Genetic characterisation of non-descript cattle populations in communal areas of South Africa
M. D. Mamogobo A B D , N. O. Mapholi C , K. A. Nephawe A , T. L. Nedambale A , T. J. Mpofu A , Y. P. Sanarana B and B. J. Mtileni A
A B D , N. O. Mapholi C , K. A. Nephawe A , T. L. Nedambale A , T. J. Mpofu A , Y. P. Sanarana B and B. J. Mtileni A
A Tshwane University of Technology, Department of Animal Sciences, Private Bag X680, Pretoria, 0001, South Africa.
B Agricultural Research Council, Private Bag X2, Irene, 0062, South Africa.
C University of South Africa, Department of Life and Consumer Sciences, Private Bag X6, Florida, Roodepoort, 1709, South Africa.
D Corresponding author. Email: mamogobomd@hotmail.com
Animal Production Science 61(1) 84-91 https://doi.org/10.1071/AN20030
Submitted: 23 January 2020 Accepted: 24 July 2020 Published: 31 August 2020
Journal Compilation © CSIRO 2021 Open Access CC BY-NC-ND
Abstract
Context: Indigenous cattle breeds represent an important genetic resource for livelihood of communal-area inhabitants. Indigenous breeds have the ability to withstand harsh climatic conditions, can adapt genetically to poor-quality forages and are resistant to parasites and diseases. These unique traits possessed by indigenous breeds are under threat because of unrestrained crossing with exotic commercial breeds, and this can lead to total loss of a breed.
Aims: The study was conducted to assess the genetic diversity and population structure of South African non-descript communal beef cattle populations by using 25 microsatellite markers.
Methods: Unrelated and non-descript animals (n = 150) were sampled from communal areas from five (5) provinces of South Africa, namely, Eastern Cape, KwaZulu–Natal, Limpopo, Mpumalanga and the North West, with 30 samples per breed taken. Six (6) known cattle breeds (n = 180) were used as a reference population. This included Angus, Afrikaner, Bonsmara, Brahman, Drakensberger and the Nguni, with 30 samples per breed.
Key results: High level of genetic diversity was found across the five non-descript populations, with an average heterozygosity of 75%. The Limpopo population was found to be the most diverse population, with the highest average number of alleles (8.5) and heterozygosity (ranging between observed heterozygosity of 70% and expected heterozygosity of 79%). STRUCTURE software assigned populations (2 ≤ K ≤ 20), with the most probable cluster being at K = 7. The Eastern Cape, KwaZulu–Natal and Limpopo populations had genetic material similar to those possessed by the Nguni and Bonsmara reference populations.
Conclusions: Results from the study showed that most genetic differentiation occurred within populations rather than among populations, and this might be due to the fact that there is no selection for or against any specific production trait expressed in the populations.
Implications: The obtained information will serve as a baseline for the development and implementation of sound breeding programs that will assist in controlling the gene flow, so as to lower the possible genetic dilution of the currently available genetic material.
Additional keywords: genetic diversity, heterozygosity, microsatellite markers.
Introduction
Indigenous breeds are important genetic resources for the livelihood of rural societies (Ramsay et al. 2000) to meet nutritional, economic and socio-cultural requirements (Zulu 2009). These indigenous breeds have an ability to withstand harsh climatic conditions (Thornton et al. 2009), resistance to parasites (Mapholi et al. 2016), diseases (Marufu et al. 2011, 2014) and genetic adaptation to poor-quality forages. The unique gene pool is under threat because of alterations in farming systems (Kristensen et al. 2015; Marsoner et al. 2017), such as crossbreeding occurring in small population sizes, the absence of breed societies (whose role is to keep records of pedigrees, promote and encourage the conservation, breeding and genetic improvement of the production potential among other things) and inbreeding depression (Ollivier and Foulley 2002; FAO 2007). These factors can lead to possible extinction or a total loss of a breed (Ollivier and Foulley 2002); therefore, it is important that the farm-animal genetic resources are managed effectively for future use.
Genetic-diversity studies conducted in South Africa have focused on stud and commercial herds of the Afrikaner cattle (Pienaar et al. 2014, 2018), six cattle breeds from research stations (Makina et al. 2014) and Nguni cattle ecotype (Sanarana et al. 2016). The genetic status of cattle populations existing in communal areas of South Africa is unknown and implementation of appropriate conservation measures must be considered to ensure the effective management of indigenous-animal genetic resources (Taberlet et al. 2008; Boettcher et al. 2010; Makina et al. 2014). With the constant decline in farm-animal genetic resources (Rege 1999; Nyamushamba et al. 2017; Gwaze et al. 2009), there is a need to characterise beef cattle populations of the communal area, so as to understand the existing diversity to facilitate the development of rational conservation strategies and sustainable utilisation for the breeds (Hanotte and Jianlin 2006). Genetic characterisation of these populations will provide a better understanding of breed formation that can be used in selection program for potential animals for present and future genetic resources (Sunnucks 2000; Toro and Caballero 2005; Tixier-Boichard 2014).
Traditionally, characterisation of the non-descript populations was based on their phenotypic information such as, for example, body frame and coat colour (Teneva 2009). However, as a result of the recent advancement in genomics technology, characterisation is now conducted using genetic information to improve their production. In recent years, the use of genetic markers such as microsatellites to determine the genetic diversity of populations, has gained popularity (Vignal et al. 2002; DeSalle and Amato 2004), including mapping of genes controlling economically important traits has been made possible by the use of microsatellite markers (Beuzen et al. 2000). With the use of genetic markers, it is possible to test for parentage, relatedness of a population, an individual’s identification, as well as to determine whether any migration has occurred (Hanotte and Jianlin 2006; Toro et al. 2009). Microsatellite markers have been used to characterise the diversity of South African chickens (Van Marle-Köster et al. 2008; Mtileni et al. 2011), cattle (Pienaar et al. 2014; Sanarana et al. 2016), sheep (Soma et al. 2012; Qwabe and Van Marle-Köster 2013) and goat (Mdladla et al. 2017). Therefore, the study was conducted to assess the genetic diversity and population structure of South African non-descript communal beef cattle populations, by using 25 microsatellite markers.
Materials and methods
Sampling sites
In total, 150 hair samples were randomly collected from different farmers from five non-descript cattle populations. This was from the Eastern Cape (EC, n = 30), KwaZulu–Natal (KZN, n = 30), Limpopo (LP, n = 30), Mpumalanga (MP, n = 30) and the North West (NW, n = 30) provinces of South Africa. Since there is no well structured recording system in smallholder farms, unrelated female animals (one per farmer) were selected and sampled. The sampled hair was collected from the tail end of each animal. Each sample was placed in an individual bag, sealed and labelled as per animal identification, so as to avoid any contamination. Six known beef cattle breeds were obtained from the Agricultural Research Council database, and used as reference populations for genetic admixture evaluation. The reference populations included Angus (ANG), Afrikaner (AFR), Bonsmara (BON), Brahman (BRA), Drakensberger (DRA) and the Nguni (NGU), with these being 30 animals per breed. The study was conducted in accordance with the approval of the Ethics Committee (AEC) of the Agricultural Research Council, South Africa (APIEC17/08).
DNA extraction, amplification and genotyping
DNA was extracted from each hair sample with visible roots, by using phenol–chloroform, following the protocol of Sambrook et al. (1989). The concentration (260/280 nm) and the purity (260/230 nm) of the genomic DNA was measured using a spectrophotometer (Nanodrop 2000c; Thermo Fisher Scientific Inc., Waltham, MA, USA; and Nanodrop 2000c). DNA polymorphisms were determined using a set of 25 autosomal microsatellite markers (Table 1) recommended by the Food and Agricultural Organisation of the United Nations and the International Society for Animal Genetics Advisory Group (FAO 2011) for genetic-diversity studies. These markers were selected on the basis of their high level of polymorphism reported by several studies (Kim et al. 2004; Pienaar et al. 2014; Sanarana et al. 2016). The DNA amplification was performed using a Perkin Elmer Gene Amp PCR System 9700 Thermo cycler (Applied Biosystems, Foster city, CA, USA). Amplicons were separated by capillary electrophoresis using an ABI 3130xl automatic sequencer (Applied Biosystems). Fluorescently labelled fragments were detected and sized using GeneMapper software (version 4.0, Applied Biosystems).
Statistical analyses
Genetic variation
The genetic diversity per locus and across the populations was estimated using Microsatellite Toolkit (Park 2001). Arlequin version 3.1 was used to perform locus by locus analysis of molecular variance (AMOVA), so as to determine the differentiation within and among the populations, genetic measures per locus and population (Excoffier et al. 2005). Genetic relationship among communal beef cattle populations were determined according to Nei’s standards (Nei 1987).
Cluster analysis
The genetic population-structure analysis of the communal cattle was performed using Bayesian admixture procedure implemented in STRUCTURE 2.3.4 (Pritchard et al. 2000), to infer the most likely number of clusters. The most probable number of populations was determined according to Evanno et al. (2005). A length of burning period was set at 10 000, with the number of Markov-chain Monte Carlo (MCMC) reps after burning being 20 000 and the number of iterations being 14 (for 2 ≤ number of clusters (K) ≥ 20). The most probable K value that reasonably describes the substructure of the populations under study was determined from the log probability of the data (Ln Pr (X|K)), using the STRUCTURE Harvester software (Earl and Von Holdt 2012), which implements Evanno’s method (Evanno et al. 2005). All other parameters in STRUCTURE were left as default (the default initial value of α was 1.0, so the value of λ was 1.0). The clustering pattern was visualised using DISTRUCT 1.1 (Rosenburg 2004). The factorial correspondence analysis (FCA) was computed using DARwin ver. 6 software package (Perrier and Jacquemound-Collet 2006). POPTREE2 was used to construct the phylogeny of the populations (Takezaki et al. 2010).
Results
Of 25 microsatellite markers used to study the genetic diversity of five non-descript cattle populations from five provinces of South Africa, 24 were found to be polymorphic, except one marker (HAUT27). Table 2 shows measures of genetic diversity among and within the populations. The average number of alleles ranged from 7.68 ± 2.10 for MP to 8.48 ± 2.40 for KZN. The expected heterozygosity (HE) ranged from 0.72 ± 0.03 (EC) to 0.79 ± 0.02 (LP), while observed heterozygosity (HO) ranged from 0.56 ± 0.02 (EC) to 0.70 ± 0.02 (LP and NW). All HE values were higher than the HO values in all populations. Moderate levels of inbreeding were observed in all populations, ranging from 0.02 to 0.20.
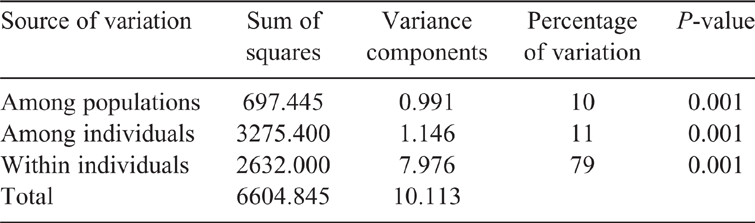
The AMOVA showed 10% of variation among populations and 11% of the variation among individuals, with the remaining 79% accounting for the differences within individuals in the population (Table 3).
|
Nei’s genetic distances among populations are shown in Table 4. The highest genetic differentiation of 0.42 was observed between MP and EC, while the lowest genetic differentiation of 0.02 was between EC and KZN.
Genetic clustering based on STRUCTURE analysis of the five populations is shown in Fig. 1. This was performed following Rosenberg et al. (2004), by using a Bayesian approach that inferred the K present in the population, permitting the detection of differences among populations and the hidden substructure within them. Each population was represented by one column, each with a different colour. The most probable cluster was found at K = 7. The non-descript populations formed three clusters, namely, the first cluster (NW), the second cluster (MP) and the third cluster (EC, KZN and LP). The clustering of EC, KZN and LP populations suggests that these populations are very similar; it is also noted that they had genetic material similar to those possessed by the NGU and BON reference populations. The DRA, AFR, ANG and the BRA reference populations each formed separate clusters.
The FCA was performed to determine the genetic relationship among the studied populations and the results are presented in Fig. 2. FCA showed very clear separation between the NW and MP. This analysis also showed an overlap of the EC, KZN and LP populations, suggesting a genetic relationship among the three populations.
The rooted neighbour-joining dendogram illustrated the genetic divergence between the five non-descript populations and the six reference populations (Fig. 3). The EC, KZN, LP shared some genotypes with the reference populations (NGU, DRA, BON, AFR and ANG) as they clustered together. The NW, MP and BRA populations formed two separate clusters. The BRA was the only reference population that formed a separate cluster.

|
Discussion
Genetic characterisation is an important tool in the implementation of appropriate breeding programs. It is important to genetically characterise farm animals so as to conserve the existing genetic material (Arora et al. 2008). In the current study, the focus was on genetic characterisation of non-descript-type cattle populations in communal farming in South Africa. This was achieved by assessing the level of genetic variation and inbreeding in the communal beef cattle populations by using autosomal microsatellite markers and by determining the population structure of beef cattle population, by using a reference dataset consisting of other South African purebred commercial lines.
The genetic diversity within the populations is indicated by the average number of alleles observed over a range of loci in different populations (Machugh et al. 1997; Hassen et al. 2012). Average number of alleles is, therefore, a strong indicator for the evolutionary potential of a population (Allendorf et al. 2012; Caballero and Garcıa-Dorado 2013), and it has been suggested that this measure is of key importance in population management and conservation (Simianer 2005; Foulley and Ollivier 2006). The overall average number of alleles in the populations of 8.13 observed in the current study was lower than that obtained in South African Nguni cattle (9; Sanarana et al. 2016) and that in Ankole longhorn cattle in the African Great Lakes Region (13.8; Ndumu et al. 2008). Moreover, the average number of alleles in the present study was higher than that observed in Afrikaner cattle in South Africa (4.8; Pienaar et al. 2018) and that in Yellow cattle in Taiwan (3.8) (Tu et al. 2014). The high average number of alleles in KZN and LP (8.48) is an indication of a higher genetic variation, and the low average number of alleles in MP (7.68) is an indication of a reduction in the population’s potential to adapt to future environmental changes, since this diversity is the raw material for evolution by natural selection (Fisher 1930). Barker (1994) suggested that the required minimum number of alleles for a microsatellite study is four and, thus, this study complied with that number as a higher number of alleles were observed, therefore justifying the selected markers from the FAO panel of microsatellite markers.
Nei (1987) and Barker (2001) reported that gene diversity (average HE) is a great measure of genetic variation within a population. High genetic variation occurs when there is a high number of alleles and this can be achieved by having a large population size. Genetic diversity ranges between 0 and 1 and this means that the historic admixture of different populations and a long-term natural selection for adaptation are indicated by high gene-diversity levels (Ojango et al. 2011; Paiva et al. 2011). The HE mean value of 0.75 observed in the current study was lower than the 0.92 observed in Vechur cattle in India (Radhika et al. 2018). The HO values ranged between 0.56 and 0.70, with a mean value of 0.66, and this was higher than 0.52 observed in Lidia bovine breed in Spain (Canon et al. 2008), but lower than the value of 0.67 for Nguni cattle populations in Mozambique (Bessa et al. 2009) and the value of 0.69 for Kankrej Cattle in India (Sodhi et al. 2007). Therefore, the difference between the HO and HE could be due to non-random mating among the individuals of the population (Sharma et al. 2016). The HE values in the present study were higher than that observed across all populations, and this may be attributed to forces such as inbreeding (Mburu and Hanotte 2005), isolation or genetic drift resulting in loss of genetic diversity (Lacy 1987). The LP population was found to be the most diverse population in the study, with the highest HE (79%) and HO (70%), and with an average number of alleles of 8.48.
It was observed that there was a higher genetic variation within individuals in the population (79%) than among populations (10%) or among individuals (11%). These low levels of differentiation among populations suggested common historical origins and a high level of inter-population gene flow (Radhika et al. 2018). These findings are in line with Groeneveld et al. (2010), who stated that most of the genetic diversity is present within a breed and not among breeds. For a breed to adapt to different environments and to respond to selection, genetic diversity is a requirement (Frankham et al. 2002) and AMOVA describes the genetic differentiation within and among populations (Toro and Caballero 2005). AMOVA showed that there was more genetic differentiation that occurred within populations than among populations and this might be due to the fact that there is no selection for or against any specific production trait expressed in the populations.
Nei’s genetic-distance pairwise matrix estimates were used to estimate the genetic relationship among the studied populations and they indicated that the EC and KZN were much closer to each other than they were to other populations. From these results, it can be concluded that farmers in those two provinces could be buying cattle from one another as these provinces are neighbours and transport costs can be kept at a minimum.
The results of STRUCTURE, FCA as well as the neighbour-dendogram analyses were in line with each other as they all supported the genetic distinctiveness of the NW and the MP populations. The NW and MP populations distinctively clustered away from each other and from any of the studied populations in all the methods that were used. The population STRUCTURE allows the evaluation of a population’s admixture from individuals, migrants in a population and also evaluates whether crossbreeding has occurred (Pritchard et al. 2000). The EC, KZN and LP populations had genetic material similar to that of the NGU and BON reference populations. Reasoning behind the EC, KZN and LP sharing genetic material similar to that of the NGU and BON is because these populations (EC, KZN and LP) are being reared in the communal areas, where there is no proper breeding programs, wherein any dominant male has a chance to breed (Scholtz et al. 2008). Most cattle in the rural areas are non-descript crossbreds with small populations of local breeds, such as Afrikaner, and locally developed Bonsmara and Drakensberger (Palmer and Ainslie 2006). The NGU and BON populations clustering together could be due to the fact that they are admixed with a distinct genome component that is shared with the Kuri (a hybrid between indicine populations and African taurine) and Ankole–Watusi, Boran, Sheko, short-horned Zebu and zebu Bororo (African zebus; Gautier et al. 2009). This was expected as most of the cattle in the country, although not being genetically verified, are regarded as NGU, BON or AFR and this could be due to the fact that most communal farmers practice uncontrolled mating programs and the animals graze together with those of NGU, BON or AFR farmers in the regions. Steinfeld et al. (2006) stated that strong management systems and agricultural practices are required for the conservation of livestock biodiversity. Another reason could be that farmers often prefer farming with the Nguni cattle, mainly because of the characteristics they possess (Baker and Rege 1994; Nyamushamba et al. 2017; Shabtay 2015). Correspondingly, the factorial analysis method showed a close genetic relationship among the EC, KZN and LP populations as these populations were mixed but the NW and MP populations formed separate clusters respectively. It is evident that the non-descript populations have an admixture of genetic material in common with those of the reference populations (the NW and MP excluded). The EC, KZN and LP populations had genetic material similar to that of any reference population, namely, the NGU and BON populations. This suggested that the EC, KZN and LP populations possessed similar genetic material.
Conclusions
The present study was the first to attempt to evaluate the existing genetic diversity of non-descript cattle populations in the communal areas of South Africa. The studied populations that were found to have genetic material similar to that of any of the reference populations were the EC, KZN and LP. The NW and MP populations did not possess any genetic material that was similar to that of the reference populations. Correct breeding programs should be put in place to conserve the current genetic material. The genetic diversity in the studied areas can be used as a baseline in the conservation, development and implementation of breeding programs. Programs that are directed to control indiscriminate crossbreeding practices in smallholder systems because an admixture of animals has been shown to suppress production, due to the mismatch among the production, environment and genotype, so as to lower the possible genetic dilution of the current available genetic material.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The National Research Foundation (NRF), Technology Innovation Agency (TIA), Southern African Science Service Centre for Climate Change and Adapted Land Use (SASSCAL), Agricultural Research Council (ARC) in South Africa are acknowledged for their financial support. We also thank farmers from the studied provinces for their cooperation during data collection.
References
Allendorf FW, Luikar TGH, Aitken SN (2012) ‘Conservation and the genetics of populations.’ (Wiley-Blackwell: West Sussex UK)Arora R, Bhatia S, Sehrawat A, Maity SB, Kundu SS (2008) Genetic variability in Jalauni sheep of India inferred from microsatellite data. Livestock Research for Rural Development 20, 1–8.
Baker RL, Rege JEO (1994) Genetic resistance to disease and other stresses in improvement of ruminant livestock in the tropics. In ‘Proceedings of the 5th World Congress on Genetics Applied to Livestock Production, 7–12 August 1994, University of Guelph, Guelph, Canada, pp. 405–412.
(a) Barker JSF (1994) A global protocol for determining genetic distances among domestic livestock breed. In ‘Proceedings of the 5th world congress on genetics applied to livestock production,University of Guelph, Guelph, Canada, vol. 21’, pp. 501–508.
Barker JSF (2001) Conservation and management of genetic diversity: a domestic animal perspective. Canadian Journal of Forest Research 31, 588–595.
| Conservation and management of genetic diversity: a domestic animal perspective.Crossref | GoogleScholarGoogle Scholar |
Bessa I, Pinheiro I, Matola M, Dzama K (2009) Genetic diversity and relationship among indigenous Mozambican cattle breeds. South African Journal of Animal Science 39, 61–72.
Beuzen ND, Stear MJ, Chang KC (2000) Molecular markers and their use in animal breeding. Veterinary Journal 160, 42–52.
| Molecular markers and their use in animal breeding.Crossref | GoogleScholarGoogle Scholar |
Boettcher PJ, Tixier-Boichard M, Toro MA, Simianer H, Eding H, Gandini G, Joost S, Garcia D, Colli L, Ajmone-Marsan P, the GLOBALDIV Consortium (2010) Objectives, criteria and methods for using molecular genetic data in priority setting for conservation of animal genetic resources. Animal Genetics 41, 64–77.
| Objectives, criteria and methods for using molecular genetic data in priority setting for conservation of animal genetic resources.Crossref | GoogleScholarGoogle Scholar | 20500756PubMed |
Caballero A, Garcia-Dorado A (2013) Allelic diversity and its implications for the rate of adaptation. Genetics 195, 1373–1384.
| Allelic diversity and its implications for the rate of adaptation.Crossref | GoogleScholarGoogle Scholar | 24121776PubMed |
Canon J, Tupac-Yupanqui I, Garcia-Atance MA, Cortes O, Garcia D, Fernandez J, Dunner S (2008) Genetic variation within the Lidia bovine breed. International Society for Animal Genetics Animal Genetics 39, 439–445.
| Genetic variation within the Lidia bovine breed. International Society for Animal GeneticsCrossref | GoogleScholarGoogle Scholar | 18492131PubMed |
DeSalle R, Amato G (2004) The expansion of conservation genetics. Nature Reviews. Genetics 5, 702–712.
| The expansion of conservation genetics.Crossref | GoogleScholarGoogle Scholar | 15372093PubMed |
Earl DA, Von Holdt BM (2012) Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4, 359–361.
| Structure harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method.Crossref | GoogleScholarGoogle Scholar |
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using software STRUCTURE: a simulation study. Molecular Ecology 14, 2611–2620.
| Detecting the number of clusters of individuals using software STRUCTURE: a simulation study.Crossref | GoogleScholarGoogle Scholar | 15969739PubMed |
Excoffier L, Laval G, Schneider S (2005) Arlequin version 3.1: an intergrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1, 47–50.
| Arlequin version 3.1: an intergrated software package for population genetics data analysis.Crossref | GoogleScholarGoogle Scholar |
FAO (2007) ‘The state of the world’s animal genetic resources for food and agriculture.’ (Eds B Rischkowsky, D Pilling) (FAO: Rome, Italy)
FAO (2011) ‘Molecular genetic characterization of animal genetic resources. FAO animal production and health guidelines. No. 9.’ (FAO: Rome, Italy)
Fisher R (1930) ‘The genetical theory of natural selection.’ (Oxford University Press: Oxford, UK)
Foulley J, Ollivier L (2006) Estimating allelic richness and its diversity. Livestock Science 101, 150–158.
| Estimating allelic richness and its diversity.Crossref | GoogleScholarGoogle Scholar |
Frankham R, Ballou JD, Briscoe DA (2002) ‘I: introduction to conservation genetics.’ (Cambridge University: Cambridge, UK)
Gautier M, Flori L, Riebler A, Jaffrézic F, Laloé D, Gut I, Moazami-Goudarzi K, Foulley J-L (2009) A whole genome Bayesian scan for adaptive genetic divergence in West African cattle. BMC Genomics 10, 550
| A whole genome Bayesian scan for adaptive genetic divergence in West African cattle.Crossref | GoogleScholarGoogle Scholar | 19930592PubMed |
Groeneveld LF, Lenstra JA, Eding H, Toro MA, Scherf B, Pilling D, Negrini R, Finlay EK, Jialin H, Groeneveld E, Weigend S, the GLOBALDIV Consortium (2010) Genetic diversity in farm animals: a review. Animal Genetics 41, 6–31.
| Genetic diversity in farm animals: a review.Crossref | GoogleScholarGoogle Scholar | 20500753PubMed |
Gwaze FR, Chimonyo M, Dzama K (2009) Communal goat production in southern Africa: a review. Tropical Animal Health and Production 41, 1157–1168.
| Communal goat production in southern Africa: a review.Crossref | GoogleScholarGoogle Scholar |
Hanotte O, Jianlin H (2006) Genetic characterization of livestock populations and its use in conservation decision-making. The role of biotechnology in exploring and protecting agricultural genetic resources. pp. 89–96. (Food and Agriculture Organization of the United Nations: Rome, Italy).
Hassen H, Lababidi S, Rischkowsky B, Baum M, Tibbo M (2012) Molecular characterization in Ethiopian indigenous goat populations. Tropical Animal Health and Production 44, 1239–1246.
| Molecular characterization in Ethiopian indigenous goat populations.Crossref | GoogleScholarGoogle Scholar | 22237413PubMed |
Kim KS, Min MS, An JH, Lee H (2004) Cross-species amplification of Bovidae microsatellite and low diversity of the endangered Korean goral. The Journal of Heredity 95, 521–525.
| Cross-species amplification of Bovidae microsatellite and low diversity of the endangered Korean goral.Crossref | GoogleScholarGoogle Scholar | 15475399PubMed |
Kristensen TN, Hoffmann AS, Pertoldi C, Stronen AV (2015) What can livestock breeders learn from conservation genetics and vice versa? Frontiers in Genetics 6, 38
| What can livestock breeders learn from conservation genetics and vice versa?Crossref | GoogleScholarGoogle Scholar | 25713584PubMed |
Lacy R (1987) Loss of genetic diversity from managed populations: interacting effects of drift, mutation, immigration, selection, and population subdivision. Conservation Biology 1, 143–158.
Machugh DED, Shriver MD, Loftus RTR, Cunningham PP, Bradley DGD (1997) Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 146, 1071–1086.
Makina SO, Muchadeyi FC, Van Marle-Köster E, Macneil MD, Maiwashe A (2014) Genetic diversity and population structure among six cattle breeds in South Africa using a whole genome SNP panel. Frontiers in Genetics 5, 1–7.
| Genetic diversity and population structure among six cattle breeds in South Africa using a whole genome SNP panel.Crossref | GoogleScholarGoogle Scholar |
Mapholi NO, Maiwashe A, Matika O, Riggio V, Bishop SC, Macneil MD, Banga CB, Taylor JF, Dzama K (2016) Genome-wide association study of tick resistance in South African Nguni cattle. Ticks and Tick-Borne Diseases 7, 487–497.
| Genome-wide association study of tick resistance in South African Nguni cattle.Crossref | GoogleScholarGoogle Scholar | 26897394PubMed |
Marsoner T, Vigla LE, Mancka F, Jaritzb G, Tappeinera U, Tasser E (2017) Indigenous livestock breeds as indicators for cultural ecosystem services: a spatial analysis within the Alpine space. Ecological Indicators 94, 55–63.
| Indigenous livestock breeds as indicators for cultural ecosystem services: a spatial analysis within the Alpine space.Crossref | GoogleScholarGoogle Scholar |
Marufu MC, Qokweni L, Chimonyo M, Dzama K (2011) Relationships between tick counts and coat characteristics in Nguni and Bonsmara cattle reared on semiarid rangelands in South Africa. Ticks and Tick-Borne Diseases 2, 172–177.
| Relationships between tick counts and coat characteristics in Nguni and Bonsmara cattle reared on semiarid rangelands in South Africa.Crossref | GoogleScholarGoogle Scholar | 21890073PubMed |
Marufu MC, Dzama K, Chimonyo M (2014) Cellular responses to Rhipicephalus microplus infestations in pre-sensitised cattle with differing phenotypes of infestation. Experimental & Applied Acarology 62, 241–252.
| Cellular responses to Rhipicephalus microplus infestations in pre-sensitised cattle with differing phenotypes of infestation.Crossref | GoogleScholarGoogle Scholar |
Mburu D, Hanotte O (2005) ‘A practical approach to microsatellite genotyping with special reference to livestock population genetics. ILRI Biodiversity project.’ A manual prepared for the IAEA/ILRI training course of molecular characterisation of small ruminant genetic resources of Asia. (Eds HH Montaldo, Meza-Herrera) (ILRI: Nairobi, Kenya)
Mdladla K, Dzomba EF, Muchadeyi FC (2017) Characterization of the village goat production systems in the rural communities of the Eastern Cape, KwaZulu–Natal, Limpopo and North West Provinces of South Africa. Tropical Animal Health and Production 49, 515–527.
| Characterization of the village goat production systems in the rural communities of the Eastern Cape, KwaZulu–Natal, Limpopo and North West Provinces of South Africa.Crossref | GoogleScholarGoogle Scholar | 28150112PubMed |
Mtileni BJ, Muchadeyi FC, Maiwashe A, Groeneveld E, Dzama K, Weigend S (2011) Genetic diversity and conservation of South African indigenous chicken populations. Journal of Animal Breeding and Genetics 128, 209–218.
| Genetic diversity and conservation of South African indigenous chicken populations.Crossref | GoogleScholarGoogle Scholar | 21554415PubMed |
Ndumu DB, Baumung R, Hanotte O, Wurzinger M, Okeyo MA, Jianlin H, Kibogo H, Solkner J (2008) Genetic and morphological characterization of the Ankole Longhorn cattle in the African Great Lakes region. Genetics, Selection, Evolution 40, 467–490.
Nei M (1987) ‘Molecular evolutionary genetics.’ (Columbia University Press: New York, NY, USA)
Nyamushamba GB, Mapiye C, Tada O, Halimani TE, Muchenje V (2017) Conservation of indigenous cattle genetic resources in southern Africa’s smallholder areas: turning threats into opportunities: a review. Asian-Australasian Journal of Animal Sciences 30, 603–621.
| Conservation of indigenous cattle genetic resources in southern Africa’s smallholder areas: turning threats into opportunities: a review.Crossref | GoogleScholarGoogle Scholar | 27004814PubMed |
Ojango JM, Mpofu N, Marshall K, Andersson-Eklund L (2011) Quantitative methods to improve the understanding and utilization of animal genetic resources. Animal genetics training resource, version 3. Available at http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.690.7966&rep=rep1&type=pdf [Verified 9 August 2020]
Ollivier L, Foulley JL (2002) Some suggestions on how to preserve both within- and between-breed genetic diversity. In: Book of Abstracts of the 53rd Annual Meeting of the European Association for Animal Production, Cairo, Egypt, 1–4 September 2002 (Ed. Y van der Honing). (Wageningen Academic Publishers Wageningen, The Netherlands)
Paiva SR, Facó O, Faria DA, Lacerda T, Barretto GB, Carneiro PLS, Lobo RNB, Mcmanus C (2011) Molecular and pedigree analysis applied to conservation of animal genetic resources: the case of Brazilian Somali hair sheep. Tropical Animal Health and Production 43, 1449–1457.
| Molecular and pedigree analysis applied to conservation of animal genetic resources: the case of Brazilian Somali hair sheep.Crossref | GoogleScholarGoogle Scholar | 21533896PubMed |
Palmer T, Ainslie A (2006) Country pasture/forage resource profiles: South Africa. FAO Publishing Policy and Support Branch: Rome, Italy.
Park SDE (2001) Trypanotolerance in West African and population genetic effects of selection. PhD Thesis, University of Dublin, Dublin, Ireland.
Perrier X, Jacquemound-Collet JP (2006) ‘DARwin software.’ Available at http://darwin.cirad.fr/darwin. [Verified 19 April 2017]
Pienaar L, Grobler JP, Neser FWC, Scholtz MM, Swart H, Ehlers K, Marx M (2014) Genetic diversity in selected stud and commercial herds of the Afrikaner cattle breed. South African Journal of Animal Science 44, 80–84.
Pienaar L, Grobler JP, Scholtz MM, Swart H, Ehlers K, Marx M, Macneil MD, Neser FWC (2018) Genetic diversity of Afrikaner cattle in southern Africa. Tropical Animal Health and Production 50, 399–404.
| Genetic diversity of Afrikaner cattle in southern Africa.Crossref | GoogleScholarGoogle Scholar | 29043474PubMed |
Pritchard JK, Stephens M, Donnely P (2000) Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Qwabe SO, Van Marle-Köster E (2013) Genetic diversity and population structure of the endangered Namaqua Afrikaner sheep. Tropical Animal Health and Production 45, 511–516.
| Genetic diversity and population structure of the endangered Namaqua Afrikaner sheep.Crossref | GoogleScholarGoogle Scholar | 22930466PubMed |
Radhika G, Aravindakshan TV, Jinty S, Ramya K (2018) Evaluation of genetic diversity, population structure, and relationship between legendary vechur cattle and crossbred cattle of Kerala State, India Animal Biotechnology 29, 50–58.
| Evaluation of genetic diversity, population structure, and relationship between legendary vechur cattle and crossbred cattle of Kerala State, IndiaCrossref | GoogleScholarGoogle Scholar | 28358589PubMed |
Ramsay K, Harris L, Kotze A (2000) Landrace breeds: South Africa’s indigenous and locally developed farm animals. Farm Animal Conservation Trust, Pretoria. FAO/UNEP Animal Genetic Resources Information Bulletin 27, 9–16.
| Landrace breeds: South Africa’s indigenous and locally developed farm animals. Farm Animal Conservation Trust, Pretoria.Crossref | GoogleScholarGoogle Scholar |
Rege JEO (1999) The state of African cattle genetic resources I. Classification framework and identification of threatened and extinct breeds. Animal Genetic Resources 25, 1–25.
| The state of African cattle genetic resources I. Classification framework and identification of threatened and extinct breeds.Crossref | GoogleScholarGoogle Scholar |
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Molecular Ecology Notes 4, 137–138.
| Distruct: a program for the graphical display of population structure.Crossref | GoogleScholarGoogle Scholar |
Sambrook J, Fritsch EF, Maniati ST (1989) ‘Molecular cloning: a laboratory manual, vol. 3.’ (Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA)
Sanarana Y, Visser C, Bosman L, Nephawe K, Maiwashe A, Van Marle-Köster E (2016) Genetic diversity in South African Nguni cattle ecotypes based on microsatellite markers. Tropical Animal Health and Production 48, 379–385.
| Genetic diversity in South African Nguni cattle ecotypes based on microsatellite markers.Crossref | GoogleScholarGoogle Scholar | 26611262PubMed |
Scholtz MM, Bester J, Mamabolo JM, Ramsay KA (2008) Results of the national cattle survey undertaken in South Africa, with emphasis on beef. Applied Animal Husbandry & Rural Development 1, 1–9.
Shabtay A (2015) Adaptive traits of indigenous cattle breeds: the Mediterranean Baladi as a case study. Meat Science 109, 27–39.
| Adaptive traits of indigenous cattle breeds: the Mediterranean Baladi as a case study.Crossref | GoogleScholarGoogle Scholar | 26025652PubMed |
Sharma R, Kumar B, Arora R, Ahlawat S, Mishra AK, Tantia MS (2016) Genetic diversity estimates point to immediate efforts for conserving the endangered Tibetan sheep of India. Meta Gene 8, 14–20.
Simianer H (2005) Using expected allele number as objective function to design between and within breed conservation of farm animal biodiversity. Journal of Animal Breeding and Genetics 122, 177–187.
| Using expected allele number as objective function to design between and within breed conservation of farm animal biodiversity.Crossref | GoogleScholarGoogle Scholar | 16130469PubMed |
Sodhi M, Mukesh M, Prakash B, Mishra BP, Sobti RC, Singh KP, Ahlawat SPS (2007) Microsatellite marker based characterization of genetic diversity in Kankrej cattle. Journal of Applied Animal Research 31, 153–158.
| Microsatellite marker based characterization of genetic diversity in Kankrej cattle.Crossref | GoogleScholarGoogle Scholar |
Soma P, Kotze A, Grobler JP, Van Wyk JB (2012) South African sheep breeds: population genetic structure and conservation implications. Small Ruminant Research 103, 112–119.
| South African sheep breeds: population genetic structure and conservation implications.Crossref | GoogleScholarGoogle Scholar |
Steinfeld H, Gerber P, Wassenaar T, Castel V, Rosales M, De Haan C (2006) ‘Livestock’s long shadow.’ (FAO: Rome, Italy)
Sunnucks P (2000) Efficient genetic markers for population biology. Trends in Ecology & Evolution 15, 199–203.
| Efficient genetic markers for population biology.Crossref | GoogleScholarGoogle Scholar |
Taberlet P, Valentini A, Rezaei HR, Naderi S, Pompanon F, Negrini R, Ajmone-Marsan P (2008) Are cattle, sheep, and goats endangered species? Molecular Ecology 17, 275–284.
| Are cattle, sheep, and goats endangered species?Crossref | GoogleScholarGoogle Scholar | 17927711PubMed |
Takezaki N, Nei M, Tamura K (2010) POPTREE2: software for constructing population trees from allele frequency data and computing other population statistics with Windows interface. Molecular Biology and Evolution 27, 747–752.
| POPTREE2: software for constructing population trees from allele frequency data and computing other population statistics with Windows interface.Crossref | GoogleScholarGoogle Scholar | 20022889PubMed |
Teneva A (2009) ‘Molecular markers in animal genome analysis, biotechnology in animal husbandry.’ (Institute for Animal Husbandry, Belgrade-Zemun: Belgrade, Serbia)
Thornton PK, Van De Steeg J, Notenbaert A, Herrero M (2009) The impacts of climate change on livestock and livestock systems in developing countries: a review of what we know and what we need to know. Agricultural Systems 101, 113–127.
| The impacts of climate change on livestock and livestock systems in developing countries: a review of what we know and what we need to know.Crossref | GoogleScholarGoogle Scholar |
Tixier-Boichard M (2014) Status and gaps in characterization of animal genetics resources. In ‘Proceedings of the 10th world congress of genetics applies to livestock production’, Vancouver, BC, Canada, 17–22 August.
Toro MA, Caballero A (2005) Characterization and conservation of genetic diversity in subdivided populations. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 360, 1367–1378.
| Characterization and conservation of genetic diversity in subdivided populations.Crossref | GoogleScholarGoogle Scholar | 16048780PubMed |
Toro MA, Fernández J, Caballero A (2009) Molecular characterization of breeds and its use in conservation. Livestock Science 120, 174–195.
| Molecular characterization of breeds and its use in conservation.Crossref | GoogleScholarGoogle Scholar |
Tu P, Lin D, Li G, Huang J, Wang D, Wang P (2014) Characterization of the genetic diversity and population structure for the Yellow Cattle in Taiwan based on microsatellite markers. Animal Biotechnology 25, 234–249.
| Characterization of the genetic diversity and population structure for the Yellow Cattle in Taiwan based on microsatellite markers.Crossref | GoogleScholarGoogle Scholar | 24813218PubMed |
Van Marle-Köster E, Hefer CA, Nel LH, Groenen MAM (2008) Genetic diversity and population structure of locally adapted South African chicken lines: Implications for conservation. South African Journal of Animal Science 38, 271–281.
Vignal A, Milan D, Sacristobal M, Eggen A (2002) SNP and other types of molecular markers and their use in animal genetics: review. Genetics, Selection, Evolution 34, 275–305.
| SNP and other types of molecular markers and their use in animal genetics: review.Crossref | GoogleScholarGoogle Scholar | 12081799PubMed |
Zulu ND (2009) Genetic characterization of Zambian Native Cattle Breeds. MSc Thesis, Faculty of the Virginia Polytechnic Institute and State University, Animal and Poultry Sciences, Blacksburg, Virginia.







