The evolutionary development of plant body plans
Karl J. Niklas A C and Ulrich Kutschera BA Department of Plant Biology, Cornell University, Ithaca, NY 14853, USA.
B Institute of Biology, University of Kassel, Heinrich-Plett-Strasse 40, D-34109 Kassel, Germany.
C Corresponding author. Email: kjn2@cornell.edu
This paper is part of an ongoing series: ‘The Evolution of Plant Functions’.
Functional Plant Biology 36(8) 682-695 https://doi.org/10.1071/FP09107
Submitted: 12 May 2009 Accepted: 12 June 2009 Published: 23 July 2009
Abstract
Evolutionary developmental biology, cladistic analyses, and paleontological insights make it increasingly clear that regulatory mechanisms operating during embryogenesis and early maturation tend to be highly conserved over great evolutionary time scales, which can account for the conservative nature of the body plans in the major plant and animal clades. At issue is whether morphological convergences in body plans among evolutionarily divergent lineages are the result of adaptive convergence or ‘genome recall’ and ‘process orthology’. The body plans of multicellular photosynthetic eukaryotes (‘plants’) are reviewed, some of their important developmental/physiological regulatory mechanisms discussed, and the evidence that some of these mechanisms are phyletically ancient examined. We conclude that endosymbiotic lateral gene transfers, gene duplication and functional divergence, and the co-option of ancient gene networks were key to the evolutionary divergence of plant lineages.
Additional keywords: apogamy, apospory, auxin, endosymbiosis, euphyllophytes, floral identity genes, homeodomain genes, MADS-box genes, plant evolution, TIR1.
Introduction
In his Origin of Species, Charles Darwin (1859) wrote that ‘… all organic beings have been formed on two great laws – Unity of Type and the Conditions of Existence. By unity of type is meant that fundamental agreement in structure, which we see in organic beings of the same class, and which is quite independent of their habits of life. The expression of conditions of existence… is fully embraced by the principle of natural selection [which] acts by either now adapting the varying parts of each being to its organic conditions of life; or by having adapted them in long-past periods of time’. This musing focuses on an important evolutionary phenomenon, viz. the conservation of body plans within major clades (Darwin’s unity of type) despite numerous, often dramatic phenotypic divergence in different environments (Darwin’s conditions of existence). The chordate body plan is thus identifiable by its metameric myotomes, notochord, dorsal hollow nervous system, and many other character states. Yet, its unity of type has evolutionarily diversified under the influence of random genomic changes and directional natural selection to yield a constellation of animals, ranging in size and appearance from the aquatic lancelet (Amphioxus) and the burrowing acorn worm (Balanoglossus) to the tiger (Felis tigris) and our species (Homo sapiens). Likewise, the angiosperm body plan is characterised by carpels, reduced microgametophytes, polyploid endosperm, and a variety of other features. Yet, it encompasses 270 000 extant species, ranging from the parasitic Indian pipe (Monotropa) and carnivorous Pitcher plant (Nepenthes) to the garden varieties of corn (Zea mays L.) and tomato (Solanum lycopersicum L.). It is not surprising, therefore, that Darwin pondered the then unknown mechanisms that simultaneously permit body plans to achieve their uniquely organised growth and development and yet permit evolutionarily adaptive modifications.
Darwin’s body plan dilemma
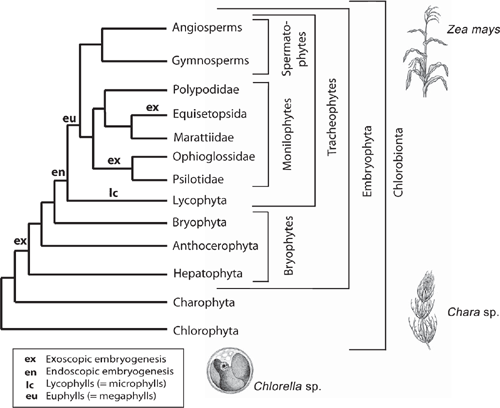
How body plans are simultaneously conserved and yet remain adaptively plastic is a long-unanswered question. However, recent insights from evolutionary developmental biology, cladistic methodology, and paleontology are providing new insights, particularly for plants, which have emerged as preferred model organisms. Our understanding of dedicated genes (e.g. transcription factors) and regulatory systems (e.g. auxin signal perception and transport mechanisms) has inspired new approaches to dissecting the genomics driving morphogenesis. The operation of MADS-box genes has been used to infer the evolution of key seed plant characteristics including the evolution of the flower. Other studies have focussed on the roles played by homeodomain, MYB, and phytochrome genes during vegetative growth. Most of the model systems under investigation are from crown plant groups (e.g. the moss Physcomitrella, the fern Ceratopteris, the dicot Arabidopsis, and the monocot Zea) (Fig. 1). This narrow phyletic sampling limits the phyletic depth to which the evolutionary origins of developmental mechanisms can be traced. Nevertheless, stringent cladistic methods employing new data have corrected early misconceptions about phyletic relationships and reveal developmental evolutionary patterns with greater accuracy. New discoveries in the fossil record have also shed light on ancient phenotypic character transformations, which provide more informed speculation about the relationship between ontogeny and phylogeny. When viewed collectively, the melding of data from developmental genomics, cladistics, and paleontology offers hope that Darwin’s dilemma will be resolved.
|
The goal of this article is to review the body plans of the various photosynthetic eukaryotic lineages (‘plants’), the major developmental evolutionary transformations that produced them, the regulatory mechanisms that might have chaperoned these transformations, and the evidence that some of these mechanisms are very ancient. Given its scope, our review cannot be synoptic. Rather, it focuses on a few better known regulatory systems of the land plants (embryophytes), particularly the vascular plants (tracheophytes), which are the most extensively studied lineages.
An important caveat in this enterprise is that a limited number of molecular components have been evolutionarily redeployed to yield different morphogenetic systems. This is particularly true for the regulators of central developmental processes such as signal receptors, signal transduction components, and transcription factors. This ‘redeployment’ strategy to augment subsequent evolutionary innovations makes it difficult to adduce developmental homologies based on genomic comparisons. MADS-box transcription factor genes, which likely evolved before the great Cambrian explosion, present a good example. Over 100 such genes are characterised for plants. Among the best known are the MIKC-type MADS-box genes found in mosses, ferns, gymnosperms, and angiosperms, which regulate various developmental processes, including floral morphogenesis. The amplification of this subfamily and its recruitment for reproductive development likely occurred late in embryophyte evolution. Expression of these genes in angiosperms occurs only after the specification of the vegetative to inflorescence meristem transition, which is mediated through the transcription factor encoded by FLORICAULA/LEAFY. In Pinus, the FLO/LFY gene homologue shows meristem-specific expression and can compliment Arabidopsis lfy mutants. In ferns, FLO/LFY homologues are expressed predominantly in sporogenous meristematic tissues but MADS-box gene expression is not closely correlated, suggesting that these genes have not yet been subordinated to FLO/LFY regulation. In Physcomitrella, there are two FLO/LFY paralogues (PpLFY-1 and PpLFY-2), which are required for the first division of the zygote and subsequent early sporophyte development (see Henschel et al. 2002; Tanahashi et al. 2005). Thus, an ancient gene function has been recruited to perform new functions among embryophytes, culminating in the specification of floral organ identity.
Developmental processes and plant body plans
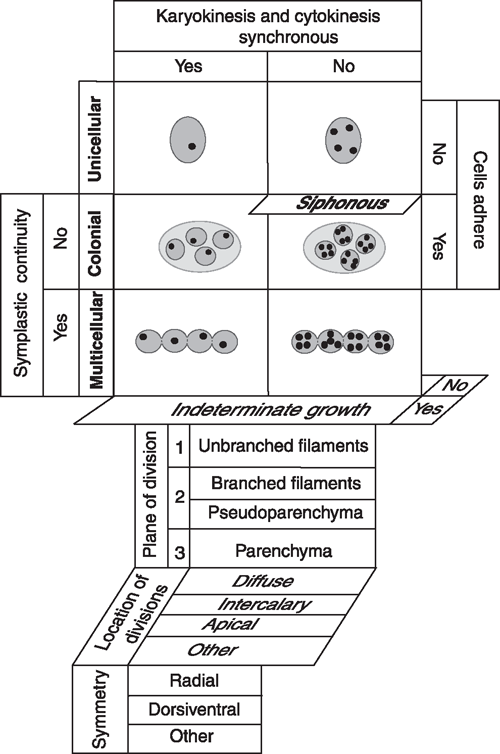
Five developmental processes generate all plant body plans (Niklas 2000): (1) the degree to which cyto- and karyokinesis are synchronised, (2) the extent to which dividing cells remain adjoined, (3) whether cytoplasmic continuity is maintained after cell division, (4) whether growth in size is determinate (‘closed’) or indeterminate (‘open’), and (5) the number and orientation of the planes of cell division (Fig. 2). Concatenation of these processes yields four basic body plans (the unicellular, colonial, multicellular, and siphonous) of which one (the siphonous) may be a variant of either the unicellular or multicellular plan. A comparatively small number of additional developmental processes can be added to this matrix to yield a plethora of body plan variants, e.g. differences in the duration, location, and planes of cell division (Fig. 2).
|
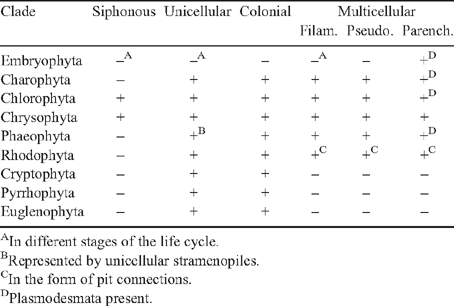
The fossil record shows that the unicellular eukaryotic body plan is the most ancient (Niklas 1997). It occurs in each of the major algal lineages, and it is represented in the form of embryophyte meiospores and motile gametes (Table 1). This body plan has two variants – the uninucleate and the multinucleate cell (e.g. Chlamydomonas and Vaucheria, respectively). Colonial body plans result when cells remain aggregated within a common matrix but lack cytoplasmic (symplastic) connections. This body plan is represented in each major algal lineage (although it is rare in some) and wholly absent among the embryophytes (Table 1). The multicellular body plan is characterised by the presence of plasmodesmata among most adjoining cells. Four variants exist: unbranched filaments, branched filaments, interweaving filaments (i.e. pseudoparenchymatous construction), and the parenchymatous body plan. Differences in the planes of cell division dictate which variant is achieved. Unbranched and branched filamentous construction results when the plane of cell division is confined to one or two planes (e.g. the green algae Ulothrix and Stigeoclonium, respectively). The pseudoparenchymatous construction requires two planes of cell division, whereas parenchymatous tissues require three. Pseudoparenchymatous and parenchymatous body plans permit the construction of specialised tissues (e.g. sieve-cell filaments in the brown alga Macrocystis and phloem in Arabidopsis). The transition from cleaving cells in three as opposed one or two planes (as mirrored by the transition from moss protonema to gametophore) was a key developmental innovation.
|
Multicellularity, plasmodesmata and cellulose
The elementary nature of the developmental processes underlying the siphonous and colonial body plans and the recognition that the unicellular body plan is the ancestral condition for each algal clade direct our attention on the multicellular plant body and plasmodesmata. These symplastic conduits facilitate intercellular transport (of high as well as low molecular weight solutes), help to establish large-scale supracellular molecular signalling systems, and may buffer somatic mutation by allowing non-mutant cell physiology to compensate for the metabolic defects of adjoining mutant cells. Unfortunately, little to nothing at all is known about the genomics of plasmodesmata in part because these structures form continuously during cell division.
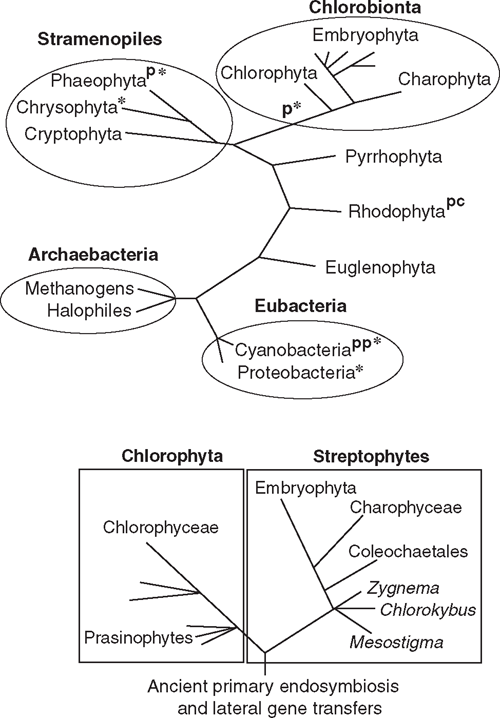
Plasmodesmata occur in all embryophytes and in many species of the Phaeophyta and the Chlorophyta (Table 1, Fig. 3). Molecular, cytological, and phylogenetic analyses show that the Phaeophyta and Chlorophyta evolved intracellular organelles (mitochondria, chloroplasts), cellulosic walls and multicellularity independently due to ancient primary endosymbiotic events (see Raven 1997; Kutschera and Niklas 2005, 2008; Archibald 2009). Thus, in the absence of lateral gene transfer between host and endosymbionts, multicellularity with plasmodesmata evolved independently at least twice. The plasmodesmata of the Phaeophyta are similar in size and general structure to those of the Chlorobionta (= Chlorophyta, Charophyta, and Embryophyta), but differ in a variety of ways that suggest independent origins. Bhattacharya and Medlin (1995) postulate that the Chlorophyta and Rhodophyta have chloroplasts derived from endosymbiotic cyanobacteria and are thus sister groups at this level of biological organisation. Chlorophyta evolved after the loss of cyanobacterial (phycobilin) light-harvesting pigments and the acquisition of chlorophyll b; the Rhodophyta evolved after the replacement of cyanobacterial ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) by a β-proteobacterial variety. Molecular analyses further suggest that plasmodesmata evolved independently within the Chlorophyta, once in the Chlorophyceae and again in the Charophyceae (Fig. 3). Although they may serve the same purpose as plasmodesmata, the intercellular connections of multicellular red algae are acidic polysaccharide trans-cell-wall ‘plugs’ surrounded by a lipoprotein bilayer membrane continuous with the plasmalemma. These ‘pit connections’ may have evolved twice in the Rhodophyta (Raven 1997). Although pit connections permit the transport of low molecular weight solutes, evidence for the transport of high molecular weight solutes, such as those that can pass through plasmodesmata, is equivocal.
|
The supposition that the Rhodophyta, Phaeophyta, and the Chlorobionta independently evolved multicellularity and plasmodesmata (Fig. 3) neglects the possibility of lateral gene transfers during endosymbiotic events and subsequent ‘developmental recall’ and ‘process orthology’. Although the ability to re-express long-silent genes decreases with time, Marshall et al. (1994) estimate that successful gene reactivation may occur after tens of millions of years; Fryer (1999) suggests even longer time spans. Regardless, there is good evidence that lateral gene transfers from chloroplasts to nuclei occurred during the history of all algal clades (Kutschera and Niklas 2005, 2008; Archibald 2009). For example, remarkable molecular homologies among the functionally non-redundant cellulose synthase genes (CesA) exist across diverse clades. Ultrastructural comparisons of the trans-membrane complexes containing CesA proteins support this hypothesis (Delmer 1999; Richmond and Somerville 2000; Nobles et al. 2001; Roberts et al. 2002; Römling 2002). All members of the CesA gene family isolated from embryophytes encode for integral membrane proteins with one or two trans-membrane helices in the N-terminal protein region, three to six trans-membrane helices in the C-terminal region, and an N-terminal domain structure that includes a cytoplasmic loop of four conserved regions (U1–U4), each of which contains a D residue or the QXXRW sequence, which is predicted to code for glycosyltransferase functionality (Richmond and Somerville 2000). Three additional shared features are a CR-P region between the U1 and U2 conserved regions, an N-terminal LIM-like zinc-binding domain, and a region between U2 and U3 (Delmer 1999) that is conserved within specific clades (Vergara and Carpita 2001).
Molecular comparisons indicate that the CR-P insertion and the D-D-D-QXXRW motif evolved before the appearance of the embryophytes – indeed, before that of eukaryotes – because both features have been identified in CesA proteins from the green alga Mesotaenium caldariorum (Roberts et al. 2002) and in CesA-like proteins from cyanobacteria (Nobles et al. 2001). Thus, the genome for cellulose biosynthesis may be traceable to the endosymbiotic origins of chloroplasts, a key event in the history of life (Niklas 2004). Subsequent gene duplication and functional divergence occurred after ancient CesA-like genes were integrated within eukaryote genomes, because eukaryotic CesA proteins are functionally non-redundant and are arranged in structurally well defined trans-membrane structures, called terminal complexes, which are invariably involved in cellulose assembly and deposition into the cell walls of the Phaeophyta, Chrysophyta, Chlorophyta, and Embryophyta (Kutschera 2008).
Lateral gene transfers may also be responsible for the evolution of plasmodesmata. Multicellular cyanobacteria exchange metabolites and regulatory molecules among adjoining cells. The mechanism of exchange is unknown. No structures comparable to plasmodesmata are known for multicellular cyanobacteria. Whether the ‘micro-plasmodesmata’ reported for some species represent analogues to animal gap junctions remains problematic. However, ultrastructural studies of cyanobacterial cell walls suggest that the outer membrane of filaments is a continuous structure; the existence of a supracellular periplasm may serve as a communication conduit among cells that provided a template for plasmodesmata and pit connections (Flores et al. 2006).
Auxin and morphogenesis
Multicellularity requires efficient intercellular communication by chemical messengers (growth hormones) capable of polar transport. Traditionally, five major phytohormones were thought necessary to control embryophyte development (i.e. auxins, gibberellins, cytokinins, ethylene, and abscisic acid). The capacity to synthesise these phytohormones is ancient; all five are produced by microorganisms (unicellular photoautotrophs, fungi, and bacteria, including the cyanobacteria; for a review, see Johri 2004, 2008). Although recent work implicates an increasing array of small and large molecular weight messenger molecules (e.g. polyamines and brassinosteroids), the physiology and evolution of auxin and cytokinin systems have received the most attention. No embryophyte mutant lacking either hormone has been found, suggesting that both hormones are required continuously at some level.
Here, we focus on auxin (i.e. indole-3-acetic acid, IAA) and its role in the evolutionary development of tracheophytes because it rapidly and specifically modulates gene expression at the level of transcription and is involved in cell elongation and division, photo- and gravitropism, apical dominance, and vascular tissue differentiation. The initial steps in the auxin transduction pathway are believed to involve a comparatively small group of receptors that regulate protein degradation by means of an evolutionarily ancient ubiquitin-proteasome pathway. Upon IAA activation, the receptor–enzyme complex targets specific transcriptional repressors for hydrolysis, which results in the activation of auxin-response genes. The principal auxin receptors are F-box soluble proteins belonging to the TIR1/AFB protein family, which were discovered as sub-units of ubiquitin ligase complexes (SCF). F-box proteins are found in all eukaryotic lineages and function in the regulation of protein abundance. According to a recent model (Ruegger et al. 1998; Dharmasiri et al. 2005), the TIR1 protein is a component of the SCFTIR1 ubiquitin ligase complex. In the absence of IAA, AUX/IAA repressors inhibit the transcription of auxin-induced genes by binding to and inhibiting ARF transcription factors. IAA activates SCFTIR1/ABF complexes that attach ubiquitin to AUX/IAA proteins, thereby promoting their decomposition (via 26S proteasome), unlocking ARF transcriptional activators that then bind to auxin response elements (AuxRE) and stimulate auxin-induced gene transcription.
A different type of auxin receptor binding protein (auxin-binding protein 1, ABP1) may be required for auxin-dependent cell elongation (via the mobilisation of plasmamembrane H+-ATPases and subsequent cell wall acidification). ABP1 may also play a role in angiosperm embryogenesis (Palme et al. 1992). For example, in Arabidopsis, ABP1 is encoded by a single gene (At-ERabp1). Knockout embryos harbouring a T-DNA insertion in the first At-ERabp1 exon result in embryo abortion at the globular stage of development. Transgenic addition of a single functional copy of ABP1 rescues them. Examination of aborted embryos reveals anomalous cell wall patterns, although cell elongation is normal (Chen et al. 2001). The failure to mediate sustained IAA-induced cell expansion may cause embryo lethality in Arabidopsis knockout plants. Because cell expansion occurs in the absence of normal cell elongation, the At-ERabp1 transcript may interact with as yet unidentified factors influencing asymmetric cell expansion.
Intercellular IAA transport is largely basipetal in tracheophyte shoots and both acropetal and basipetal in roots. Lateral IAA transport is important to photo- and gravitropic responses. IAA polar transport involves an IAA-influx protein carrier encoded by the AUX1 gene, whereas the efflux of the IAA anion involves the activity of at least two membrane-bound proteins (Muday and DeLong 2001). One of these is a trans-membrane transport protein encoded by members of the PIN gene family. PIN auxin efflux protein facilitators are crucial to the apical–basal polarity of the plant body. Although striking differences exit between them, the PAR-polarity modules known from animals systems are suggested to share a common regulatory logic with that of PIN protein systems (Geldner 2009). Another is an IAA-inhibitor-binding protein that performs a regulatory function in response to endogenous IAA inhibitors.
The mechanism by which IAA-transport inhibitors affect IAA-efflux carrier proteins is poorly understood. The Arabidopsis gene RCN1 encodes a regulatory subunit of protein phosphatase 2A (PP2A). The rcn1 mutant exhibits a near 2-fold increase in IAA basipetal transport (Muday and DeLong 2001). Treatment of control plants with phosphatase inhibitors (e.g. cantharidin) produces the rcn1 phenotype, suggesting that PP2A regulates the IAA-polar-efflux carrier protein complex and that the regulatory effects of PP2A on basipetal and acropetal IAA transport are different. The results of inhibitor studies further suggest that genes encoding several kinases may play a key role in regulating polar IAA transport, e.g. tobacco cells treated with broad-spectrum kinase inhibitors (e.g. staurosporine) show rapid IAA efflux reduction but little or no change in IAA influx.
The model systems used to understand IAA signal perception, transduction, polar transport and mode of action are from crown embryophyte groups, e.g. Arabidopsis and Zea (Kutschera 2003, 2006; Kutschera and Edelmann 2005). Less is known about these aspects of development for basal embryophyte species and even less is known about the Charophyceae (Fig. 4). Poli et al. (2003) examined the effects of externally applied auxin and IAA antagonists on the growth of the sporophytes of the hornwort Phaeoceros personii, the liverwort Pellia epiphylla, and the moss Polytrichum ohioense. IAA movement in young hornwort sporophytes was nonpolar and insensitive to the auxin-transport inhibitor N-(1-naphthyl)phthalamic acid. It was concluded that IAA moves by simple diffusion rather than active transport (Poli et al. 2003). Liverwort sporophytes had slightly higher IAA fluxes, were sensitive to transport inhibitors, but lacked any measurable polarity, suggesting the existence of a unique type of apolar facilitated IAA diffusion. IAA movement in young moss sporophytes was predominantly basipetal (and occurred at fluxes exceeding those measured in corn coleoptiles). In older sporophytes, acropetal IAA flux exceeded basipetal flux (and had different IAA inhibitor sensitivities), suggesting that acropetal and basipetal IAA transport involves different cellular pathways, much like in angiosperms (Poli et al. 2003). In contrast, the auxin transport inhibitor NPA does not cause changes in the distribution of IAA in Physcomitrella gametophytes (Fujita et al. 2008), suggesting that the bryophyte lineages either evolved different IAA transport systems independently, or represent an evolutionary transformation series, with hornworts being the most ancient and the mosses being the most derived (Fig. 4). These data also suggest different IAA systems exist in the gametophyte and sporophyte generations.
|
Sztein et al. (1995) examined the IAA conjugation patterns in the charophycean alga Nitella and 23 embryophytes to determine whether they correlated with the presence of specialised conducting tissues (none in Nitella; problematic in some liverworts; leptoids and hydroids in some mosses; and vascular tissues in tracheophytes). Free IAA is biologically active. However, most of IAA occurs in covalently bond (conjugated) forms (e.g. esters of IAA with glucose and IAA-glucans), which are not easily degraded enzymatically and thus likely stored for future use as in IAA participation in vascular tissue differentiation. Sztein et al. (1995) report three main IAA conjugation patterns: a charophyte–liverwort pattern (consistent with no conducting tissues in the Charophyceae and the general absence of specialised water conducting cells in liverworts), a hornwort-moss pattern involving a limited number of IAA amide and ester conjugates (consistent and perhaps associated with the differentiation and subsequent maturation of specialised conducting cell in some mosses and hornworts), and a complex tracheophyte conjugation pattern that differed among the 23 embryophytes examined. Whether these data reflect a phyletic transformation series in IAA conjugation (and polar transport) remains problematic.
Phylogenetic development of the embryophyte life cycle
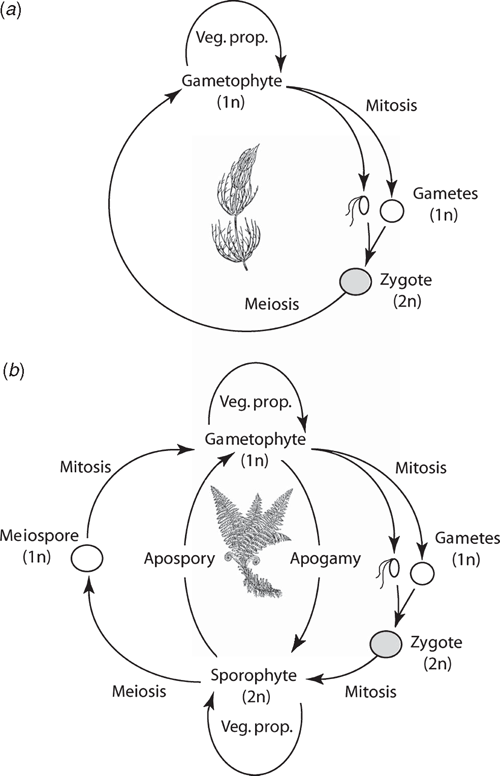
Phylogenetic analyses consistently identify the Charophyta and the Embryophyta as sister lineages (Figs 3 and 4). All charophytes have a haplobiontic-haploid life cycle, i.e. a life cycle with a single multicellular haploid generation (Fig. 5a) (Graham and Wilcox 2000; Archibald 2009). Thus, the embryophyte diplobiontic life cycle (i.e. the alternation between a multicellular haploid gametophyte and a multicellular diploid sporophyte; Fig. 5b) is likely a derived condition that evolved by means of delayed zygotic meiosis and the intercalation of one or more mitotic divisions. This transformation would have conferred a selective advantage by amplifying the reproductive dividends of rare fertilisation events in ancient aquatic/semi-aquatic algal-like plants (Graham 1993; Niklas 1997). The subsequent or concurrent evolution of meiospores with sporopollenin-rich walls (a hallmark of all embryophytes) would have conferred additional benefits in habitats prone to desiccation.
|
The haplobiontic-haploid-to-diplobiontic life cycle transformation probably involved several genomic transfers in function. Phylogenetic analyses indicate that MADS-box gene expression in the charophycean alga Chara globularis occurs during gametangium differentiation and declines after fertilisation (Tanabe et al. 2005). Similar genes are expressed in the formation of reproductive structures by embryophyte sporophytes. Thus, MADS-box genes probably originally functioned in the differentiation of haploid reproductive organs but were recruited to function in the formation of diploid reproductive organs. Indeed, combinatorial homeodomain-based transcriptional control of reproduction has deep phylogenetic roots. Ectopic expression of the homeoproteins Gsp1 and Gsm1 in the plus and minus strains of the unicellular green alga Chlamydomonas activates vegetative cells to form zygote-like structures (Lee et al. 2008). Gsp1 and Gsp2 are members of the TALE (three amino acid loop extension) homeodomain containing transcription factors, which includes the class 1 KNOX and class 2 KNOX proteins. Homeodomain gene networks, similar to those in land plants, have been reported for prasinophytes (e.g. Micromonas), which are close to the last common ancestor of all green plants (Worden et al. 2009) (Figs 3 and 4).
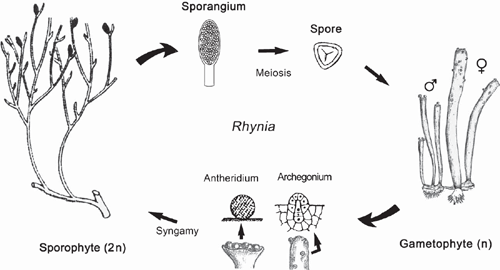
It is suggested that the most ancient embryophytes possessed an isomorphic alternation of generations (Kenrick and Crane 1997a, 1997b; Steemans et al. 2009) much like that of the Devonian tracheophyte Rhynia (Fig. 6). The gametophytes and sporophytes of the most ancient embryophytes undoubtedly shared similar genomic and developmental repertories. In the absence of gene silencing, sex chromosomes, or epigenetic effects, differences in ploidy may not have equated with significant gametophyte-sporophyte dimorphism. Indeed, among some modern day mosses and ferns, sporophyte morphologies can form directly from gametophytic cells (apogamy) and gametophyte morphologies can develop directly from sporophyte cells (apospory) (Fig. 5b). These phenomena indicate that the haploid genome provides sufficient information to construct the gametophyte and sporophyte body plans. Apogamy can be induced by cell trauma, low light intensities, suitable concentrations of sugar, or IAA. It can also be induced by the deletion of the CURLY LEAF orthologue in Physcomitrella (PpCLF). Okano et al. (2009) report that gametophytic cells that usually form protonema or gametophore apical cells generate meristematic apical cells that form branched morphologies, which can be induced to form sporangium-like structures with the exogenous application of PpCLF. The resulting morphologies are reported to be similar to very ancient tracheophytes, e.g. Cooksonia (Fig. 7), suggesting that PpCLF regulatory gene networks may have participated in the evolution of the vascularised polysporangiate sporophyte (Okano et al. 2009).
|
|
However, it is doubtful that the most ancient embryophyte life cycles were truly isomorphic. Monoploid embryophytes do not develop apogamous sporophytes, suggesting that gene duplication and subsequent functional divergence presaged the evolution of multicellular sporophytes. Further, sporophytes and gametophytes normally develop in very different biological contexts. Young sporophytes develop within archegonia; ‘free-living’ embryophyte gametophytes develop from dispersed meiospores. In both cases, numerous epigenetic factors influence early morphogenesis that fosters dimorphism (Sinnott 1960). If the most ancient sporophytes represent an ‘intercalated’ multicellular generation, it is difficult to imagine that they were morphologically elaborate – indeed, they may have been nothing more than the functional equivalent of a sporangium (Niklas 1997). The different functional obligations of the gametophyte and sporophyte generations would have quickly propelled dimorphism. Under any circumstances, the so-called ‘isomorphic’ life cycle of ancient plants like Rhynia (Kenrick and Crane 1997a, 1997b) is clearly dimorphic (Fig. 6) as may be that of even more ancient tracheophytes (Gerrienne et al. 2006).
Another important evolutionary transformation relates to embryo polarity. With few exceptions, the first zygotic division among embryophytes is transverse (giving rise to an epi- and a hypobasal cell) and establishes one of two types of polarity: exoscopic polarity, wherein the major embryonic growing point develops from the epibasal cell, and endoscopic polarity, in which the apical embryonic pole develops from the hypobasal cell. The former characterises all extant bryophytes; endoscopic polarity occurs in most tracheophytes (Niklas 2008). The phyletic position of the bryophytes suggests that exoscopic embryogenesis is the ancestral condition (Scherp et al. 2001).
Embryo polarity, however, does not establish the plane of the first zygotic division (e.g. in the hornwort Anthoceros and the moss Funaria, the first division is longitudinal), suggesting that polarity is established by cytoplasmic factors or under the epigenetic influence of the archegonium. In Arabidopsis, Long et al. (2006) suggest that polarity is a two-stage phenomenon with axis formation occurring during the first cell divisions (perhaps relying on axisymmetric polar auxin transport; also see Friml et al. 2003) and subsequent axis fixation requiring a chromatin-mediated transcriptional repression system for axis stabilisation. A similar scenario is posited for polarity establishment in the zygotes of the brown alga Fucus, where axis formation and fixation are temporally distinct events. Whether this is a generalised developmental phenomenon remains unclear. The recent discovery of the paternally inherited Arabidopsis mutant short suspensor (SSP) may provide insights. SSP regulates the YODA mitogen-activated protein kinase pathway, which promotes normal embryo elongation. Zygotes with the SSP allele divide asymmetrically and elongate to form normal suspensors and basal cells. Zygotes inheriting the SSP allele fail to elongate and have defective suspensor development (Bayer et al. 2009), suggesting that paternal inheritance influences embryo polarity.
Recent studies of endogenous small RNAs, including microRNAs (miRNAs), characterised for Arabidopsis, Physcomitrella, and the lycopod Selaginella moellendorffii also indicate that ancient, highly conserved RNA regulatory interactions are disproportionately involved in development and that these units of post-transcriptional gene control were indispensable during embryophyte diversification (see Axtel et al. 2007). For example, the targets of conserved Physcomitrella miRNAs are homologous with the known targets of the same miRNAs in Arabidopsis control transcriptional regulators influencing multicellular development and morphology. Diverse, lineage-specific, small RNAs that perform common biological functions in plants therefore may have played a role in the evolution of gametophyte-sporophyte dimorphism.
Most tracheophyte sporophytes manifest indeterminate growth in size as a result of continued apical meristematic activity. In contrast, all bryophyte sporophytes lack a vegetative apical meristem. It is hypothesised that the gene networks controlling the apical meristems of tracheophyte sporophytes (such as the class 1 KNOX transcription factors) functioned similarly in the gametophytes of basal embryophytes and that these networks were co-opted for expression in the apical meristems of tracheophyte sporophytes. However, Sakakibara et al. (2008) report that class 1 KNOX orthologues in Arabidopsis do not function in the (determinate) meristems of Physcomitrella gametophytes and suggest that the gene networks governing the haploid indeterminate meristem with the class 1 KNOX genes evolved de novo in tracheophytes. This hypothesis would gain much more credibility if 1 KNOX functionality is examined for bryophyte indeterminate (as opposed to determinate) apical meristems.
Unequal meristem fate and the evolution of leaves
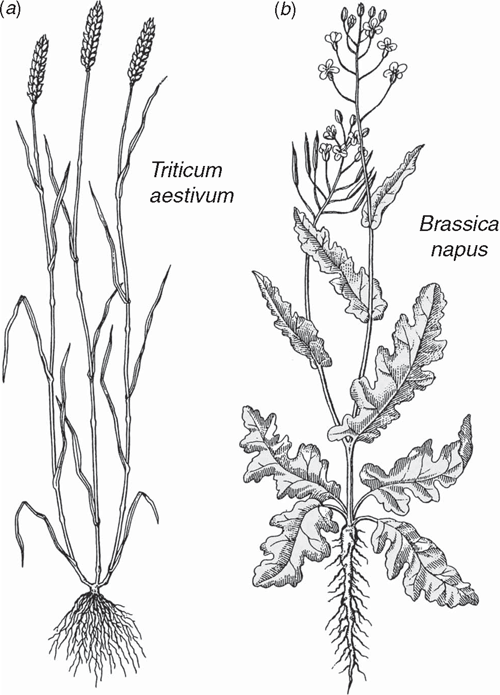
The sporophytes of most tracheophytes have leafy stems (Fig. 1). Developmental genetic analyses have begun to reveal the mechanisms responsible for the formation of leaves and related aspects of sporophyte morphology but for only a few model systems dominated by angiosperms (e.g. Solanum and Zea). The lack of broader phyletic sampling is troubling because leaves (defined here as appendicular asymmetric structures produced by apical meristems) have evolved independently as ‘phyllids’ in mosses and liverworts, as ‘lyco(micro)phylls’ in the Lycophyta, and numerous times as ‘eu(mega)phylls’ in other plant lineages (Niklas 1997) (Fig. 4). This cautions against the assumption that a single ancestral genomic system underlies leaf development for all embryophytes. This caveat is warranted in light of the deep phyletic divide separating the lycopods from other tracheophyte lineages well before sporophytes evolved leaf/stem/root organographic distinctions (Fig. 6).
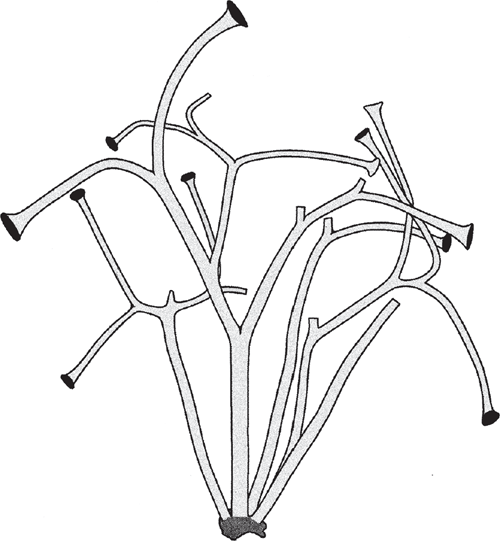
Little is currently known about the developmental genetics of lycopod leaf development. Preliminary investigations of ARP genes indicate that they are expressed in the lateral primordia of micro- and megaphylls (Harrison et al. 2005), although HD-ZIP class III genes carry out very different roles in the development of the two leaf-types (Floyd and Bowman 2006). One of the initial developmental transformations prefiguring the leaves of all monilophyte lineages likely involved meristematic inequality attending apical bifurcation. The most ancient vascular sporophytes, such as Cooksonia (Fig. 7), had more or less equal branching. The sporophytes of more derived taxa had unequal or pseudomonopodial branching, resulting from meristem fate inequality (Fig. 6) as predicted by the telome theory (Zimmermann 1930, 1952; Beerling and Fleming 2007) (Fig. 8).
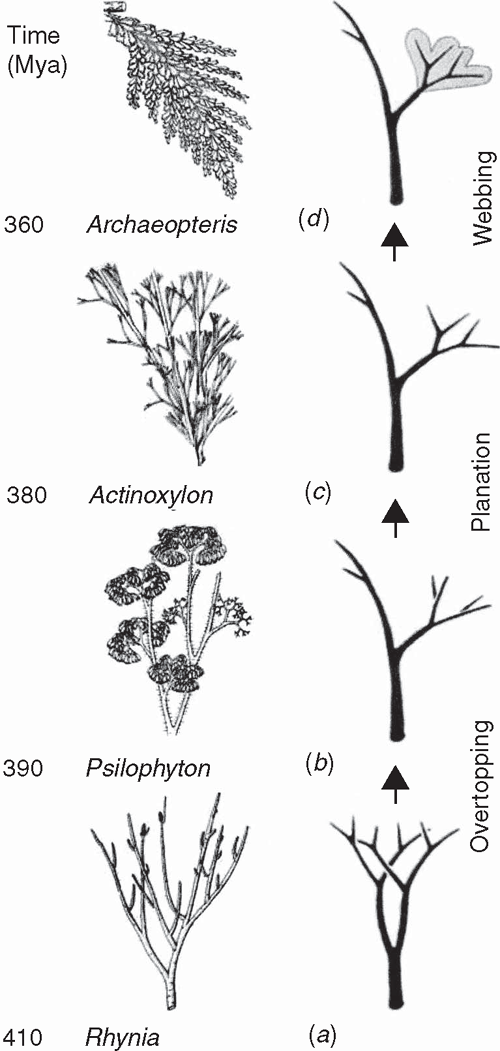
|
Model angiosperm systems provide examples of how meristem inequality can be enforced or repressed. For example, the precise mode of expression of knotted1-like homeobox (KNOX) transcription factors influences both shoot apical meristem function and the acquisition of leaf identity. Among simple-leafed dicots and monocots (e.g. Arabidopsis and Zea), KNOX proteins are strongly expressed during the indeterminate growth of the apical meristem, but rapidly downregulated in cells flanking the apical dome in regions defined by leaf primordia. Although this pattern holds true for some species with compound leaves, such as pea, it is not conserved across all angiosperms. Data for tomato indicate that leaf initiation does not involve KNOX downregulation and that the formation of marginal leaf lobbing is associated with KNOX expression, suggesting that gene regulation might involve the secondary recruitment of mechanisms originally operating in the shoot apex. The determinancy of leaf growth observed for many, but not all angiosperms may also be the result of downregulating genetic systems operating in shoot apices, because most genetic systems known to promote or repress apical meristem activity function similarly in axillary bud meristems. The only known exception is the Teosinte branched1 (Tb1) gene, which is expressed during the indeterminate growth of axillary bud meristems and thus determines much of the maize body plan (Doebley 2004) (Fig. 9).
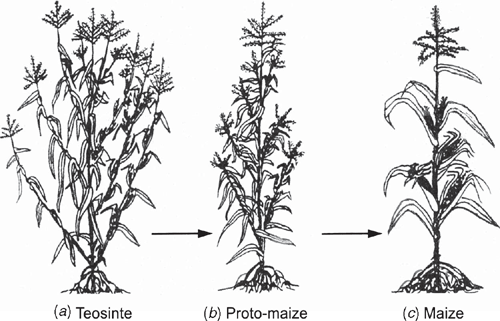
|
Most megaphylls manifest tissue asymmetry and dorsiventrality (either perpendicular or parallel to the shoot axis) (Figs 1 and 8), both of which may rely on developmental signals from the apical meristem that provide cues for adaxial/abaxial identity. Whether apical meristem polarity provides a common mechanism for defining the characteristics of megaphylls remains problematic. However, support for this hypothesis comes from the Arabidopsis homeodomain-leucine zipper (HD-ZIP) III gene subfamily that includes the PHABULOSA (PHB) gene (Ratcliffe et al. 2000; McConnell et al. 2001). When activated, PHB genes appear to specify adaxial organ fate and repress genes required for abaxial fate (Kerstetter et al. 2001), suggesting that the activating ligand may come from the shoot apex. If adaxial organ fate is a response to apical meristem asymmetry, the appendicular status of the leaf primordium is a requisite for dorsiventrality. HD-ZIP III genes have been identified in Physcomitrella (Sakakibara et al. 2001) and in the fern Ceratopteris (Aso et al. 1999). They are also expressed during normal vascular tissue development in Arabidopsis (Zhong and Ye 1999), suggesting that they are multifunctional as well as very ancient.
Little is known about the upstream or downstream components of the KNOX pathway, and most of what is known comes from angiosperm model systems that are not representative of the early stages in the developmental evolution of leafy shoots. The rough sheath2 (RS2) gene of Zea, the phantastica (PHAN) gene of Antirrhinum, and the asymmetric1 (AS1) gene in Arabidopsis (all of which are known to encode related MYB transcription factors) are each required for the normal compartmentalisation of KNOX gene expression in their respective species. Although KNOX genes are misrepresented in rs2/phan/as1 mutants, the early downregulation of KNOX associated with incipient primordium development remains intact. Thus, KNOX repression may involve separate, possibly redundant pathways. An additional complexity is that AS1, PHAN, and RS2 proteins can have equivalent action at the molecular level, but evoke different phenotypes, e.g. phan mutants have radial leaves, whereas rs2 and as1 mutant leaves are dorsiventral. In terms of downstream KNOX pathway effectors, ectopic expression of KNOX genes in some systems (e.g. Nicotiana) results in phenotypes similar to those produced by increased cytokinin levels (Li et al. 1992). However, increased cytokinin levels do not fully account for the phenotypic spectrum observed for KNOX gene overexpression, suggesting that these genes function globally to coordinate phytohormones other than cytokinin that influence leaf size and shape (e.g. gibberellin and IAA).
Floral evolution
Darwin (1859) was perplexed by the apparently abrupt evolutionary origin of the angiosperms, which violated his belief that natura non-facit saltum. Despite significant progress in reconstructing spermatophyte phylogeny and the insights gleaned from floral identity genes, the evolutionary origin of the angiosperms remains problematic (Crepet and Niklas 2009). Nevertheless, recent developments cast light on how flowers may have evolved (for a review, see Soltis et al. 2009).
For example, the classic combinatorial ABC model depicts the activities of transcription factors regulating floral organ identity and thus the structure of certain flowers. In this model, the A function specifies sepals, A + B functions give petals, B + C functions produce stamens, and that the C function specifies carpels. In Arabidopsis, APETALA1(AP1) and APETALA2(AP2) are A-function genes; APETALA3(AP3) and PISTILLATA(PI) are B-function genes, and AGAMOUS (AG) is a C-function gene. More recently, the role of SEEDSTICK(STK) has been identified to participate in ovule identity (designated as the D function), whereas SEPALLATA(SEP) participates in the specification of other floral organ identities (E function). These elaborations have lead to the ABCDE model. With the exception of AP2, all of the aforementioned genes are MADS-box genes (Fig. 10). As noted, MADS-box genes have been identified in mosses and ferns (e.g. Physcomitrella, Ophioglossum and Ceratopteris) but are not orthologues of the ABC genes. Tanahashi et al. (2005; see also Henschel et al. 2002) have identified two FLORICAULA/LEAFY genes, PpLFY1 and PpLFY2, in Physcomitrella that regulate the first zygotic cell division and are thus involved in vegetative growth, whereas LFY genes among seed plants function during the vegetative to reproductive transition.
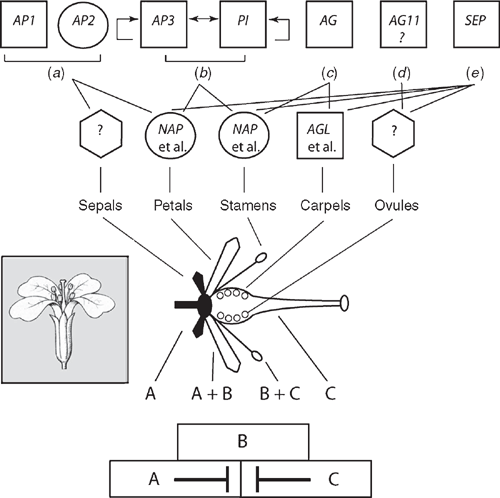
|
Orthologues of class B, C and D organ identity genes have been identified in gymnosperms. In addition, sister genes to class B genes (denoted as Bsister), which are involved in ovule and carpel development, have been identified in gymnosperms (Theißen et al. 2002). The presence of B, Bsister, C, and D orthologues in gymnosperms suggests that the floral organ identity gene network was recruited from the last common ancestor of all spermatophytes (Fig. 4) and may have been involved in the specification of sexual identity. Class B (and possibly Bsister) genes in spermatophytes appear to distinguish between ‘male’ (microsporangia) function where B and Bsister gene expressions are ‘on and off’ and ‘female’ function (megasporoangia) where B and Bsister gene expressions are ‘off and on’. This differential gene expression may represent the sex determination system of the angiosperm ancestor (Winter et al. 1999; Theißen et al. 2002). The male-to-female identity switch could have involved changes in the activity of a few genes and may have been the developmental basis for transforming a unisporangiate into a bisporangiate (‘proto-flower’) reproductive structure as posited by the Mostly Male theory (Frohlich 2002) and the Bsister hypothesis (Winter et al. 1999; Theißen et al. 2002).
Concluding remarks
It is obvious that ancient primary endosymbiosis (symbiogenesis) and lateral gene transfers, followed by duplication and functional gene divergences were key events in the evolutionary history of plant life. However, much of this article deals with how the ‘transcription factor’ paradigm helps to unravel the developmental macroevolution of plants (embryophytes). This paradigm has implicated at least six molecular mechanisms for phenotypic evolution: (1) gene array duplication and subsequent sub-functionalisation, (2) changes in the spatial expression patterns of pre-existing arrays, (3) homeodomain protein sequence alterations, (4) modifications of DNA binding domains, (5) alterations in downstream regulated gene-networks, and (6) changes in upstream regulatory genes. Thus, even when the mode of action and the spatial domain of gene expression remain unchanged, modifications in the interactions between regulatory and downstream target genes can participate in significant phenotypic evolutionary changes. Nevertheless, this paradigm needs to be approached cautiously. Consider the ectopic expression of the normal form of the ey fruit fly gene and the normal Sey mouse gene within the fruit fly genome (Halder et al. 1995). Because these genes retain their participatory function in the development of photoreceptors, the similarities of the phenotypes resulting from their expression indicates that the regulatory ey and Sey gene sequences have evolved little since the divergence of arthropods and chordates hundreds of millions of years ago. However, the gene networks targeted by ey and Sey have changed profoundly as have the morphologies resulting from their participation. This single example illustrates that molecular homology at the level of regulatory genes guarantees neither developmental nor phenotypic homology.
This caveat is reinforced by studies of LEAFY (LFY) genes in mosses, ferns, and spermatophytes. Among angiosperms, a single LFY gene product binds to sequences in the enhancers of several homeotic floral genes (e.g. APETALA1). Among non-flowering plants, several LFY gene products control more general and numerous life cycle features. The LFY DNA binding domain is strongly conserved across all taxa. However, the LFY protein as a whole has diverged in activity across lineages, as indicated by the ability of LFY cDNAs (isolated from mosses, ferns, and gymnosperms linked to the Arabidopsis LFY promoter) to progressively recover the lfy Arabidopsis mutant with decreasing phyletic distance (Maizel et al. 2005). Two scenarios can explain this phenomenology: either LFY controls similar gene networks that have coevolved with target genes that have themselves become modified during plant diversification, or the function of LFY in basal (moss) and derived lineages has changed completely as a result of the recruitment or intercalation of new target genes (Maizel et al. 2005). In either case, understanding phenotypic macroevolution requires thinking both within and outside the transcription-factor paradigm.
Evo-devo stands at the same precipice genetics did before the Modern Synthesis when it was difficult to reconcile Mendelian quantitative genetics with the Darwinian supposition that evolution involves gradualistic genomic changes (Kutschera and Niklas 2004). The rapidly expanding insights gained from developmental genomics have not been integrated with those provided by more traditional evolutionary disciplines. This situation will change, but only when both disciplines consider all levels of biological organisation simultaneously, from the molecular to the ecosystem. It will also require exploring taxa from deeper nodes in phylogenetic trees. The task is intimidating, but if left unaccomplished we run a risk best described in T. S. Eliot’s Four Quartets:
It seems, as one becomes older,
That the past has another pattern, and ceases to be a mere sequence–
Or even development: the latter a partial fallacy
Encouraged by superficial notions of evolution,
Which becomes, in the popular mind, a means of disowning the past.
Archibald JM
(2009) Green evolution, green revolution. Science 324, 191–192.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Aso K,
Kato M,
Banks JA, Hasebe M
(1999) Characterization of homeodomain-leucine zipper genes in the fern Ceratopteris richardii and the evolution of the homeodomain-leucine zipper gene family in vascular plants. Molecular Biology and Evolution 16, 544–552.
| PubMed |

Axtell MJ,
Snyder JA, Bartel DP
(2007) Common functions for diverse small RNAs of land plants. The Plant Cell 19, 1750–1769.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Bayer M,
Nawy T,
Giglione C,
Galli M,
Meinnel T, Lukowitz W
(2009) Paternal control of embryonic patterning in Arabidopsis thaliana. Science 323, 1485–1488.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Beerling DJ
(2005) Leaf evolution: gases, genes and geochemistry. Annals of Botany 96, 345–352.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Beerling DJ, Fleming AJ
(2007) Zimmermann’s telome theory of megaphyll leaf evolution: a molecular and cellular critique. Current Opinion in Plant Biology 10, 4–12.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Bhattacharya D, Medlin L
(1995) The phylogeny of plastids: a review based on comparisons of small-subunit ribosomal RNA coding regions. Journal of Phycology 31, 489–498.
| Crossref | GoogleScholarGoogle Scholar |

Chen J-G,
Shimomura S,
Sitbon F,
Sandberg G, Jones AM
(2001) The role of auxin-binding protein 1 in the expansion of tobacco cell. The Plant Journal 28, 607–617.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Crepet WL, Niklas KJ
(2009) Darwin’s second “abominable mystery”: why are there so many angiosperms? American Journal of Botany 96, 366–381.
| Crossref | GoogleScholarGoogle Scholar |

Delmer DP
(1999) Cellulose biosynthesis: exciting times for a difficult field. Annual Review of Plant Physiology and Plant Molecular Biology 50, 245–276.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Dharmasiri N,
Dharmasiri S, Estelle M
(2005) The F-box protein TIR1 is an auxin receptor. Nature 435, 441–445.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Doebley J
(2004) The genetics of maize evolution. Annual Review of Genetics 38, 37–59.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Flores E,
Herrera A,
Wolk CP, Maldener I
(2006) Is the periplasm continuous in filamentous multicellular cyanobacteria. Trends in Microbiology 14, 439–443.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Floyd SK, Bowman JL
(2006) Distinct developmental mechanisms reflect the independent origins of leaves in vascular plants. Current Biology 16, 1911–1917.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Friml J,
Vieten A,
Sauer M,
Weijers D,
Schwarz H,
Hamann T,
Offringa R, Jürgens G
(2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Fryer G
(1999) The case of the one-eyed shrimp: are ancient atavisms possible? Journal of Natural History 33, 791–798.
| Crossref | GoogleScholarGoogle Scholar |

Fujita T,
Sakaguchi H,
Hiwatashi Y,
Wagstaff SJ,
Ito M,
Deguchi H,
Sato T, Hasebe M
(2008) Convergent evolution of shoots in land plants: lack of auxin polar transport in moss shoots. Evolution & Development 10, 176–186.
| PubMed |

Geldner N
(2009) Cell polarity in plants – a PARspective on PINs. Current Opinion in Plant Biology 12, 42–48.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gerrienne P,
Dilcher DL,
Bergamaschi S,
Milagres I,
Pereira E, Rodrigues MAC
(2006) An exceptional specimen of the early land plant Cooksonia paranensis, and a hypothesis on the life cycle of the earliest eutracheophytes. Review of Palaeobotany and Palynology 142, 123–130.
| Crossref | GoogleScholarGoogle Scholar |

Halder G,
Callaerts P, Gehring WJ
(1995) Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science 267, 1788–1792.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Harrison CJ,
Corley SB,
Moylen EC,
Alexander DL,
Scotland RW, Langdale JA
(2005) Independent recruitment of a conserved developmental mechanism during leaf evolution. Nature 434, 509–514.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Henschel K,
Kofuji R,
Hasebe M,
Saedler H,
Münster T, Theißen G
(2002) Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Molecular Biology and Evolution 19, 801–814.
| PubMed |

Johri MM
(2004) Possible origin of hormonal regulation in green plants. Proceedings of the Indian National Science Academy Part B Biological Sciences 70, 335–365.

Johri MM
(2008) Hormonal regulation in green plant lineage families. Physiology and Molecular Biology of Plants 14, 23–38.
| Crossref | GoogleScholarGoogle Scholar |

Kenrick P, Crane PR
(1997a) The origin and early evolution of plants on land. Nature 389, 33–39.
| Crossref | GoogleScholarGoogle Scholar |

Kerp H,
Trewin NH, Hass H
(2004) Rhynie Chert gametophytes. Transactions of the Royal Society of Edinburgh. Earth Sciences 94, 411–428.

Kerstetter RA,
Bollman K,
Taylor RA,
Bombles K, Poethig RS
(2001)
KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kutschera U
(2003) Auxin-induced cell elongation in grass coleoptiles: a phytohormone in action. Current Topics in Plant Biology 4, 27–46.

Kutschera U
(2006) Acid growth and plant development. Science 311, 952–954.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kutschera U
(2008) The outer epidermal wall: design and physiological role of a composite structure. Annals of Botany 101, 615–621.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kutschera U, Edelmann HG
(2005) Osmiophilic nanoparticles in epidermal cells of grass coleoptiles: implications for growth and gravitropism. Recent Research Developments in Plant Science 3, 1–14.

Kutschera U, Niklas KJ
(2004) The modern theory of biological evolution: an expanded synthesis. Naturwissenschaften 91, 255–276.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kutschera U, Niklas KJ
(2005) Endosymbiosis, cell evolution, and speciation. Theory in Biosciences 124, 1–24.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Kutschera U, Niklas KJ
(2008) Macroevolution via secondary endosymbiosis: a Neo-Goldschmidtian view of unicellular hopeful monsters and Darwin’s primordial intermediate form. Theory in Biosciences 127, 277–289.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Lee JH,
Lin HW,
Joo S, Goodenough U
(2008) Early sexual origins of homeodomain heterodimerization and evolution of the plant KNOX/BELL family. Cell 133, 829–840.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Li Y,
Hagen G, Guilfoyle TJ
(1992) Altered morphology in transgenic tobacco plants that overproduce cytokinins in specific tissues and organs. Developmental Biology 153, 386–395.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Long JA,
Ohio C,
Smith ZR, Meyerowitz EM
(2006)
TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Maizel A,
Busch MA,
Tanahashi T,
Perkovic J,
Kato M,
Hasebe M, Weigel D
(2005) The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science 308, 260–263.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Marshall CR,
Raff EC, Raff RA
(1994) Dollo’s law and the death and resurrection of genes. Proceedings of the National Academy of Sciences of the United States of America 91, 12283–12287.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

McConnell J,
Emery J,
Eshed Y,
Bao N,
Bowman J, Barton MK
(2001)
PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Muday GK, DeLong A
(2001) Polar auxin transport: controlling where and how much. Trends in Plant Science 6, 535–542.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Niklas KJ
(2000) The evolution of plant body plans – a biomechanical perspective. Annals of Botany 85, 411–438.
| Crossref | GoogleScholarGoogle Scholar |

Niklas KJ
(2004) The cell walls that bind the tree of life. Bioscience 54, 831–841.
| Crossref | GoogleScholarGoogle Scholar |

Nobles DR,
Romanovicz DK, Brown RM
(2001) Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiology 127, 529–542.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Palme K,
Hesse T,
Campos N,
Garbers C,
Yanofsky MF, Schell J
(1992) Molecular analyses of an auxin binding protein gene located on chromosome 4 of Arabidopsis. The Plant Cell 4, 193–201.
| Crossref |
PubMed |

Poli DB,
Jacobs M, Cooke TJ
(2003) Auxin regulation of axial growth in bryophyte sporophytes: its potential significance for the evolution of early land plants. American Journal of Botany 90, 1405–1415.
| Crossref | GoogleScholarGoogle Scholar |

Ratcliffe OJ,
Riechman JL, Zhang JZ
(2000)
INTERFASCICULAR FIBERLESS1 is the same gene as REVOLUTA. The Plant Cell 12, 315–317.
| Crossref |
PubMed |

Raven J
(1997) Multiple origins of plasmodesmata. European Journal of Phycology 32, 95–101.
| Crossref | GoogleScholarGoogle Scholar |

Richmond TA, Somerville CR
(2000) The cellulose synthase superfamily. Plant Physiology 124, 495–498.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Roberts AW,
Roberts EM, Delmer DP
(2002) Cellulose synthase (CesA) genes in the green alga Mesotaenium caldariorum. Eukaryotic Cell 1, 847–855.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Römling U
(2002) Molecular biology of cellulose production in bacteria. Research in Microbiology 153, 205–212.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ruegger M,
Dewey E,
Gray WM,
Hobbie L,
Turner J, Estelle M
(1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes & Development 12, 198–207.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Sakakibara K,
Nishiyama T,
Kato M, Hasebe M
(2001) Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants. Molecular Biology and Evolution 18, 491–502.
| PubMed |

Sakakibara K,
Nishiyama T,
Deguchi H, Hasebe M
(2008) Class 1 KNOX genes are not involved in shoot development in the moss Physcomitrella patens but do function in sporophyte development. Evolution & Development 10, 555–566.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Scherp P,
Grotha R, Kutschera U
(2001) Occurrence and phylogenetic significance of cytokinesis-related callose in green algae, bryophytes, ferns an seed plants. Plant Cell Reports 20, 143–149.
| Crossref | GoogleScholarGoogle Scholar |

Soltis PS,
Brochington SF,
Yoo M-Y,
Piedrahita A,
Latvis M,
Moore MJ,
Chanderbali AS, Soltis DE
(2009) Floral variation and floral genetics in basal angiosperms. American Journal of Botany 96, 110–128.
| Crossref | GoogleScholarGoogle Scholar |

Steemans P,
Le Hérissé A,
Melvin J,
Miller MA,
Paris F,
Verniers J, Wellmann CH
(2009) Origin and radiation of the earliest vascular land plants. Science 324, 353.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Sztein AE,
Cohan JD,
Slovin JP, Cooke TJ
(1995) Auxin metabolism in representative land plants. American Journal of Botany 82, 1514–1521.
| Crossref | GoogleScholarGoogle Scholar |

Tanabe Y,
Hasebe M,
Sekimoto H,
Nishiyama T,
Kitani M,
Henschel K,
Munster T,
Theißen G,
Nozaki H, Ito M
(2005) Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proceedings of the National Academy of Sciences of the United States of America 102, 2436–2441.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Tanahashi T,
Sumikawa N,
Kato M, Hasebe M
(2005) Diversification of gene function: homologs of the floral regulator FLO/LFY control the first zygotic division in the moss Physcomitrella patens. Development 132, 1727–1736.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Vergara CE, Carpita NC
(2001) β-D-glycan synthases and the CesA gene family: lessons to be learned from the mixed-linkage (1–3), (1–4) β-D-glucan synthase. Plant Molecular Biology 47, 145–160.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Winter K-U,
Becker A,
Münster T,
Kim JT,
Saedler H, Theißen G
(1999) MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proceedings of the National Academy of Sciences of the United States of America 96, 7342–7347.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Worden AZ,
Lee J-H,
Mock T,
Rouzé P, Simmons MP ,
et al
.
(2009) Green evolution and dynamic adaptations revealed by the genomes of the marine picoeukaryotes Micromonas. Science 324, 268–272.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Zhong R, Ye ZH
(1999)
IFL1, a gene regulating interfascicular fibre differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. The Plant Cell 11, 2139–2152.
| Crossref |
PubMed |

Zimmermann W
(1952) Main results of the telome theory. The Paleobotanist 1, 456–470.



