, 77–86.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Martin GB,
Bogdanove AJ, Sessa G
(2003) Understanding the functions of plant disease resistance proteins. Annual Review of Plant Biology 54, 23–61.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

McDonald BA, Linde C
(2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annual Review of Phytopathology 40, 349–379.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Melotto M,
Underwood W,
Koczan J,
Nomura K, He SY
(2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Melotto M,
Underwood W, He SY
(2008) Role of stomata in plant innate immunity and foliar bacterial diseases. Annual Review of Phytopathology 46, 101–122.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Morrissey JP, Osbourn AE
(1999) Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiology and Molecular Biology Reviews 63, 708–724.
| PubMed |

Naum M,
Brown EW, Mason-Gamer RJ
(2009) Phylogenetic evidence for extensive horizontal gene transfer of type III secretion system genes among enterobacterial plant pathogens. Microbiology 155, 3187–3199.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Nurnberger T,
Brunner F,
Kemmerling B, Piater L
(2004) Innate immunity in plants and animals: striking similarities and obvious differences. Immunological Reviews 198, 249–266.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ochman H, Moran NA
(2001) Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science 292, 1096–1099.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Oldroyd GE,
Harrison MJ, Paszkowski U
(2009) Reprogramming plant cells for endosymbiosis. Science 324, 753–754.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Oliver RP, Solomon PS
(2008) Recent fungal diseases of crop plants: is lateral gene transfer a common theme? Molecular Plant–Microbe Interactions 21, 287–293.
| Crossref | GoogleScholarGoogle Scholar |

Oliver RP,
Rybak K,
Shankar M,
Loughman R,
Harry N, Solomon PS
(2008) Quantitative disease resistance assessment by real-time PCR using the Stagonospora nodorum–wheat pathosystem as a model. Plant Pathology 57, 527–532.
| Crossref | GoogleScholarGoogle Scholar |

Papadopoulou K,
Melton RE,
Leggett M,
Daniels MJ, Osbourn AE
(1999) Compromised disease resistance in saponin-deficient plants. Proceedings of the National Academy of Sciences of the United States of America 96, 12 923–12 928.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Parniske M
(2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nature Reviews Microbiology 6, 763–775.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Peleg-Grossman S,
Volpin H, Levine A
(2007) Root hair curling and rhizobium infection in Medicago truncatula are mediated by phosphatidylinositide-regulated endocytosis and reactive oxygen species. Journal of Experimental Botany 58, 1637–1649.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Penmetsa RV,
Uribe P,
Anderson JP,
Gish JC, Nam YW ,
et al
.
(2008) The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. The Plant Journal 55, 580–595.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Pii Y,
Crimi M,
Cremonese G,
Spena A, Pandolfini T
(2007) Auxin and nitric oxide control indeterminate nodule formation. BMC Plant Biology 7, 21.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Plummer KM, Howlett BJ
(1995) Inheritance of chromosomal length polymorphisms in the ascomycete Leptosphaeria maculans. Molecular & General Genetics 247, 416–422.
| Crossref | GoogleScholarGoogle Scholar |

Radutoiu S,
Madsen LH,
Madsen EB,
Felle HH, Umehara Y ,
et al
.
(2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ramu SK,
Peng H-M, Cook DR
(2002) Nod factor induction of reactive oxygen species production is correlated with expression of the early nodulin gene rip1 in Medicago truncatula. Molecular Plant–Microbe Interactions 15, 522–528.
| Crossref | GoogleScholarGoogle Scholar |

Rehmany AP,
Gordon A,
Allen RL,
Armstrong MR,
Whisson SC,
Kamoun S,
Tyler BM,
Birch PRJ, Beynon JL
(2005) Differential recognition of highly divergent downy mildew avirulence gene alleles by RPP1 resistance genes from two Arabidopsis lines. The Plant Cell 17, 1839–1850.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Remy W,
Taylor TN,
Hass H, Kerp H
(1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proceedings of the National Academy of Sciences of the United States of America 91, 11 841–11 843.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Robert-Seilaniantz A,
Navarro L,
Bari R, Jones JD
(2007) Pathological hormone imbalances. Current Opinion in Plant Biology 10, 372–379.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Rosenblueth M, Martinez-Romero E
(2006) Bacterial endophytes and their interactions with hosts. Molecular Plant–Microbe Interactions 19, 827–837.
| Crossref | GoogleScholarGoogle Scholar |

Sato M,
Mitra RM,
Coller J,
Wang D,
Spivey NW,
Dewdney J,
Denoux C,
Glazebrook J, Katagiri F
(2007) A high-performance, small-scale microarray for expression profiling of many samples in Arabidopsis–pathogen studies. The Plant Journal 49, 565–577.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Schardl CL,
Leuchtmann A, Spiering MJ
(2004) Symbioses of grasses with seedborne fungal endophytes. Annual Review of Plant Biology 55, 315–340.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Schulz B, Boyle C
(2005) The endophytic continuum. Mycological Research 109, 661–686.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Schwessinger B, Zipfel C
(2008) News from the frontline: recent insights into PAMP-triggered immunity in plants. Current Opinion in Plant Biology 11, 389–395.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Shan LB,
He P, Sheen J
(2007) Endless hide-and-seek: dynamic co-evolution in plant–bacterium warfare. Journal of Integrative Plant Biology 49, 105–111.
| Crossref | GoogleScholarGoogle Scholar |

Shan L,
He P,
Li J,
Heese A,
Peck SC,
Nurnberger T,
Martin GB, Sheen J
(2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host & Microbe 4, 17–27.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Soltis DE,
Soltis PS,
Morgan DR,
Swensen SM,
Mullin BC,
Dowd JM, Martin PG
(1995) Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proceedings of the National Academy of Sciences of the United States of America 92, 2647–2651.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Soriano IR,
Riley IT,
Potter MJ, Bowers WS
(2004) Phytoecdysteroids: a novel defense against plant-parasitic nematodes. Journal of Chemical Ecology 30, 1885–1899.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Soto MJ,
Sanjuan J, Olivares J
(2006) Rhizobia and plant–pathogenic bacteria: common infection weapons. Microbiology 152, 3167–3174.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Soto MJ,
Dominguez-Ferreras A,
Perez-Mendoza D,
Sanjuan J, Olivares J
(2009) Mutualism versus pathogenesis: the give-and-take in plant–bacteria interactions. Cellular Microbiology 11, 381–388.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Spaepen S,
Vanderleyden J, Remans R
(2007) Indole-3-acetic acid in microbial and microorganism–plant signaling. FEMS Microbiology Reviews 31, 425–448.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Spiers AG, Hopcroft DH
(1994) Comparative studies of the poplar rusts Melampsora medusae, M. larici-populina and their interspecific hybrid M. medusae-populina. Mycological Research 98, 889–903.
| Crossref | GoogleScholarGoogle Scholar |

Sprent JI
(2007) Evolving ideas of legume evolution and diversity: a taxonomic perspective on the occurrence of nodulation. New Phytologist 174, 11–25.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Sprent JI, James EK
(2007) Legume evolution: where do nodules and mycorrhizas fit in? Plant Physiology 144, 575–581.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Staskawicz BJ,
Mudgett MB,
Dangl JL, Galan JE
(2001) Common and contrasting themes of plant and animal diseases. Science 292, 2285–2289.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Stukenbrock EH, McDonald BA
(2009) Population genetics of fungal and oomycete effectors involved in gene-for-gene interactions. Molecular Plant–Microbe Interactions 22, 371–380.
| Crossref | GoogleScholarGoogle Scholar |

Takai R,
Isogai A,
Takayama S, Che FS
(2008) Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice. Molecular Plant–Microbe Interactions 21, 1635–1642.
| Crossref | GoogleScholarGoogle Scholar |

Thatcher LF,
Anderson JP, Singh KB
(2005) Plant defence responses: what have we learnt from Arabidopsis? Functional Plant Biology 32, 1–19.
| Crossref | GoogleScholarGoogle Scholar |

Thomma B,
Penninckx I,
Broekaert WF, Cammue BPA
(2001) The complexity of disease signaling in Arabidopsis. Current Opinion in Immunology 13, 63–68.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Thomma B,
Cammue BPA, Thevissen K
(2002) Plant defensins. Planta 216, 193–202.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Thordal-Christensen H
(2003) Fresh insights into processes of nonhost resistance. Current Opinion in Plant Biology 6, 351–357.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Thrall PH,
Bever JD,
Mihail J, Alexander HM
(1997) The population dynamics of annual plants and soil-borne pathogens. Journal of Ecology 85, 313–328.
| Crossref | GoogleScholarGoogle Scholar |

Tsuda K,
Sato M,
Glazebrook J,
Cohen JD, Katagiri F
(2008) Interplay between MAMP-triggered and SA-mediated defense responses. The Plant Journal 53, 763–775.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Van Alfen NK,
Janes RA,
Anagnostakis SL, Day PR
(1975) Chestnut blight: biological control by transmissible hypovirulence in Endothia parasitica. Science 189, 890–891.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

van den Burg HA,
Westerink N,
Francoijs KJ,
Roth R,
Woestenenk E,
Boeren S,
de Wit PJGM,
Joosten MHAJ, Vervoort J
(2003) Natural disulfide bond-disrupted mutants of AVR4 of the tomato pathogen Cladosporium fulvum are sensitive to proteolysis, circumvent Cf-4-mediated resistance, but retain their chitin binding ability. The Journal of Biological Chemistry 278, 27 340–27 346.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

van Esse HP,
Bolton MD,
Stergiopoulos L,
de Wit P, Thomma B
(2007) The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Molecular Plant–Microbe Interactions 20, 1092–1101.
| Crossref | GoogleScholarGoogle Scholar |

VanEtten H,
Funnell-Baerg D,
Wasmann C, McCluskey K
(1994) Location of pathogenicity genes on dispensible chromosomes in Nectria haematococca MPVI. Antonie Van Leeuwenhoek 65, 263–267.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Vasse J,
Billy F, Truchet G
(1993) Abortion of infection during the Rhizobium meliloti–alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. The Plant Journal 4, 555–566.
| Crossref | GoogleScholarGoogle Scholar |

Wan J,
Zhang XC,
Neece D,
Ramonell KM,
Clough S,
Kim SY,
Stacey MG, Stacey G
(2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. The Plant Cell 20, 471–481.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Wellings CR, McIntosh RA
(1990)
Puccinia striiformis f. sp. tritici in Australia – pathogenic changes during the first ten years. Plant Pathology 39, 316–325.
| Crossref | GoogleScholarGoogle Scholar |

Wilson DJ,
Gabriel E,
Leatherbarrow AJ,
Cheesbrough J,
Gee S,
Bolton E,
Fox A,
Hart CA,
Diggle PJ, Fearnhead P
(2009) Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Molecular Biology and Evolution 26, 385–397.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Zambryski PC
(1992) chronicles from the agrobacterium-plant cell-DNA transfer story. Annual Review of Plant Physiology and Plant Molecular Biology 43, 465–490.
| Crossref | GoogleScholarGoogle Scholar |

Zarate SI,
Kempema LA, Walling LL
(2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiology 143, 866–875.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Zhang X,
Dai Y,
Xiong Y,
DeFraia C,
Li J,
Dong X, Mou Z
(2007) Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. The Plant Journal 52, 1066–1079.
| Crossref |
PubMed |

Zhang W,
He SY, Assmann SM
(2008) The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. The Plant Journal 56, 984–996.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Zhao YF,
Thilmony R,
Bender CL,
Schaller A,
He SY, Howe GA
(2003) Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. The Plant Journal 36, 485–499.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Zipfel C,
Kunze G,
Chinchilla D,
Caniard A,
Jones JDG,
Boller T, Felix G
(2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125, 749–760.
| Crossref | GoogleScholarGoogle Scholar | PubMed |



