Unravelling methanogenesis in ruminants, horses and kangaroos: the links between gut anatomy, microbial biofilms and host immunity
R. A. Leng AA Emeritus Professor, University of New England, Armidale, NSW 2351, Australia. Email: rleng@ozemail.com.au
Animal Production Science 58(7) 1175-1191 https://doi.org/10.1071/AN15710
Submitted: 8 October 2015 Accepted: 31 January 2018 Published: 4 May 2018
Journal compilation © CSIRO 2018 Open Access CC BY-NC-ND
Abstract
The present essay aims to resolve the question as to why macropod marsupials (e.g. kangaroos and wallabies, hereinafter termed ‘macropods) and horses produce much less methane (CH4) than do ruminants when digesting the same feed. In herbivores, gases produced during fermentation of fibrous feeds do not pose a major problem in regions of the gut that have mechanisms to eliminate them (e.g. eructation in the rumen and flatus in the lower bowel). In contrast, gas pressure build-up in the tubiform forestomach of macropods or in the enlarged tubiform caecum of equids would be potentially damaging. It is hypothesised that, to prevent this problem, evolution has favoured development of controls over gut microbiota that enable enteric gas production (H2 and CH4) to be differently regulated in the forestomach of macropods and the caecum of all three species, from the forestomach of ruminants. The hypothesised regulation depends on interactions between their gut anatomy and host-tissue immune responses that have evolved to modify the species composition of their gut microbiota which, importantly, are mainly in biofilms. Obligatory H2 production during forage fermentation is, thus, captured in CH4 in the ruminant where ruminal gases are readily released by eructation, or in acetate in the macropod forestomach and equid caecum–colon where a build-up in gas pressure could potentially damage these organs. So as to maintain appropriate gut microbiota in different species, it is hypothesised that blind sacs at the cranial end of the haustral anatomy of the macropod forestomach and the equid caecum are sites of release of protobiofilm particles that develop in close association with the mucosal lymphoid tissues. These tissues release immune secretions such as antimicrobial peptides, immunoglobulins, innate lymphoid cells and mucin that eliminate or suppress methanogenic Archaea and support the growth of acetogenic microbiota. The present review draws on microbiological studies of the mammalian gut as well as other microbial environments. Hypotheses are advanced to account for published findings relating to the gut anatomy of herbivores and humans, the kinetics of digesta in ruminants, macropods and equids, and also the composition of biofilm microbiota in the human gut as well as aquatic and other environments where the microbiota exist in biofilms.
Additional keywords: acetogenesis, blind sacs, haustra, innate lymphoid cells, mucin.
Introduction
Strategies to inhibit enteric methane (CH4) production in herbivorous animals have been targeted as major areas for research funding because of the perceived need to curb the release of greenhouse gases into the atmosphere (Hristov et al. 2013). Because of their importance as major sources of food and fibre production for humans, ruminants have been studied more closely than other herbivores, especially in relation to aspects of their gut microbiology, gut physiology and gaseous emissions (Annison and Bryden 1998).
All animals host a large and complex biomass of microbes in their gastrointestinal tract. The microbiota present include viruses, Archaea, bacteria, protozoa and fungi that have evolved in communities that exhibit intimate between-species and species–host tissue associations. Considerable advances have been made in our understanding of the ecology of the gut microbiota of mammals and mutualistic interactions among the microbiota with the host’s immune systems have been recognised and explored (for recent reviews, see Eberl 2010; Bevins and Salzman 2011; Sansonetti 2011; Schluter and Foster 2012; Bang et al. 2014; Dishaw et al. 2014). These associations control many aspects of the physiology and biochemistry of the host, to such an extent that they are often regarded as an integral component of what has been dubbed the ‘super organisms’ (Eberl 2010). Mutualism is a two-way process and host secretions may be responsible for differences in the fermentation processes and, therefore, the end products of feed and fibre digestion in different animal species.
When ruminants digest forage diets, they produce three to four times more CH4 (a potent greenhouse gas) per unit of digested fibre (digestible neutral detergent fibre) than do macropods (von Engelhardt et al. 1978; Madsen and Bertelsen 2012) or equids (92 ± 15 versus 28 ± 9 L/kg; Franz et al. 2010). Methane emissions represent between 6.7 ± 1.7% of the gross energy and 12.3 ± 3.1% of the digestible energy of feed ingested by ruminants, whereas these emissions represent, respectively, ~1.5 ± 0.2 of gross energy and 3.2 ± 0.7% of digestible energy for equids (Franz et al. 2010), and similarly small amounts for ‘macropods’ (Kempton et al. 1976; Madsen and Bertelsen 2012).
The among-species variability in CH4 emissions in herbivores has been attributed to various factors. It is assumed there is an evolutionary connection between the level of CH4 production per unit of digested feed and the site of CH4 production in the gut. The differences in the anatomy of the fermentation organs and their position in the gut also appear to be of major significance.
The digestive tract of horses is different from that of macropods and ruminants as feed first enters the acidic stomach and then the small intestine, before being subjected to the fermentative digestion in the hind-gut (caecum–colon). Digestion of feed by host enzymes in the small intestine would not be associated with the production of gases. Therefore, on the same diet, equids will have a lower production of CH4 per unit of dry matter digested. However, these differences can be calculated to be almost negligible with animals fed roughage-based diets without added concentrates. For instance when fed a poor-quality hay, sheep produced three-fold more CH4 per unit of digested fibre (digestible neutral detergent fibre) than did ponies (Franz et al. 2010).
The need to explain the among-species differences in fermentation processes in the gut led to the concept for the present review that focusses on the microbial ecology of the gut of ruminants and macropods (both foregut fermenters) and horses (hind-gut fermenters). It appears that the genomes (and the resulting immune systems) of macropods and equids determine the ecology of the microbiota in their forestomach and caecum–colon respectively (Hackstein and van Alen 2010; Bevins and Salzman 2011). However, the precise nature of the physiological interactions between the host and its gut microorganisms is still not fully understood.
The intention when compiling the review was to generate and test hypotheses on the basis of currently available knowledge, but also to point to areas in which knowledge is lacking, so as to stimulate further research on the gut microbial ecology of mammals in general, and herbivores in particular. The hypotheses advanced rely on recent developments in our knowledge of the factors controlling the microbial ecology of the gut, in particular, the host’s involvement in determining the fermentative end products (gases in particular) produced in the forestomach of ruminants and macropods, and the hind-gut of horses. Consideration is focussed on the biotic mechanisms that have evolved to enable an efficient and sufficiently rapid rate of solubilisation and breakdown of complex structural components of plants (in particular lignin-associated polysaccharides) to enable herbivores to survive and grow (Wang and Chen 2009), while, at the same time, restricting gas production from fermentation in digestive organs where this could be damaging.
To establish the background information leading to these hypotheses, the opening sections of the review emphasise the major anatomical differences among the fermentative regions of the gut of ruminants, those of macropods and those of horses because these affect the microbial species composition in their gut biofilms. The importance of the abundance of different microbial species in these biofilms is emphasised, particularly of those involved with the disposal of H2 into either CH4 or acetate. With this background, the principal hypothesis is advanced to explain why H2 from fermentation is necessarily directed into acetate rather than CH4 in tubiform regions of the macropod forestomach and the hind-gut of most mammals; the reasoning, which in accord with the danger theory of Matzinger (2012), is that these fermentative organs lack physical mechanisms to remove gases that might otherwise accumulate and damage the host’s gut integrity, so the host uses its immune system to alter its gut microbiota, so as to minimise gas production. Current evidence for a two-way interaction between host immune factors and the abundance of different microbial species in biofilms involved in feed digestion, and, therefore, the acetogenic or methanogenic nature of the fermentation, is then presented.
Anatomical differences between the guts of herbivores
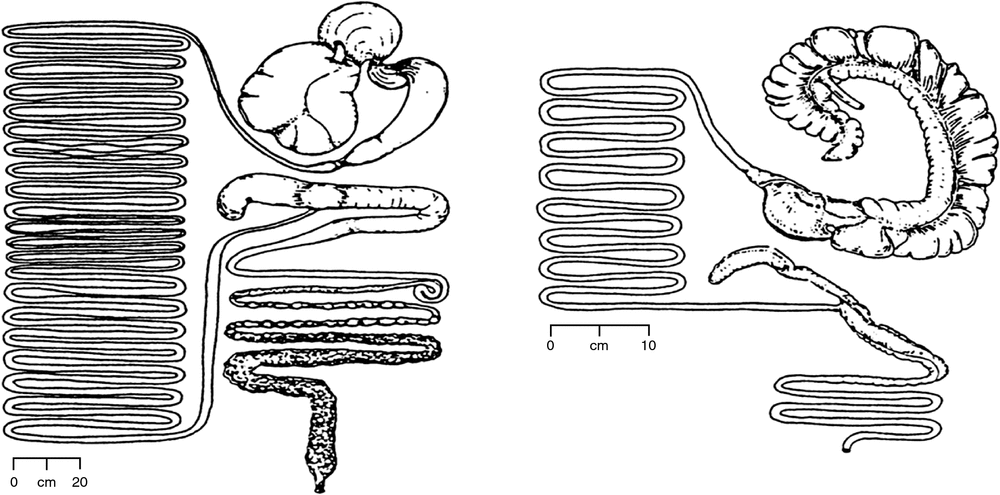
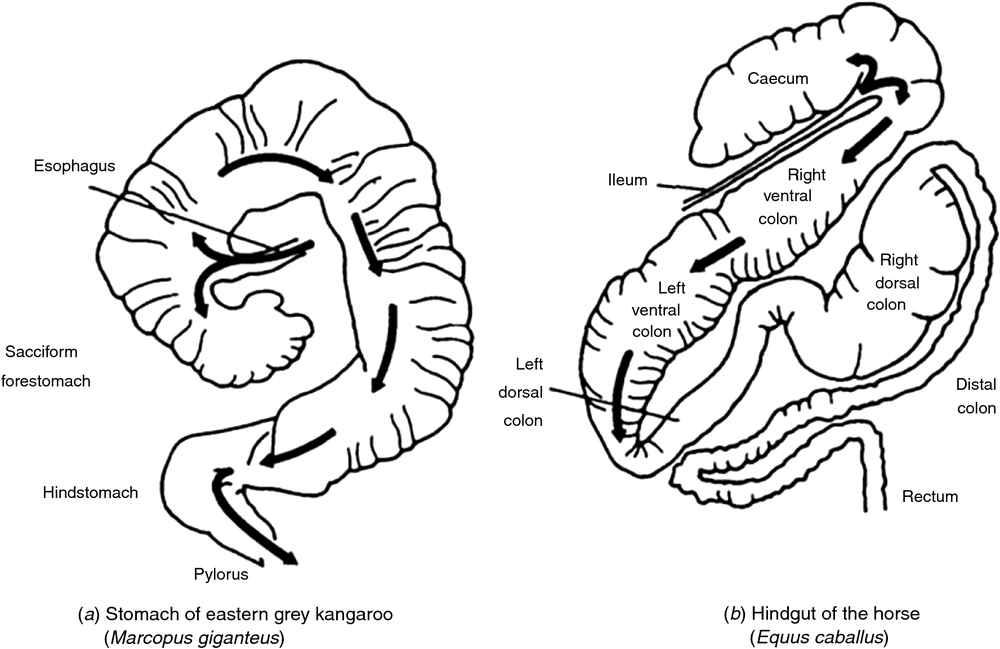
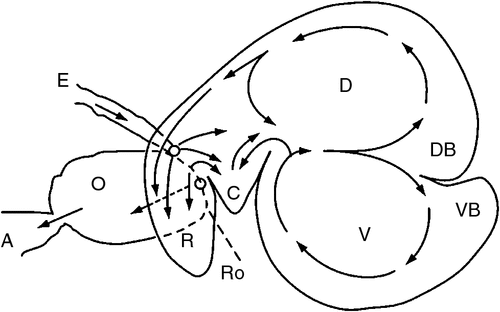
The differences in gut anatomy between two foregut fermenters, the ruminant and the macropod, are shown in Fig. 1. Similarities between the foregut of macropods and the caecum of horses are illustrated in Fig. 2.
|
|
Ruminant gut anatomy, feed processing and digesta kinetics
A ruminant uses its mouth and tongue to harvest forages during grazing or to ingest feeds offered under more intensive conditions. During the day, grazing animals typically spend more than one-third of their time foraging and at least one-third of their time ruminating. The roof of the ruminant mouth is a hard palate without incisors. The lower-jaw incisors are wide with a shovel-shaped crown and work against this hard dental pad. Premolars and molars match between upper and lower jaws. These teeth crush and grind plant material during initial chewing and also during rumination. In the mouth, forage mixes with saliva and chewing forms it into a bolus. Muscle contractions and pressure differences move the bolus down the oesophagus and on into the reticulo-rumen (the main foregut compartments, hereinafter referred to as the ‘rumen’), where contractions of the gut wall mix the contents. Newly ingested feed materials and larger particles that are less dense than the other rumen digesta often rise to form a mat of fibrous materials in close association with a gas cap on the surface of the rumen contents.
The bi-directional function of the oesophagus of ruminants allows them to regurgitate boluses mainly derived from the larger feed particles in the rumen, for further chewing and particle-size reduction. The re-chewed materials are then swallowed again with new feed added; eventually, the materials return to the rumen where the highest proportion of feed is solubilised, with the production of fermentative end products (see video, Gookin et al. 2011).
It has been proposed that, during rumination, partially digested forage is returned to the mouth where biofilm fragments that are abraded during chewing, innoculate newly gathered feed (Leng 2011); microbial fermentation in the rumen is efficient and mainly confined to the matrices of biofilm-encased microbes associated with the feed particles (Leng 2014, and references therein) and it can be likened to a continuous-flow, stirred-tank reactor (CFSTR; Hume 2002).
Macropod gut anatomy, feed processing and digesta kinetics
Macropod dentition is distinctively different from that of ruminants. The larger species of macropods have complex, high-crowned teeth. The four permanent molars on each side of both jaws erupt in sequence from front to back and move forward in the jaw with age, eventually being pushed out at the front. The molars possess cross-cutting ridges, so that tough grass or herbs are sheared between opposing teeth. The molars of smaller macropods are much simpler, so their mechanisms for grinding of feed materials into smaller units are inferior to those of ruminants. In addition, they have no mechanism similar to rumination. Nevertheless, regurgitation of food caused by violent contractions of the sacculated forestomach can occur occasionally during the day. The process has been termed ‘mycerism’ by Baker et al. (1963). The macropod eats more slowly and chews feed into much finer particles (Hume 2002). This strategy may increase the rate of feed colonisation by biofilms and enhance fermentative digestion (Langer 1984).
The wall of the forestomach of macropods is organised into three longitudinal bands of muscle, termed taeniae. Folds between the taeniae form the haustra. Contractions of the haustra propel feed boluses distally, while, at the same time, the folds selectively retain particles and release fluid. The overall effect is to squeeze fluid through the ingested feed particles, so that the fluid passes through the digestive tract more quickly than do the solids (Dellow 1982; Hume 2002; Figs 1, 2).
Haustral churning is the sequential movement of gut contents from one haustrum to the next; one haustrum expands as materials fill it, causing the muscles to contract, and the contents are pushed to the next haustrum. In this manner, remnant digesta is moved through these tubiform organs, with minimal mixing among batches. It is proposed that solid digesta is solubilised by the concerted effects of the syntrophic microbes in biofilms attached to the particles. The mechanisms may be the same wherever haustra are present in the gut compartment, including in the forestomach of the macropod and the caecum–colon of the horse and other mammals.
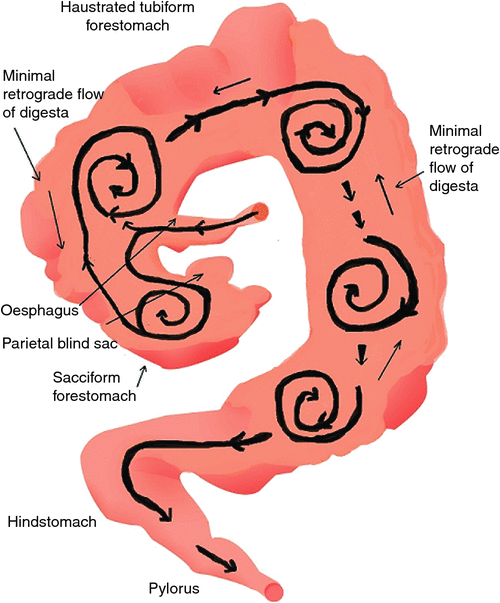
As the digesta move distally, particles decrease in size as a result of microbial fermentation. Radiographic studies indicate that ingested food is initially retained in the cranial region (Dellow 1979; Langer et al. 1980; Richardson 1980) and, although localised mixing of a single dose of a marker is effective, the marker does not mix with the entire contents of the forestomach. Digesta markers were observed radiologically to be transported slowly along the length of the forestomach, so that, in the frequently fed animal, after a period of 6 h, newly ingested food was not marked with previously administered contrast medium (Dellow 1979). These observations are evidence of bolus or tubular flow, perhaps commencing in the region of the second haustral pouch, as it appears that markers are mixed in sacculated forestomach and the first tubular components.
The finding of an accelerated solute marker excretion from the forestomach of macropods (Dellow 1979) is highly suggestive of a propulsive (aborad) peristalsis of the forestomach haustra. The forestomach of the macropods appears, therefore, to be analogous to a series of CFSTRs preceded by a continuously stirred reactor (Hume 2002), which is the sacciform component at the junction of the oesophagus and the tubiform stomach. The sacculated stomach has two small blind sacs, viz. the parietal sac and medial blind sac (Langer et al. 1980; Shoeib et al. 2015). The structure of the macropod’s foregut resembles that of the caecum in the horse (see Fig. 2). In the discussion that follows, it is speculated that the parietal blind sac of the macropod forestomach has a function similar to that ascribed by Bollinger et al. (2007) to the human appendix. It is a source of mucosal protobio-films containing immune agents (discussed in more detail below) that, having developed on the gut mucosal surface, adhere to and inoculate incoming feed materials with appropriate species of microorganisms.
Hume (1999) suggested that, on the basis of chemical-reactor theory, digesta mixing and movement in the macropod forestomach is best understood by viewing the forestomach as a series of CFSTRs, whereas the foregut of the ruminant is better documented as a single CFSTR. The CFSTR model, which features continuous flow of material into and out of a spherical reaction vessel, certainly describes the rumen well. In an ideal CFSTR, mixing is continuous and reactant concentrations are uniform throughout the vessel and, at steady-state, inflow rate equals outflow rate.
The presence of a sequence of CFSTRs in the macropod foregut suggests that very different digesta flow patterns must occur in their fermentative compartments (as envisaged in Fig. 3), from those in the forestomach of ruminants, which have been well documented (Fig. 4).
|
|
The sacciform morphology of the rumen, and the separation of small particles and return of large particles during rumination, increases the retention time of feed particles and improves the digestibility of structural plant cell-wall materials. Vigorous mixing, together with the rumination cycle (Gookin et al. 2011), causes feed particles to fragment, increasing the surface area available to microbial colonisation, and allows biofilm particles abraded from other feed particles to attach to newly ingested feed materials. This, in turn, reinforces the establishment of biofilms on newly arriving feed material.
Fermentation in the gut of herbivores
Digestion of fibrous feeds
Herbivores obtain the majority of their energy needs by digesting simple and complex polysaccharides, proteins and fats in feed materials. The organs in which fermentation occurs retain large amounts of cellulosic biomass for extended periods, enabling the feed organic matter (OM) to be more extensively solubilised and then fermented to volatile fatty acids (VFAs) that are the main source of absorbed energy substrates for the host (Hungate 1966). In foregut fermenters, most fermentation occurs before digesta reaches the host’s gastric stomach and small intestine and there is only subsidiary OM fermentation in the caecum and colon. In hindgut fermenters, digesta passes through the gastric stomach and small intestine, enabling sugars, fats and amino acids to be absorbed, before entering an enlarged fermentative region of the hindgut (caecum and colon) where the majority of the fermentative microbes are present. In forestomach fermenters, the microorganisms formed during fermentation are rich sources of protein (amino acids) for the host after they move into the lower gut and are digested. However, in hindgut fermenters, microbial proteins are lost to the animal when they are voided in faeces.
The potential for damage from fermentation gases in the mammalian gut
In ruminants and macropods, fermentation end products provide the major source of energy to the host in the form of VFA and microbial proteins are the major source of essential amino acids. Anaerobic microorganisms digest the majority of the feed OM via the Embden–Meyerhof–Parnas pathway of metabolism to pyruvate, and then to acetyl CoA and VFA (Hungate 1966). For this pathway to be efficient, there are the following two main requirements:
-
The adherent microbes must be closely associated in syntrophic associations within biofilms to facilitate efficient digestion of fibrous materials by hydrolysing their complex carbohydrates to soluble sugars and then to VFAs (Stoodley et al. 2002; De Mulder et al. 2016).
-
Glycolysis and oxidative decarboxylation of pyruvate result in the release of NADH+ +H+, which must then be oxidised to NAD+ with the release of metabolic hydrogen, [H], to permit fermentation to be continuous. Metabolic [H] is released as gaseous H2 or removed by methanogenesis or acetogenesis.
If the [H] is converted to gaseous H2, large volumes of gas are produced (although the volume is lower by a factor of 4 if [H] is oxidised to CH4). The solubilities of both H2 and CH4 are low, so these gases must be removed quickly to prevent gut distension, e.g. by eructation or as flatus. If methanogenesis is suppressed, the [H] must be removed in other reactions, usually by acetogenesis, to avoid possible damage to the gut (discussed below). Importantly, no gas is produced when acetogenesis is the [H] sink.
An example of the amounts of H2 or CH4 that can be produced (in a stable rumen at pH 7 where CO2 is mostly present as bicarbonate and therefore not problematic) follows. A steer fed a high-roughage, low concentrate-based diet, weighing 320 kg liveweight and growing at 1 kg/day produces ~135 g or 8.43 mol of CH4 daily. In gaseous form, this represents 189 L of CH4. If the gas produced had been H2, the volume of gas excreted would have been 756 L. If either of these gases were released into a rumen capable of holding ~30 L of liquid, in the absence of a release mechanism (the eructation reflex), the pressure generated would have distended the rumen and caused damage to its epithelium, as well as potentially also damaging other vital organs. Accordingly, the ruminant forestomach has evolved with anatomical and physiological adaptations to prevent gaseous distension.
Gas that accumulates in the apical part of the rumen can be quickly eliminated by eructation (Gookin et al. 2011) and eructation seems to be effective for even high rates of H2 production, e.g. when CH4 production is inhibited by selective toxins (Mitsumori et al. 2012). Nevertheless, when the eructation mechanism is inhibited by the formation of foam, gases build up and massive distension (bloat) of the organ can result (Clarke and Reid 1974). This is a rare phenomenon, mostly restricted to cattle on starch-rich feeds or on legume-based pastures subject to certain plant growth conditions; these situations would have been unlikely to occur during evolution and, so, protective mechanisms have not developed.
Where there is no gas-escape mechanism, the build-up of gas could damage cells by impairing NAD+ regeneration and inhibiting respiration; it could also disrupt the gut mucosa and, when extreme, gas production can lead to pneumatosis intestinalis (Gibson et al. 1993). This condition is characterised by gas cysts in the bowel wall. Excessive gas pressure is, therefore, a major threat to the integrity of the gut. The onset of physical damage to the gut mucosa may provoke an immune reaction according to the danger theory of Matzinger (1994, 2012). This theory is an extension of the conventional understanding of the immune system that allows the host to differentiate ‘self’ from ‘non-self’; Matzinger and colleagues have argued that the immune system not only reacts to foreign substances but also responds to alleviate situations that are potentially ‘dangerous’ to the animal. Pradeu and Cooper (2012) preferred ‘damaging’ as a more appropriate description.
Although macropods do have a form of eructation, this occurs only a few times per day and so is not a gas-release mechanism. Even so, it may allow the microbial mix in the forestomach to be inoculated with microbes from the more distal haustra. Similar retrograde movements of gut contents occur in the rabbit where reverse (orad) peristalsis of the colonic haustra separates very fine particles, including bacteria, from other digesta and concentrates these in the caecum (Ehrlein et al. 1983).
Metabolic H2 disposal when methanogens are absent from, or suppressed in, biofilm microbiota
In the rumen, most metabolic [H] is removed by CH4 generation. If methanogenic activity is suppressed, metabolic [H] has to be oxidised in other ways to enable fermentation of OM to continue. This is achieved mainly by acetogenic microbes that use [H] to reduce CO2 to acetate and have a higher H2 threshold than do methanogens.
The H2 threshold is the minimum H2 concentration in an anaerobic microbial ecosystem during the reduction of only one specific terminal electron acceptor (Lovley 1985; Cord-Ruwisch et al. 1988; Lovley and Godwin 1988; Conrad 1996). For methanogenesis, the H2 threshold in parts per million by volume (ppmv) is 6–120 (Conrad et al. 1983; and, for acetogenesis it is 430–4660. In mixed populations, homo-acetogens are, therefore, disadvantaged and often prevented from establishing in biofilms on feed particles. Owing to the presence of multiple hydrogenases with distinct properties, production and uptake of H2 can occur simultaneously within a single bacterial species or between microbial colonies within the biofilm (Fauque et al. 1988). The release of H2 in the biofilm matrix can be discontinuous and the steady-state partial pressure of H2 is potentially maintained by the ability of some acetogens to increase acetate production at high H2 pressures and to produce H2 from acetate whenever the partial pressure falls below a certain level. The acetyl-CoA or Wood–Ljungdahl pathway is the major pathway of acetogenesis and appears to be totally reversible (Ragsdale and Pierce 2008). Acetogens are metabolically versatile and almost all known acetogens can utilise alternative terminal electron acceptors such as nitrate. Some acetogens utilise the acetyl-CoA pathway to grow autotrophically on H2 and CO2; others grow heterotrophically or mixotrophically on a variety of organic compounds (Ragsdale and Pierce 2008).
The ability of homoacetogens to grow mixotrophically by metabolising sugars to generate ATP in the early stages of development of the biofilm would allow a larger pool of acetogens to develop so that, when the availability of sugars declines as the digesta remnants move through the haustra, the acetogens could revert to using H2 as their main energy source. The higher partial pressure of H2 needed to support homo-acetogenesis as against that required by methanogens could be maintained in biofilms by the ability of acetogens to reverse the acetogenesis reactions as digesta move distally in the macropod forestomach. In contrast, methanogens and reductive acetogens grow at close to their thermodynamic limits and close to the minimum quantum of free energy needed to sustain microbial growth (Schink 1997; McInerney et al. 2009).
Overview of biofilms
The nature of biofilms
The majority of microbes in nature, including those that ferment carbohydrates in the gut of mammals, exist in biofilms attached to solid surfaces (see Box 1).
| Box 1. Biofilms |
| Universally, the majority of microbes, including those in the gut of animals, exist in biofilms attached to solid surfaces. The microorganisms in biofilms are distributed as sessile colonises in a self-produced matrix of hydrated extracellular polymeric substances (EPS) that form their immediate environment. The EPS consist mainly of polysaccharides, proteins, nucleic acids and lipids that provide the mechanical stability of biofilms, mediate their adhesion to surfaces and form a cohesive, three-dimensional polymer network that interconnects and transiently immobilises microbial cells. In addition, the biofilm matrix acts as an external digestive system by keeping extracellular enzymes such as cellulase and hemicellulase close to feed surfaces, enabling them to degrade dissolved, colloidal and solid biopolymers. The closeness and distribution of the microbial colonies within the biofilm promotes syntrophic growth (outlined in Box 2) where the end products of one group of organisms become substrates for adjacent microbes. This is particularly important in controlling the concentrations of H2 in the vicinity of the organisms that ferment soluble carbohydrates to volatile fatty acids. Distances between cooperating species are small and, therefore, reaction times are short, resulting in high rates of degradation of plant organic matter in the fermentative areas of the gut. |
The critical role of biofilms in the fermentation of complex OM in the rumen has been reviewed by Craig et al. (1987), McAllister et al. (1994), Cheng et al. (1995), McAllister and Cheng (1996), Mayorga et al. (2007), Edwards et al. (2008), Leng (2014) and de Mulder et al. (2016). Many reviewers have emphasised the point that microbes must be attached to, or closely associated with feed particles, so as to enable efficient degradation of feed materials to occur. However, the concept that attached rumen microorganisms are organised in complex biofilms that facilitate mutualistic associations among co-located microbial species that also facilitate feed degradation has only recently been highlighted (Leng 2014; De Mulder et al. 2016, and references therein; summarised in Box 2).
| Box 2. Syntrophic growth of microbes |
| Syntrophism is a mutualistic interaction between two or more metabolically different organisms that are linked by the need to maintain an exchange of metabolites at low concentrations, making their overall metabolism feasible. The cooperation between fermentative microorganisms is based, in part, on the transfer of H2, formate, or acetate from fermentative bacteria to methanogens/acetogens, which ensures that the degradation of electron-rich substrates is thermodynamically favourable. Syntrophic metabolism proceeds at very low Gibbs’ free energy changes, close to the minimum free energy change needed to conserve energy biologically, i.e. the energy needed to transport one proton across the cytoplasmic membrane (Schink 1997). |
Much more is known about the biofilms in the gut of ruminants than of those in the guts of the other herbivores. The suggestion that biofilms promote a sufficiently high rate of solubilisation of fibrous feeds to meet the protein and energy requirements of ruminants (Weimer et al. 2009) supports the concept that dense biofilms must be responsible for the efficient digestive activity in the forestomach of macropods and the caecum–colon of equids. Pope et al. (2011) indicated that muramidase found in the foregut microbiome of the Tammar wallaby can direct cell aggregation and biofilm formation and this provides some indication that fermentative digestion is undertaken mainly by biofilm microbiota. However, in omnivorous or carnivorous mammals, a major ecological niche for gut microbiota is in biofilms attached to gut-wall mucosal cells (Sommer and Bäckhed 2013).
Useful information about biofilm-like structures can be obtained from many sources. Most aquatic ecosystems contain planktonic transparent exopolymer particles (Bar-Zeev et al. 2012). These organic microgels are highly surface-active (Bar-Zeev et al. 2012) and, when they contain microbial colonies, they are termed proto-biofilms. The transparent exopolymer particles in seawater originate from dissolved polymeric OM or from preformed biofilms and have most of the characteristics of ‘fledgling’ biofilms; they attach to surfaces and provide habitat for planktonic microbes to become sessile and proceed to develop colonies and to self-produce a matrix of hydrated extracellular polymeric substances that form their immediate environment. In the rumen, similar exopolymer particles seem to originate as sloughed biofilm fragments released from the surface of feed particles during rumination (Leng 2011, 2014), which is in accord with observations that 80–90% of the microbes in the rumen appear to be associated with particulate matter (Rodríguez et al. 2003). In addition, 80% of the microbes leaving the rumen have been particle-associated (Krebs 1987), indicating that, before leaving the rumen, a high percentage of bacteria and Archaea have moved between feed particles, without apparently mixing in the planktonic microbial pool (Krebs 1987). However, this does not preclude the likelihood that planktonic bacteria can adhere to the solid feed particles as a focal point for biofilm development (Huws et al. 2013). Both processes probably occur simultaneously and are complimentary.
The above arguments suggest there are significant differences between the way biofilms develop in the rumen and in the forestomach of macropods, and the caecum–colon of mammals in general. In the human appendix, for example, biofilms initially develop in association with epithelial cells linked to lymphoid tissue (Bollinger et al. 2003, 2007) and are thought to detach from the host epithelial tissue and then attach to digesta particles on which they mature as they move distally through the caecum (Bollinger et al. 2007). Accordingly, it appears probable that mucosal biofilms develop in the blind sacs, proximal to tubiform areas of the mammalian gut, where fermentative digestion of feed particles and other solids occurs. On this basis, it is likely that these proto-biofilms could develop in all blind sac structures containing lymphatic tissues. In macropods, the site of proto-biofilm development would be cranial to the forestomach and, in equids and ruminants, before the caecum.
In support of the above discussion, Bollinger et al. (2003, 2007) showed in humans that, following the collapse of the caecal microbiota from antibiotic treatment, mucosa-produced biofilms that had apparently been protected in the appendix were able to re-inoculate the caecum with normal microbiota. On this basis, the blind sac has been termed a ‘safe house’.
In summary, a crucial part of the hypothesis advanced later in this essay to explain the observed differences in fermentation characteristics and gas production between herbivores is that microgels containing immune factors formed in the parietal blind sac of the foregut of macropods, and in the blind sacs of the caecum of horses and ruminants, may inoculate proto-biofilms before they attach to particles and develop into mature biofilms as digesta moves through the tubiform forestomach and colon. These rudimentary biofilms, which form in close association with a host’s mucosal epithelial cells, would have the same function as proto-biofilms that can quickly mature to form ‘hotspots ‘of biofilm microbes on solid particles (Bar-Zeev et al. 2012). In the rumen, no such mechanisms are necessary and are not present.
Dynamic interactions between the animal and its gut microbiota
The hypothesis advanced herein involves a dynamic interplay between the animal and its gut microbiota; this interplay has been a focus of considerable recent research, particularly where the colonising bacteria play important roles in the digestion of food/feed and play a major role in the health of animals (reviewed by Bevins and Salzman (2011), Schluter and Foster (2012) and Dishaw et al. (2014) and summarised in Box 3).
| Box 3. Antimicrobial peptides |
| Antimicrobial peptides (AMPs), also called ‘host defence peptides’ are part of the innate immune response and are found among all classes of life. The AMPs are diverse and evolutionarily ancient molecules produced by all living organisms. They are polypeptides with fewer than 100 amino acids that are found in host defence settings (e.g. the gut of mammals) and have anti-microbial activity at the physiological concentrations prevailing in their tissues of origin (Ganz 2003). In humans and other mammals, the two main AMP families are defensins and cathelicidins. |
| Peptides belonging to these gene families exhibit an immune strategy as they defend against infection or potential tissue damage by targeted killing of bacteria, Archaea, viruses and fungi. They modify hosts by triggering tissue-specific defence and repair events. A variety of processes have evolved in microbes to evade the action of AMPs, including their ability to degrade or inactivate them, or to suppress their production by the host in response to infection (Ganz 2003). |
The term, microbiome, has been coined to describe all of the genes in the many different species found in the gut microflora. The abundance of individual species in the gut can alter markedly, in a matter of days, such as, for example, in response to changes in diet and the kinds of genes expressed (Ha et al. 2014). Currently, researchers are trying to understand the links between the dynamic activities of microbial communities and host biology and pathobiology (e.g. Brugman and Nieuwenhuis 2010). Even though the composition of these mutualistic communities of the digestive tract is variable, in the human at least, it appears that the overall microbiome is established and actually entrenched during birth, or shortly thereafter. This is apparently also so for the macropods (Peel et al. 2013) that give birth to partially developed fetuses (with a well developed head and front legs but an under-developed gut); these fetuses climb from the vagina into the pouch and attach to a teat at a very early stage of development (Tyndale-Biscoe 2005). During this period, a gut microbiome may partially establish before the animal is fully exposed to the external environment.
Molecular analysis based on bacterial 16S rRNA sequences has revealed that more than 1000 different species of bacteria are represented in the human gut microbiome (Eckburg et al. 2005) and most of these species have not been able to be cultured in vitro. Dominant bacteria are being identified in humans by using these new tools that are now also being applied to the rumen (Bath et al. 2013; de Mulder et al. 2016) and the macropod forestomach (Godwin et al. 2014). Recent molecular analyses of the composition of rumen ecosystems have demonstrated that there is great complexity within the myriad of species present in biofilms in any one location (de Mulder et al. 2016). The factors that are involved in the establishment of a specific microbiome in the gut, and those that influence the microbiota at the various anatomical sites in the gut are major areas for research (Ley et al. 2008).
Immuno-modulation of biofilm consortia in the forestomach of macropods
The biofilm mode of fermentation in the macropod forestomach has received little study but, as argued by Wang and Chen (2009) and Weimer et al. (2009) for ruminants, it would not be possible for macropods to digest plant OM at a sufficient rate to meet nutrient requirements without syntrophic growth of microbes in close association with feed particles. In macropods, biofilms probably develop on the mucosal lining of the parietal blind sac or possibly on the apical structures of the sacculated forestomach. The medial blind sac is not likely to be a site of biofilm development, as its epithelium consists of non-glandular mucosa (Shoeib et al. 2015).
In the present essay, it is speculated that, when the proto-biofilms detach from the mucosa, they contain host immune secretions that may include antimicrobial peptides (AMPs), immunoglobulins (IGs), innate lymphoid cells (ILCs) and mucin (Dishaw et al. 2014). The presence of these secretions either prevents the establishment of methanogens or kills them, creating a niche for acetogens to become the major hydrogenotrophs present in the biofilm community. The immune cells responsible for these secretions appear to recognise ‘non-self’-molecules by numerous membrane-bound, cytosolic or secreted receptors (pattern-recognition receptors) and are the first effectors of the population balance of Archaea, bacteria, fungi or viruses (Banchereau and Steinman 1998). By recognition and binding of microbe-associated molecular patterns on the surface of microorganisms, the innate immune responses are activated so that there is the release of AMPs or production of cytokines or activation of the complement cascade (Hart and McKenzie 1990; Banchereau and Steinman 1998; Lipscomb and Masten 2002; Bang et al. 2014).
The potential for hydrogenotrophic organisms in biofilms to be recognised by the immune system will be related to their specific chemical structures, or to other factors such as disturbances to the mucous layer in the intestines. The absence of methanogenic Archaea would be likely to allow the level of H2 to increase in the biofilm matrix and the ubiquitous acetogenic population to grow and expand, removing excess H2 and maintaining its partial pressure at levels favouring acetogenesis without compromising the rate of fermentative digestion. If this is true, then blind sacs are more than just ‘safe houses’ that inoculate the gut digesta following a disturbance in the ecosystem The biofilms from the blind sac continuously control the nature of the microbial associations in biofilms on solids in the tubular fermentative area of the gut and limit CH4 production at sites where gas is not easily expelled. This is in accord with Smith et al. (2009) who suggested that the driving force for the evolution of biofilm development in the blind sac of the proximal large bowel of vertebrates provided a means of maintenance of their normal bacterial populations.
In macropods, the development, subsequent growth and maturation of the mucosal-derived biofilm may be similar to that of biofilms in rumen digesta, except that acetogens are more numerous than methanogens in their layered structures. It is suggested that the biofilm communities on particulate digesta develop and mature as they move through the haustrated gut, progressively losing their immune agents as these are hydrolysed by proteolytic bacteria (Ganz 2003). In the terminal part of the macropod forestomach, secretion of acid prevents any local intrusion of methanogens. However, in the distal colon, there is greater potential for methanogens to develop.
The hypotheses proposed to explain these differences are outlined in Box 4.
| Box 4. Blind sacs in the gut may be where initiation of mucosal biofilms occurs and host’s immune secretions modulate their microbial composition |
| The caecum and its appendages, where present in vertebrates, are associated with rich lymphoid tissues and have been associated with vital immune modulation of the gut microbiota and maintenance of a ‘normal’ microbial flora (Smith et al. 2009). This modulation of gut flora was potentially the driving force for the evolution of blind sacs in the proximal large bowel of humans and other invertebrates (Smith et al. 2009). There are at least three morphotypes of the caecal appendix, as well as appendix-like structures in some animal species that lack a true caecal appendix. Immuno modulation of the gut microbial profile was potentially the driving force for the evolution of blind sacs in the proximal large bowel of humans and other invertebrates (Smith et al. 2009). The kangaroo has a small sacculated component in the forestomach with associated blind sacs, including one basal, one apical. The apical segment, although not characterised as an appendix, apparently has many of the attributes of the human appendix. |
| In the mammalian species that have been evaluated, mucosal biofilms tend to be most prominent in the appendix (when present) and in the apex of the caecum in animals without an appendix. In contrast, biofilms are generally absent in the distal large bowel and in the small intestine (Smith et al. 2009). Microbial biofilms are not only strengthened by secretary immunoglobulin A (Sig A) or antimicrobial peptides (AMPs) but can also be supported by mucin secreted by epithelial cells (Bollinger et al. 2006). |
| Biofilm consortia are essential for efficient digestion of feed organic matter containing complex carbohydrates. In the rumen, the composition of biofilms that form on particulate matter is mainly controlled by the partial pressure of H2. These H2 pressures are normally lower than that needed to support acetogenesis, but are adequate to support methanogenesis by Archaea. However, such methanogenic fermentation patterns might generate methane (CH4) gas pressures in the macropod forestomach or the equid caecum that would damage the gut lining. In light of Matzinger’s damage theory, it is suggested that macropods and equids have evolved immune-regulation mechanisms in these gut regions that suppress methanogenesis; proto-biofilms are probably formed in close association with the lymphoid tissues of blind sacs where various immune factors are secreted to specifically target and kill Archaea. This would encourage ubiquitous acetogens to develop in the mature biofilms where most of the organic-matter degradation takes place, thereby greatly reducing the release of fermentation gases and the risk of gut-wall damage. |
Factors affecting the nature and microbial composition of biofilms in ruminants and other herbivores
The fermentation process in the haustrated forestomach of macropods and the hind-gut of the horse differs from that in the rumen because, as discussed already, batches of digesta move as individual boluses towards the lower gut. At any particular distance from the sacculated forestomach in the tubiform organ (or haustral pouch), biofilm consortia can be expected to be at the same stage of development, growth, maturation and detachment. This expectation is reinforced by the fact that differently sized particles travel at the same rate during digesta passage through the forestomach of macropods (Munn et al. 2012).
Biofilms with a high microbial digestive ability are always composed of complex multi-species layers of microbes or associated aggregates of each species (Stoodley et al. 2002). In the absence of information to the contrary, acetogens in the forestomach of macropods are assumed to be associated with the biofilms in or on feed particles and probably also attached to the epithelial wall of the forestomach. In humans, biofilms on gut mucosal surfaces adhere to the mucosa, with cell-to-cell adhesion leading to multi-structure formation. These biofilms consist of colonies of microbes that are embedded in a matrix of self-produced extracellular polymeric substances that mediate protective functions and nutrient supply and, most importantly, facilitate H2 transfer (Bang et al. 2014).
Acetogens would need to be distributed in both the outer layers of these biofilms (to remove H2) and close to sites of attachment on the solid digesta where substrates such as sugars are produced by enzymatic hydrolysis of the structural carbohydrates. In the rumen, the biofilms present on feed particles would be at varying stages of maturation. This is because, as already noted, the rumen is essentially a continuous fermentative reactor where digesta from sequential meals are well mixed.
Summary of the proposed control of acetogens in the absence of methanogens in the forestomach of macropods
In the rumen, the dominant means of removal of H2 is by methanogenesis, even when ruminal microbes capable of reductive acetogenesis are present (Morvan et al. 1994; Pinder and Patterson 2012). In contrast, reductive acetogenesis appears to dominate as the major mechanism for H2 removal in the macropod forestomach (Kempton et al. 1976; Madsen and Bertelsen 2012; Godwin et al. 2014) and methanotrophic Archaea are virtually absent (Ouwerkerk et al. 2009; Klieve et al. 2012).
In anaerobic regions of the gut, and in the absence of dietary nitrate or sulfate, it appears that acetogenesis can become significant only if CH4 and formate syntheses by methanogens are inhibited by AMPs and the partial pressure of H2 is elevated. The latter may be maintained at the threshold pressure needed to support acetogenesis by switching the direction of the Wood–Ljungdahl pathway (Hattori et al. 2005; Zinder and Koch 1984). This pathway is found in all known acetogens and methanogens and is used in both the oxidative and reductive directions (Ragsdale and Pierce 2008). These organisms apparently still generate ATP when the pathway is reversed, but how they do this is unknown (Zinder and Koch 1984; Hoehler et al. 2002). Leng and Leonard (1965) found that when labelled acetate (methyl carbon labelled with tritium (3H) and carboxyl carbon with 14C) was infused into the rumen of sheep, the 3H : 14C ratio in rumen fluid acetate declined substantially over time, indicating that the methyl tritium was being replaced with unlabelled H. The labile nature of the methyl group of acetate is evidence that microbes that can utilise the Wood–Ljungdahl pathway are present in the rumen and may play an important role in controlling the H2 concentrations in the biofilms.
Acetogens appear to be present in the digestive tracts of all mammals. However, they appear to be competitive with the hydrogenotrophic Archaea only in certain sites where acetate is the electron sink. In environmental biofilms, sulfate- or nitrate-reducing bacteria are able to lower H2 partial pressure (Cord-Ruwisch et al. 1988) and these bacteria partially or totally out-compete reductive acetogens. Under most feeding conditions, sulfate or nitrate are present in only small amounts in the diet but, when concentrations of sulfate or nitrate are elevated by direct dietary supplementation, these salts become the dominant H2 sinks ahead of CO2 (Lee and Beauchemin 2014).
Host immune secretions that alter the microbial composition of mucosal-derived biofilms in the gut
Immunomodulation of the gut microbiome of herbivores
Recent research has indicated that unknown genetic factors are involved in determining the predominant hydrogenotrophic organisms in the gut of mammals (Hackstein and van Alen 2010; Dishaw et al. 2014). There appears to be a strong reciprocal and mutually beneficial interaction between the immune system and the microbial profile in the colon of humans (Bollinger et al. 2007; Dishaw et al. 2014). Because syntrophic associations of microbes in biofilms are essential for efficient and rapid digestion in ruminants, it is assumed here that biofilm formation is also essential for the efficient digestion of feed in the forestomach of macropods and hind-gut of horses. It is envisaged that these mucosa-associated biofilms contain immune agents obtained in a way similar to that described by Bollinger et al. (2007) for the human appendix. The parietal blind sac of the macropod forestomach may have a secretory function and also be the site where these proto-biofilms develop before being released into the sacculated forestomach. Once detached, these proto-biofilms may then re-attach to feed particles and continue to develop in the sacculated part of the foregut and as the digesta bolus proceeds through the haustrated forestomach. There is increasing knowledge of both host-mediated biofilms in the human colon (Bollinger et al. 2003; Bang et al. 2014) and of the role of biofilms in the rumen (see Costerton 2007; de Mulder et al. 2016); however, no studies have compared and contrasted the development of host-mediated biofilms in other mammals. It was, therefore, necessary to extrapolate from the available literature when developing the hypotheses advanced in the present essay.
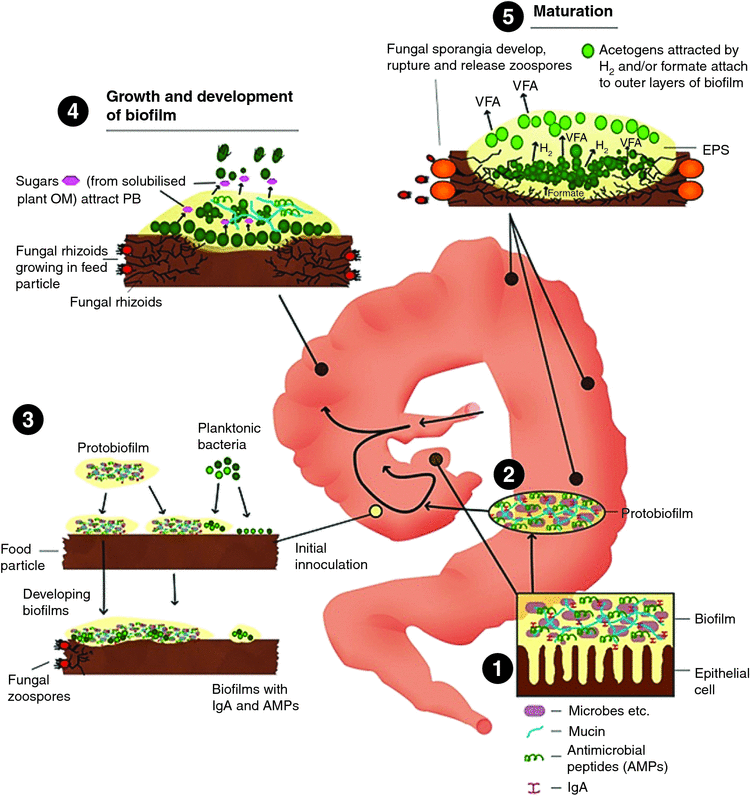
Development of biofilms on feed particles in the sacculated forestomach of macropods
Conditioning of a feed-particle surface for biofilm and microbial attachment begins immediately after the feed enters the mouth. During chewing, feed is mixed with submaxillary salivary mucin. The key characteristic of mucin is its ability to form gels and it is, therefore, a component in most gel-like secretions. The proposed mechanisms for mucosal biofilm formation take into account the recently recognised role of mucosal-associated microgel particles resembling proto-biofilm in facilitating and accelerating this process. It is proposed here that this is analogous to the inoculation of inert surfaces with biofilms in aquatic environments, as discussed by Bar-Zeev et al. (2012).
It is further argued that mucosal biofilms (as microgel particles) develop on, and are released from the uppermost mucosal layer of the parietal blind sac (see Box 4), and that these immature biofilms harbour host secretions containing mucin and possibly immune elements such as the innate lymphoid cells (ILCs) that produce antimicrobial peptides (AMPs) and immunoglobulins (IgA; Dishaw et al. 2014). The attachment of bacteria and additional microgel particles in the released microgel probably initiates the early development of feed particle-attached biofilms that have a complement of host secretions, including substrates such as mucin (which may support acetogens) and immune agents that prevent growth of methanogens (and, therefore, predetermine the microbial mix that develops early as the biofilm matures and passes through the haustrated components of the forestomach/caecum). The envisaged series of events that occurs as the biofilm develops on feed particles and moves through the forestomach of the macropod is illustrated in Fig. 5.
Potential involvement of innate lymphoid cells in control of biofilm microbiota
In recent times, the complex interactions of the immune system that play a role in controlling gut microbiota have been revisited with the discovery of ILCs that have roles in mediating immune responses and regulating tissue homeostasis (Walker et al. 2013; see Box 5).
| Box 5. Innate lymphoid cells |
| Innate lymphoid cells (ILCs) are newly described immune cells. The ILCs secrete high concentrations of cytokines and are implicated in innate immunity, inflammation, lymphoid-tissue formation and tissue remodelling. Consistent with their potential role in immune surveillance and their involvement in early detection of microbes that may endanger or damage the host animal (as implied for methanogens in Box 4), ILCs are localised on mucosal surfaces and respond to secreted molecules from the epithelium. Because ILCs share developmental and functional similarities with T helper (Th) cells, ILCs are categorised into three groups according to the transcription factors mediating their development and the cytokines they secrete. Of these, Group-3 ILCs are of interest to this review. These are composed of lymphoid-tissues inducer cells (LTi) and ILC3 cells are required for the development of lymphoid tissues, particularly in the intestinal tract where ILC3s have a crucial role in mediating the delicate two-way interactions between the symbiotic microbiota and the intestinal immune system (Walker et al. 2013). |
The ILC family resides predominantly in mucosal issues, particularly in the intestinal tract, where ILC3s (Box 5) have a crucial role in mediating a delicate balance between the symbiotic microbiota and the intestinal immune system (Walker et al. 2013). The LTi cells, an ILC subset, have been recently recognised as important regulators of lymphoid-tissue architecture after birth; they have been shown to mediate immunity to enteric pathogens (Sonnenberg et al. 2011) and also appear to trigger release of anti-microbial peptides (Eberl et al. (2015). The scope of the present review prevents further exploration of the possible role of ILCs in the biofilm microbiota that are proposed to develop in association with the lymphoidal tissue associated with the proximal large bowel of some herbivores or the paretial blind sac of the forestomach of macropods. However, the above discussions provide some evidence for the assumption that methanogens are controlled by immune factors in the gut of some mammals.
Mucin: a potential role in mucosal biofilms
Goblet cells, epithelial cells, macrophages and dendritic cells are the major cellular constituents of the innate defence system, and the mucous layer that coats the epithelial lining of the gut compartments contains mucin (Corfield et al. 2000). Mucin is the key polymeric, visco-elastic and protective component of mucus. In addition to mucin, mucus contains molecules of the immune systems such as IgA and AMPs that facilitate the clearance of pathogenic organisms (Kim and Khan 2013; Hasnain et al. 2013) or, as postulated here, other non-pathogenic organisms capable of producing end products such as H2 and CH4 that may damage the host’s gut integrity. Communication/reaction between immunoglobulins and complexes with commensal bacteria and the epithelial cells lining the mucosal surfaces of the gut has been implicated in the symbiotic host–commensal relationship (Blais Lecours et al. 2014). Thus, mucin, as part of the endothelium-derived biofilm on digesta particles, may have an important role in shaping the microbial biome (Koropatkin et al. 2012), which appears to have been under-recognised.
Mucin is secreted as large aggregates of proteins with molecular masses of 1–10 million Da. The high content of sugars in mucin could give the biofilms considerable water-holding capacity. Mucin may also partially protect immune secretions such as AMPs and IgA from hydrolysis by resident microbes and provide a readily fermentable source of substrate to rapidly increase the rate of growth of acetogenic bacteria in the early stages of biofilm development in the tubiform regions of the gut. The mucin secreted from specialised epithelial cells exchanges Ca2+ for Na+ and expands considerably, producing a visco-elastic gel with interwoven molecules that, combined with other secretions, forms part of the mucosal biofilm (Bollinger et al. 2007; Kim and Khan 2013). Mucin secretion by the epithelial cells appears to be obligatory, providing structures for attachment of cells and host-derived chemicals for the development of biofilms on feed particles (Bollinger et al. 2007; Smith et al. 2009; Dishaw et al. 2014).
An additional speculation is that fermentation of the closely available mucin sugars within the biofilm matrix may also play a role in activating some AMPs. Schroeder et al. (2011) suggested that activation of AMPs through reduction of disulfide bonds could occur in the gut of humans, mice and probably other species (this was termed environment-dependent activation; Ostaff et al. 2013). The AMP termed human β-defensin 1 (hBD-1) is constitutively expressed by epithelia but, in comparison with other AMPs, its anti-microbial activity is low. However, Schroeder et al. (2011) found that, after reduction of the three disulfide bonds in hBD-1, its anti-microbial activity was strongly enhanced. Reduced hBD-1 is able to kill Gram-positive anaerobic bacteria of the normal gut flora of the human as well as opportunistic pathogens, whereas the oxidised peptide has not shown such activity. A possibility here is that, in the proto-biofilm stage of development, AMPs are in the oxidised form but, with movement of digesta down the tract and the commencement of fermentation, the resultant increase in the partial pressure of H2 ensures a favourable reducing environment in the biofilm matrix that activates the AMPs that kill (lyse) any Archaea that are present.
AMPs, IgA and mucin may help shape the microbial species composition of mucosa-derived biofilms
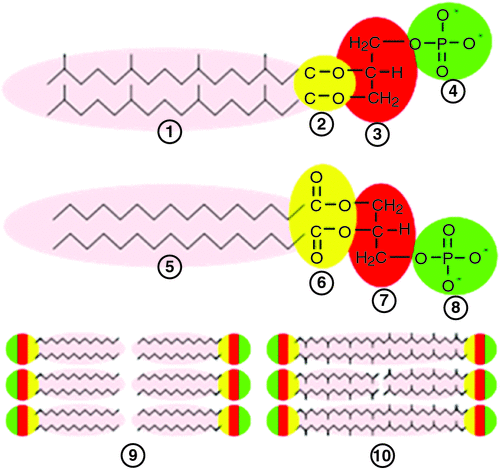
The bacterial membrane is made up of fatty acid esters. In contrast, the lipids in Archaeal membranes are derived from glycerol di-ethers of isoprenoid alcohols arranged as a bilayer or, in some Archaea, they consist of glycerol tetraethers that form mono-lamellar membrane patches (Fig. 6).
|
Archaeal cell walls lack peptidoglycans that are universally found in all bacterial cell walls and instead contain pseudo-murein (or pseudo-peptidoglycan), polysaccharide or protein (De Rosa et al. 1986). The Archaeal cell-wall polymers are structurally diverse, ranging from pseudomurein surface-layer (S-layer) proteins to methanochondroitin (Kandler and König 1998). These differences offer major clues to the roles of AMPs in shaping the microbial ecology of the gut (Sansonetti 2011). The AMPs have only recently been examined in relation to their ability to kill Archaea. Bang et al. (2014) clearly demonstrated the effects of different synthetic AMPs on the growth of Methanosarcina mazei, Methanobrevibacter smithii and Methanosphaera stadtmanae isolated from the human gut. Methanogens in general were highly sensitive to derivatives of human cathelicidin, porcine lysin and a synthetic anti-lipopolysaccharide peptide (Lpep). Sensitivities differed markedly among the methanogen species, but disruption of the unique cell envelope of Archaea occurred within minutes of exposure to all AMPs tested. This demonstrated that AMPs released by epithelial cells are a potent defence mechanism targeting methanogenic Archaea (Bang et al. 2014). The effects of the AMPs add support to the hypothesis that host-produced AMPs are secreted into the developing proto-biofilms while they are associated with the lining of the blind sac and, when AMPs are released to the lumen of the forestomach of macropods, they prevent methanogens from establishing, thus creating a more favourable environment for the growth of acetogens. This provides a plausible explanation for the differences in hydrogenotrophic microbial populations in the forestomach of ruminants and macropods, and the also the caecum of ruminants and hindgut fermenters (where AMP control of Archaea may be weakened as remnant digesta move towards the rectum).
The pouch of macropods is a rich source of AMPs (Peel et al. 2013). Considering that the under-developed fetus is attached to mother’s teats in the pouch for an extended time, it is hypothesised that it is exposed to early colonisation and so can potentially develop a wide range of AMPs. The recent whole-genome sequencing of the Tammar wallaby has allowed discovery and characterisation of novel AMPs in this species (Daly et al. 2008). Further studies have shown strong broad-spectrum activity of these AMPs (Wang et al. 2011), indicating their likely important role in the immune system of macropods in particular. From an evolutionary view point, if AMPs that specifically kill methanotrophic Archaea are responsible for the dominance of acetogenesis over methanogenesis in tubiform fermentative areas of the gut, this activity could have evolved to counteract the potential damage associated with gas pressure in tubiform organs.
The proposed hypothesis
The hypothesis advanced to explain the differences in CH4 production in ruminants versus other herbivores is that host-determined immune secretions ensure that methanogens are prevented from establishing in the fermentative chambers that would be damaged by a build-up of gas pressures; methanogenesis is suppressed and acetogenesis is promoted and supported in biofilms associated with digesta particles in these vulnerable gut regions. The hypothesis is summarised in Box 6.
| Box 6. The hypotheses proposed to explain differences in H2 utilisation in the forestomach of macropods as compared with ruminants |
| Gaseous products of fermentation (H2 and methane, CH4) are readily removed by eructation from the rumen and also via flatus from the large bowel in all mammals. However, in the forestomach of macropods and the caecum–colon of the horse, no such ‘gas release’ mechanism is available. As the volume of gas produced in the latter is large, in comparison to the volume of digesta, the pressure attained could potentially damage the organ. For this reason, these animals have evolved to support acetogenic-type fermentation; this type of fermentation is maintained by immune modulation of the species composition of microbial biofilms and differences in the gut anatomy where fermentative digestion occurs. It is postulated that mucosal cells, in close association with lymphoid tissues in the blind sacs at the proximal end of the haustral structure of the forestomach of the macropod, and the caecum of the horse and ruminants, secrete gel-like proto-biofilms containing immune agents (that suppress or kill methanogenic Archaea). These secretions also contain mucin that may promote the establishment of acetogens as the proto-biofilms develops into a mature fermentative biofilm. These biofilms solubilise the organic matter as they move distally through the forestomach and caecum. The biofilm consortia are determined by these immune secretions that may include antimicrobial peptides (AMPs), immunoglobulins A (IgAs) and possibly innate lymphoid cells (ILCs). The AMPs that lyse the distinct cell envelope of Archaea pave the way for total or partial replacement of methanogens by acetogenic bacteria. The result is that CH4 or H2 production is markedly reduced or eliminated in the caecum–colon of the equids or the foregut of macropods relative to that produced in the rumen or the lower bowel of mammals in general. |
Uncertainties with aspects of the proposed hypothesis
How do fungi complete their life cycle in the macropod forestomach?
Anaerobic fungi grow within solid components of feed (Gordon and Phillips 1998) and are found in the rumen and also the forestomach of the macropod (Dellow et al. 1988). They produce sporangia that release zoospores that actively invade insoluble plant materials, particularly in the areas where damage to the waxy surface of feed has occurred. Penetration of fungal rhizoids throughout plant particles weakens the structures and promotes more rapid reduction of particle size and greater access for other organisms. The fungi are in close contact with the biofilm consortia or can be considered as an extension of the biofilm into the solid plant particles (Leng 2014). All species of phycomycetous fungi that have been isolated from the gut of animals are obligate anaerobes. Despite this, they have been isolated successfully from the saliva and faeces of ruminants, which suggests that there is an aero-tolerant stage in their life cycle (Brookman et al. 2000). The life cycle of the anaerobic fungi is easily envisaged in the rumen; however, if the sporangia are produced and release zoospores in the more distal haustra in the macropod forestomach, back flow of digesta may be needed to continually re-inoculate the sacculated area or the first haustral pouch. There is potential for this to occur, as merycism periodically returns partially digested feed to the mouth. The vigour of the muscular contraction in merycism suggests that there may be an intermittent but forceful return of digesta from the more distal haustral pouches to the sacculated forestomach, thereby inoculating it with fungal spores and also bacteria and protozoa. Alternatively, macropods may be inoculated by ingesting aero-tolerant spores on feed; however, this appears unlikely.
How do protozoa survive and grow in ruminants and macropods?
Protozoa are present in the sacculated forestomach of macropods (Dellow et al. 1988). They may be prevented from passing distally in the liquid gut contents by attachment to larger particles or the wall of the forestomach, being released only when soluble sugars are present in the feed. A similar cycle of attachment and release is seen with protozoa in the rumen (Williams 2007). Rumen protozoa exhibit hydrogenosome activity (Williams 2007) and methanogens associate closely with the protozoa to take advantage of the release of H2 during fermentation. Whether protozoa in the forestomach of macropods also exhibit such hydrogenosome activity, but with acetogens replacing methanogens, is a possibility that requires research.
Summary of the major components of the hypothesis
Methanogenesis predominates in the forestomachs of ruminants as the eructation reflex quickly removes any potentially danger to the integrity of the fermentative organ from gases released in fermentation of feed and maximises the potential for digestion of fibrous feeds by providing a sink for disposal of metabolic [H]. Microbial inoculation of feed in the rumen appears to occur to a major extent from established biofilms associated with feed particles and there is only minor modulation by immune tissue or secretions.
In the macropod forestomach, biofilms from the blind sac may be more like biofilms that occur in the lower gut of other non-ruminant mammals; during their development, these biofilms initially may obtain immune agents from the host that control their microbial species composition. The concept is that immune recognition benefits the macropod host by promoting mutualistic gut bacteria (Everett et al. 2004) as well as selecting against microbial species that are non-pathogenic, but could cause damage or disrupt homeostasis in the foregut (as proposed by Matzinger 2012).
To explain why macropods and horses rely on acetogenesis to dispose of metabolic [H], whereas ruminants rely on methanogenesis for the same purpose, it is argued that excessive gas production from the fermentation may illicit ‘damage signals’ and activate immune secretions in potentially affected organs to suppress the microbes responsible for releasing H2 or CH4. The effect of this immune modulation is to exclude methanogens from the caecum and haustral forestomach and create opportunities for acetogenic microbes to thrive, thus benefitting the animal by eliminating the damage to the gut tissues from excessive gas, while still maintaining mechanisms for efficient digestion of fibrous feeds.
Conflicts of interest
The author declares no conflicts of interest.
Acknowledgements
I am indebted to Emeritus Professor John Nolan for his many contributions to this essay; in its structure, discussions and assistance with editing. Without his input, this paper could not have been finalised. I also thank Peter Cook and Joel Olsson who prepared the coloured diagrams.
References
Annison EF, Bryden WL (1998) Perspective on ruminant nutrition and metabolism: I. Metabolism in the rumen. Nutrition Research Reviews 11, 173–178.| Perspective on ruminant nutrition and metabolism: I. Metabolism in the rumen.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXhs12nurc%3D&md5=c6bcc71a59e1e00f8e9aa6a3bb69dee7CAS |
Baker S, Brown GD, Calaby JH (1963) Food regurgitation in the macropodiae. Australian Journal of Science 25, 430–432.
Banchereau J, Steinman RM (1998) Dendritic cells and the control of immunity. Nature 392, 245–252.
| Dendritic cells and the control of immunity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXitFWrsr8%3D&md5=76a30260383885526a925b341bdd907cCAS |
Bang C, Weidenbach K, Gutsmann T, Heine H, Schmitz RA (2014) The intestinal Archaea Methanosphaera stadtmanae and Methanobrevibacter smithii activate human dendritic cells. PLoS One 9, e99411
| The intestinal Archaea Methanosphaera stadtmanae and Methanobrevibacter smithii activate human dendritic cells.Crossref | GoogleScholarGoogle Scholar |
Bar-Zeev E, Berman-Frank I, Girshevit O, Berman T (2012) Revised paradigm of aquatic biofilm formation facilitated by microgel transparent exopolymer particles. Proceedings of the National Academy of Sciences of the United States of America 109, 9119–9124.
| Revised paradigm of aquatic biofilm formation facilitated by microgel transparent exopolymer particles.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XovF2gu70%3D&md5=70459c9dd461286a033d5a409b10267aCAS |
Bath C, Morrison M, Ross EM, Hayes BJ, Cocks BG (2013) The symbiotic rumen microbiome and cattle performance: a brief review. Animal Production Science 53, 876–881.
Bevins CL, Salzman NH (2011) The potter’s wheel: the host’s role in sculpting its microbiota. Cellular and Molecular Life Sciences 68, 3675–3685.
| The potter’s wheel: the host’s role in sculpting its microbiota.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtl2gsr%2FN&md5=9f8d9c0ab6ddd05470317e33c5d9ce73CAS |
Blais Lecours P, Marsolais D, Cormier Y, Berberi M, Haché C, Bourdages R, Duchaine C (2014) Increased prevalence of Methanosphaera stadtmanae in inflammatory bowel diseases. PLoS One 9, e87734
| Increased prevalence of Methanosphaera stadtmanae in inflammatory bowel diseases.Crossref | GoogleScholarGoogle Scholar |
Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, Parker W (2003) Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 109, 580–587.
| Human secretory immunoglobulin A may contribute to biofilm formation in the gut.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXmt1Sisbw%3D&md5=b8b5b486c82cde8e72923f3e1509d066CAS |
Bollinger RB, Everett M, Wahl S, Lee YH, Orndorff PE, Parker W (2006) Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Molecular Immunology 43, 378–387.
| Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXht1Cmu7vF&md5=3958fbbfb670df1a591f3688dfe5f690CAS |
Bollinger RR, Barbas AS, Bush EL, Lin SS, Parkera W (2007) Biofilms in the large bowel suggest an apparent function of the human vermiform appendix. Journal of Theoretical Biology 249, 826–831.
| Biofilms in the large bowel suggest an apparent function of the human vermiform appendix.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhtlGhtrfF&md5=3c73e3773b8c37fc373ebd125150c8baCAS |
Brookman JL, Ozkose E, Rogers S, Trinci PJ, Theodorou MK (2000) Identification of spores in the polycentric anaerobic gut fungi which enhance their ability to survive. FEMS Microbiology Ecology 31, 261–267.
| Identification of spores in the polycentric anaerobic gut fungi which enhance their ability to survive.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXhslamurg%3D&md5=8da44cf6cb28fbfb085ce1f55f7441b5CAS |
Brugman S, Nieuwenhuis EE (2010) Mucosal control of the intestinal microbial community. Journal of Molecular Medicine 88, 881–888.
| Mucosal control of the intestinal microbial community.Crossref | GoogleScholarGoogle Scholar |
Cheng KJ, McAllister TA, Costerton JW (1995) Biofilm of the ruminant digestive tract. In ‘Microbial biofilms’. (Eds H Lappin-Scott, JM Costerton) pp. 221–232. (Cambridge University Press: Cambridge, UK)
Clarke RTJ, Reid CSW (1974) Foamy bloat of cattle. A review. Journal of Dairy Science 57, 753–785.
| Foamy bloat of cattle. A review.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE2cXltFyrt7g%3D&md5=857434066401335412478b043c2ccc9cCAS |
Conrad R (1996) Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiological Reviews 60, 609–640.
Conrad R, Aragno M, Seiler W (1983) The inability of hydrogen bacteria to utilize atmospheric hydrogen is due to threshold and affinity for hydrogen. FEMS Microbiology Letters 18, 207–210.
| The inability of hydrogen bacteria to utilize atmospheric hydrogen is due to threshold and affinity for hydrogen.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3sXktlGrsbs%3D&md5=4bc62ed617f71dd7567d11f2c62c8b3bCAS |
Cord-Ruwisch R, Seitz HJ, Conrad R (1988) The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Archives of Microbiology 149, 350–357.
| The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1cXht1yhurc%3D&md5=090fb3a64848b975bbdaceca4fda6322CAS |
Corfield AP, Myerscough N, Longman R, Sylvester P, Arul S, Pignatelli M (2000) Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease. Gut 47, 589–594.
| Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXnsVCru7s%3D&md5=21357e906e12cac87d39806cce266245CAS |
Costerton JW (2007) The biofilm primer. In ‘Springer series on biofilms’. (Ed. C Eckey) p. 165. (Springer-Verlag: Berlin)
Craig WM, Broderick GA, Ricker DB (1987) Quantitation of microorganisms associated with the particulate phase of ruminal ingesta. The Journal of Nutrition 117, 56–62.
| Quantitation of microorganisms associated with the particulate phase of ruminal ingesta.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2sXhslGju7o%3D&md5=dd5d5d230fd8f80713019bdb01354afeCAS |
Daly KA, Digby MR, Lefevre C, Nicholas KR, Deane EM, Williamson P (2008) Identification, characterization and expression of cathelicidin in the pouch young of tammar wallaby (Macropus eugenii). Comparative Biochemistry and Physiology 149, 524–533.
| Identification, characterization and expression of cathelicidin in the pouch young of tammar wallaby (Macropus eugenii).Crossref | GoogleScholarGoogle Scholar |
de Mulder T, Goossens K, Peiren N, Vandaele L, Haegeman A, De Tender C, Ruttink T, de Wiele TV, De Campeneere S (2016) Exploring the methanogen and bacterial communities of rumen environments: solid adherent, fluid and epimural. FEMS Microbiology Ecology 93, fiw251
| Exploring the methanogen and bacterial communities of rumen environments: solid adherent, fluid and epimural.Crossref | GoogleScholarGoogle Scholar |
De Rosa M, Gambacorta A, Gliozzi A (1986) Structure, biosynthesis and physicochemical properties of Archaea bacterial lipids. Microbiological Reviews 50, 70–80.
Dellow DW (1979) Physiology of digestion in the macropodine marsupials. PhD Thesis, University of New England, Armidale, NSW.
Dellow DW (1982) Studies on the nutrition of macropodine marsupials III. The flow of digesta through the stomach and intestine of macropodines and sheep. Australian Journal of Zoology 30, 751–765.
| Studies on the nutrition of macropodine marsupials III. The flow of digesta through the stomach and intestine of macropodines and sheep.Crossref | GoogleScholarGoogle Scholar |
Dellow DW, Hume ID, Clarke RTJ, Bauchop T (1988) Microbial activity in the forestomach of free-living macropodid marsupials: comparisons with laboratory studies. Australian Journal of Zoology 36, 383–395.
| Microbial activity in the forestomach of free-living macropodid marsupials: comparisons with laboratory studies.Crossref | GoogleScholarGoogle Scholar |
Dishaw LJ, Cannon JP, Litman GW, Parker W (2014) Immune-directed support of rich microbial communities in the gut has ancient roots. Developmental and Comparative Immunology 47, 36–51.
| Immune-directed support of rich microbial communities in the gut has ancient roots.Crossref | GoogleScholarGoogle Scholar |
Eberl G (2010) A new vision of immunity: homeostasis of the superorganism. Mucosa Immunology – Nature 3, 450–460.
| A new vision of immunity: homeostasis of the superorganism.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhtVCntrrM&md5=ee796badcba461d307584888c4bbb2c8CAS |
Eberl G, Colonna M, Di-Santo MJP, McKenzie ANJ (2015) Innate lymphoid cells: a new paradigm in immunology. Science 348, aaa6566
| Innate lymphoid cells: a new paradigm in immunology.Crossref | GoogleScholarGoogle Scholar |
Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308, 1635–1638.
| Diversity of the human intestinal microbial flora.Crossref | GoogleScholarGoogle Scholar |
Edwards JE, Huws SA, Ki EJ, Lee MRF, Kingston-Smith AH, Scollan ND (2008) Advances in microbial ecosystem concepts and their consequences for ruminant agriculture. Animal 2, 653–660.
| Advances in microbial ecosystem concepts and their consequences for ruminant agriculture.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BC38vptVykuw%3D%3D&md5=2847037b89bf4eeb6f94854c9b544378CAS |
Ehrlein HJ, Reich H, Schwinger M (1983) Colonic motility and transit of digesta during hard and soft faeces formation in rabbits. The Journal of Physiology 338, 75–86.
| Colonic motility and transit of digesta during hard and soft faeces formation in rabbits.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL3s3mvVegsA%3D%3D&md5=9aff7f8577d8967c16a6421797e7a791CAS |
Everett ML, Palestrant D, Miller SE, Bollinger RB, Parker W (2004) Immune exclusion and immune inclusion: a new model of host–bacterial interactions in the gut. Clinical and Applied Immunology Reviews 4, 321–332.
| Immune exclusion and immune inclusion: a new model of host–bacterial interactions in the gut.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXlsVSitrk%3D&md5=19773999e0107b6371e52702d0b4f307CAS |
Fauque G, Peck HD, Moura JJG, Huynh BH, Berlier Y, Der Vartanian DV, Teixeira M, Przybyla AE, Lespinat PA, Moura I, LeGall J (1988) The three classes of hydrogenases from sulfate‐reducing bacteria of the genus Desulfovibrio. FEMS Microbiology Reviews 54, 299–344.
| The three classes of hydrogenases from sulfate‐reducing bacteria of the genus Desulfovibrio.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1MXhtlSru7g%3D&md5=5194fdd76639a507417786906e1f66d3CAS |
Franz R, Soliva CR, Kreuzer M, Steuer P, Hummel J, Clauss M (2010) Methane production in relation to body mass of ruminants and equids. Evolutionary Ecology Research 12, 727–738.
Ganz T (2003) The role of AMPs in innate immunity. Integrative and Comparative Biology 43, 300–304.
| The role of AMPs in innate immunity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXmslantbk%3D&md5=f803e9eaffc8c26ad90fa2aa0633606dCAS |
Gibson GR, Macfarlane GT, Cummings JH (1993) Sulphate reducing bacteria and hydrogen metabolism in the human large intestine. Gut 34, 437–439.
| Sulphate reducing bacteria and hydrogen metabolism in the human large intestine.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3sXksVGqu7s%3D&md5=45c080f7e8e0616046c6f4be984a4046CAS |
Godwin S, Kang A, Gulino LM, Manefield M, Gutierrez-Zamora ML, Kienzle M, Ouwerkerk D, Dawson K, Klieve AV (2014) Investigation of the microbial metabolism of carbon dioxide and hydrogen in the kangaroo foregut by stable isotope probing. The ISME Journal: Multidisciplinary Journal of Microbial Ecology 8, 1855–1865.
| Investigation of the microbial metabolism of carbon dioxide and hydrogen in the kangaroo foregut by stable isotope probing.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhsVWksr7E&md5=bbf1f96045492abcfecf1f49a241f32eCAS |
Gookin JL, Foster DM, Harvey AM (2011) ‘An animated model of reticulorumen motility.’ 3rd edn. North Carolina State University. Available at www.ncsu.edu/project/cvm_gookin/rumen_motility.swf [Verified July 2017]
Gordon GLR, Phillips MW (1998) The role of anaerobic gut fungi in ruminants. Nutrition Research Reviews 11, 133–168.
| The role of anaerobic gut fungi in ruminants.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1M%2FjsF2rsQ%3D%3D&md5=84506b4b3ac8ba1d6be79cfdaa0605faCAS |
Ha CW, Lam YY, Holmes AJ (2014) Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health. World Journal of Gastroenterology 20, 16498–16517.
| Mechanistic links between gut microbial community dynamics, microbial functions and metabolic health.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXitFWgsrnJ&md5=20d3df7922e746856b24823fc1bb4252CAS |
Hackstein JHP, van Alen TA (2010) Methanogens in the gastro intestinal tract of animals. In ‘Endosymbionic methanogenic Archaea’. Microbiological monographs 19, pp. 115–125. (Springer-Verlag: Berlin)
Hart DN, McKenzie JL (1990) Interstitial dendritic cells. International Reviews of Immunology 6, 127–138.
| Interstitial dendritic cells.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK3s3htVeqsA%3D%3D&md5=0b4e7eed46efac0de3fc9915a8ca320eCAS |
Hasnain SZ, Gallagher AL, Grencis RK, Thornton DJ (2013) A new role for mucins in immunity: insights from gastrointestinal nematode infection. The International Journal of Biochemistry & Cell Biology 45, 364–374.
| A new role for mucins in immunity: insights from gastrointestinal nematode infection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXosF2qtw%3D%3D&md5=11fdc01af39bd6dd7bcaf93b16d68922CAS |
Hattori S, Galushko HA, Kamagata Y, Schink B (2005) Operation of the CO dehydrogenase/acetyl coenzyme A pathway in both acetate oxidation and acetate formation by the syntrophically acetate oxidizing bacterium Thermacetogenium phaeum. Journal of Bacteriology 187, 3471–3476.
| Operation of the CO dehydrogenase/acetyl coenzyme A pathway in both acetate oxidation and acetate formation by the syntrophically acetate oxidizing bacterium Thermacetogenium phaeum.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXkt1CksLo%3D&md5=99331dba1455f0c329f3f0a16e45de6cCAS |
Hoehler TM, Alberty DB, Alperin MJ, Bebout BM, Martens CS, des Marais DJ (2002) Comparative ecology of H2 cycling in sedimentary and phototrophic ecosystems. Antonie van Leeuwenhoek 81, 575–585.
| Comparative ecology of H2 cycling in sedimentary and phototrophic ecosystems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xns1Sgtr8%3D&md5=2c0b4121c8358d35a81d7a47c3c72d69CAS |
Hristov AN, Oh J, Firkins JL, Dijkstra J, Kebreab E, Waghorn G, Makkar HPS, Adesogan AT, Yang W, Lee C, Gerber PJ, Henderson B, Tricarico JM (2013) Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options. Journal of Animal Science 91, 5045–5069.
| Mitigation of methane and nitrous oxide emissions from animal operations: I. A review of enteric methane mitigation options.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhslKktrrL&md5=0409274c066031647c58b4a732d79b18CAS |
Hume ID (1999) ‘Marsupial nutrition.’ (Cambridge University Press: Cambridge, UK)
Hume ID (2002) Digestive strategies of mammals. Dong Wu Xue Bao 48, 1–19.
Hungate RE (1966) ‘The rumen and its microbes.’ (Academic Press: New York) Available at http://www.sciencedirect.com/science/book/9781483233086 [Verified July 2017]
Huws SA, Mayorga OL, Theodorou MK, Onime LA, Kim EJ, Cookson AH, Newbold CJ, Kingston-Smith AH (2013) Successional colonization of perennial ryegrass by rumen bacteria. Letters in Applied Microbiology 56, 186–196.
| Successional colonization of perennial ryegrass by rumen bacteria.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXjtVakur8%3D&md5=bca1e5dfc91c0870dc77fcec8a2e3caeCAS |
Kandler O, König H (1998) Cell wall polymers in Archaea (Archaea bacteria). Cellular and Molecular Life Sciences 54, 305–308.
| Cell wall polymers in Archaea (Archaea bacteria).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXis1emsbo%3D&md5=66c2c4af26eedebab44bca72eff1a139CAS |
Kempton TJ, Murray RM, Leng RA (1976) Methane production and digestibility measurements in grey macropods and sheep. Australian Journal of Biological Sciences 29, 209–214.
| Methane production and digestibility measurements in grey macropods and sheep.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE28XkvFaltbk%3D&md5=373b4db28eafa17999062d025f3fe1eaCAS |
Kim JJ, Khan WI (2013) Goblet cells and mucins: role in innate defense in enteric infections. pathogens 2, 55–70.
| Goblet cells and mucins: role in innate defense in enteric infections.Crossref | GoogleScholarGoogle Scholar |
Klieve AV, Ouwekerk D, Maquire AJ (2012) Archaea in the foregut of macropod marsupials: PCR and amplicon sequence-based observations. Journal of Applied Microbiology 113, 1065–1075.
| Archaea in the foregut of macropod marsupials: PCR and amplicon sequence-based observations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsFWisrvO&md5=6f978d074c22a88445188759e66162a4CAS |
Koropatkin NM, Cameron EA, Martens EC (2012) How glycan metabolism shapes the human gut microbiota. Nature Reviews. Microbiology 10, 323–335.
| How glycan metabolism shapes the human gut microbiota.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XlsVSjtro%3D&md5=0bec5be517462ce7110f9856816c0f46CAS |
Krebs GL (1987) The rumen ecosystem: kinetics of microbial pools and effects on microbial protein yield. PhD Thesis, University of New England, Armidale, NSW.
Langer P (1984) Comparative anatomy of the stomach in mammalian herbivores. Quarterly Journal of Experimental Physiology 69, 615–625.
| Comparative anatomy of the stomach in mammalian herbivores.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaL2c3pvV2isQ%3D%3D&md5=8b206a678df95cd656e18813cc0a5c59CAS |
Langer P, Dellow DW, Hume ID (1980) Stomach structure and function in three species of Macropodine Marsupials. Australian Journal of Zoology 28, 1–18.
| Stomach structure and function in three species of Macropodine Marsupials.Crossref | GoogleScholarGoogle Scholar |
Lee C, Beauchemin KA (2014) A review of feeding supplementary nitrate to ruminant animals: nitrate toxicity, methane emissions, and production performance. Canadian Journal of Animal Science 94, 557–570.
| A review of feeding supplementary nitrate to ruminant animals: nitrate toxicity, methane emissions, and production performance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2MXhsFejurzO&md5=5a2fe135710af33d5ae8b72cebb4609dCAS |
Leng RA (2011) The rumen: a fermentation vat or a series of organized structured microbial consortia: implications for the mitigation of enteric methane production by feed additives. Livestock Research for Rural Development 23, 258
Leng RA (2014) Interactions between microbial consortia in biofilms: a paradigm shift in rumen microbial ecology and enteric methane mitigation. Animal Production Science 54, 519–543.
| Interactions between microbial consortia in biofilms: a paradigm shift in rumen microbial ecology and enteric methane mitigation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXls1ehtrw%3D&md5=a02509f02e08e1398d4e48b68d560235CAS |
Leng RA, Leonard GJ (1965) Loss of methyl tritium from [3H] acetate in rumen fluid. Nature 207, 760–761.
| Loss of methyl tritium from [3H] acetate in rumen fluid.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaF2MXks1Gmsr4%3D&md5=f27d0625f7c77d2497bf93f1173f9f94CAS |
Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI (2008) Worlds within worlds: evolution of the vertebrate gut microbiota. Nature Reviews. Microbiology 6, 776–788.
| Worlds within worlds: evolution of the vertebrate gut microbiota.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtFWitLrN&md5=7ec56dec6b0b2b412f3d05f25fd11702CAS |
Lipscomb MF, Masten BJ (2002) Dendritic cells: immune regulators in health and disease. Physiological Reviews 82, 97–130.
| Dendritic cells: immune regulators in health and disease.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XovFSisA%3D%3D&md5=ff0d1f0a67703eb1fd68d743f0978842CAS |
Lovley DR (1985) Minimum threshold for hydrogen metabolism in methanogenic bacteria. Applied and Environmental Microbiology 49, 1530–1531.
Lovley DR, Godwin S (1988) Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments. Geochimica et Cosmochimica Acta 52, 2993–3003.
| Hydrogen concentrations as an indicator of the predominant terminal electron-accepting reactions in aquatic sediments.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL1MXot1SltQ%3D%3D&md5=dee53cc3bc7033c1cbcf32064be503e4CAS |
Madsen J, Bertelsen MF (2012) Methane production by red-necked wallabies (Macropus rufogriseus). Journal of Animal Science 90, 1364–1370.
| Methane production by red-necked wallabies (Macropus rufogriseus).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XkvFanu7w%3D&md5=83c3335f31abba1ddcc9266c21637651CAS |
Matzinger P (1994) Tolerance, danger, and the extended family. Annual Review of Immunology 12, 991–1045.
| Tolerance, danger, and the extended family.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2c3otVOqsA%3D%3D&md5=63953959ba67763d7cc7c94dad0ebaedCAS |
Matzinger P (2012) The evolution of the danger theory. Expert Review of Clinical Immunology 8, 311–317.
| The evolution of the danger theory.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38Xnt1yqsrc%3D&md5=797cca84f3477bc59e924624d4b869b4CAS |
Mayorga OL, Huws SA, Kim EJ, Kingston-Smith AH, Newbold CJ, Theodorou MK (2007) Biofilm formation by rumen microbes on fresh perennial ryegrasss under in vitro anaerobic conditions. In ‘UK Molecular Microbial Ecology Group’, 13th meeting, 16–17 July 2007, University of Liverpool, UK.
McAllister TA, Cheng KJ (1996) Microbial strategies in the ruminal digestion of cereal grains. Animal Feed Science and Technology 62, 29–36.
| Microbial strategies in the ruminal digestion of cereal grains.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28Xns1Sht7o%3D&md5=a1534d2d6c254e30f77fb434803c7f1dCAS |
McAllister TA, Bae HD, Jones GA, Cheng KJ (1994) Microbial attachment and feed digestion in the rumen. Journal of Animal Science 72, 3004–3018.
| Microbial attachment and feed digestion in the rumen.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXhvFGjtrs%3D&md5=80e7bffb6509b2a74952d8faa390e4ddCAS |
McInerney MJ, Sieber JR, Gunsalus RP (2009) Syntrophy in anaerobic global carbon cycles. Current Opinion in Biotechnology 20, 623–632.
| Syntrophy in anaerobic global carbon cycles.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhsV2ks7%2FJ&md5=d37d2f9fbe5e32add0e8e5b052434242CAS |
Mitsumori M, Shinkai T, Takenaka A, Enishi O, Higuchi K, Kobayashi Y, Nonaka I, Asanuma N, Denman SE, McSweeney CS (2012) Responses in digestion, rumen fermentation and microbial populations to inhibition of methane formation by a halogenated methane analogue. British Journal of Nutrition 108, 482–491.
| Responses in digestion, rumen fermentation and microbial populations to inhibition of methane formation by a halogenated methane analogue.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhtFClu7fK&md5=68c3e4af643ee05fce05d689165c25e2CAS |
Morvan B, Dore J, Rieu-Lesme F, Foucat L, Fonty G, Gouet P (1994) Establishment of hydrogen-utilizing bacteria in the rumen of the newborn lamb. FEMS Microbiology Letters 117, 249–256.
| Establishment of hydrogen-utilizing bacteria in the rumen of the newborn lamb.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2cXis1Kmu7s%3D&md5=36b28b1f70dc44cfa6ac50084dd45848CAS |
Munn AJ, Tomlinson S, Savage T, Clauss M (2012) Retention of different-sized particles and derived gut fill estimate in tammar wallabies (Macropus eugenii): physiological and methodological considerations. Comparative Biochemistry and Physiology. Part A, Molecular & Integrative Physiology 161, 243–249.
| Retention of different-sized particles and derived gut fill estimate in tammar wallabies (Macropus eugenii): physiological and methodological considerations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhs1CqsbbO&md5=7d379018e0a20c9f01635b2c20a95a19CAS |
Ostaff MJ, Stange EF, Wehkamp J (2013) Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Molecular Medicine 5, 1465–1483.
| Antimicrobial peptides and gut microbiota in homeostasis and pathology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXhsFOgsrjE&md5=37d025a2823fe1d35b4270d0925a1f6cCAS |
Ouwerkerk D, Maguire AJ, McMillen L, Klieve AV (2009) Hydrogen utilising bacteria from the forestomach of eastern grey (Macropus giganteus) and red (Macropus rufus) macropods. Animal Production Science 49, 1043–1051.
| Hydrogen utilising bacteria from the forestomach of eastern grey (Macropus giganteus) and red (Macropus rufus) macropods.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXht1Gmtb%2FK&md5=c6eca3f0a823802b403c3ba19ea6fd2aCAS |
Peel E, Jones E, Belov K, Cheng Y (2013) Protection in the pouch: AMPs in marsupials and monotremes). In ‘Microbial pathogens and strategies for combating them’. (Ed. A Méndez-Vilas) pp. 1247–1256. Formatex. Available at http://www.formatex.info/microbiology4/vol2/1247-1256.pdf [Verified July 2017]
Pinder RS, Patterson JA (2012) Glucose and hydrogen utilization by an acetogenic bacterium isolated from ruminal contents. Agricultural Food and Analytical Bacteriology 2, 253–274.
Pope PB, Totsika M, Aguirre de Carcer D, Schembri MA, Morrison M (2011) Muramidases found in the foregut microbiome of the Tammar wallaby can direct cell aggregation and biofilm formation. The ISME Journal 5, 341–350.
| Muramidases found in the foregut microbiome of the Tammar wallaby can direct cell aggregation and biofilm formation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXmsVGjtQ%3D%3D&md5=15e6ca5d7b0dd8c8995ccc0798bd16d8CAS |
Pradeu T, Cooper EL (2012) The danger theory: 20 years later. Frontiers in Immunology 3, 287
| The danger theory: 20 years later.Crossref | GoogleScholarGoogle Scholar |
Ragsdale SW, Pierce E (2008) Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation. Biochimica et Biophysica Acta 1784, 1873–1898.
| Acetogenesis and the Wood–Ljungdahl pathway of CO2 fixation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtlaqtb7O&md5=f00168a34586ce39f2c7157d1786f241CAS |
Richardson KC (1980) The structure and radiographic anatomy of the alimentary tract of the tammar wallaby, Macropuseugenii (Marsupialia). I. The stomach. Australian Journal of Zoology 28, 367–379.
| The structure and radiographic anatomy of the alimentary tract of the tammar wallaby, Macropuseugenii (Marsupialia). I. The stomach.Crossref | GoogleScholarGoogle Scholar |
Rodríguez CA, González J, Alvir MR, Redondo R, Cajarville C (2003) Effects of feed intake on composition of sheep rumen contents and their microbial population size. British Journal of Nutrition 89, 97–103.
| Effects of feed intake on composition of sheep rumen contents and their microbial population size.Crossref | GoogleScholarGoogle Scholar |
Sansonetti PJ (2011) To be or not to be a pathogen: that is the mucosally relevant question. Mucosal Immunology 4, 8–14.
| To be or not to be a pathogen: that is the mucosally relevant question.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhs1ejtb3E&md5=b49f556681c7fe92db8ff3322294ba80CAS |
Schink B (1997) Energetics of syntrophic cooperation in methanogenic degradation. Microbiology and Molecular Biology Reviews 61, 262–280.
Schluter J, Foster KR (2012) The evolution of mutualism in gut microbiota via host epithelial selection. PLoS Biology 10, e1001424
| The evolution of mutualism in gut microbiota via host epithelial selection.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhvVWnu73L&md5=242c0c3a18857686843bc13c3b7f04b7CAS |
Schroeder BO, Wu Z, Nuding S, Groscurth S, Marcinowski M, Beisner J, Buchner J, Schaller M, Stange EF, Wehkamp J (2011) Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1. Nature 469, 419–423.
| Reduction of disulphide bonds unmasks potent antimicrobial activity of human beta-defensin 1.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXnvVKksg%3D%3D&md5=0c67011ee3b175074bfebef6fbcc26a4CAS |
Shoeib MB, Hassanin A, Elnasharty M (2015) Morphological and morphometric characteristics of gastric mucosa in western grey macropods (Macropus fuliginosus). Journal of Advanced Veterinary and Animal Research 2, 40–48.
| Morphological and morphometric characteristics of gastric mucosa in western grey macropods (Macropus fuliginosus).Crossref | GoogleScholarGoogle Scholar |
Smith HF, Fisher RE, Everett MI, Thomas AD, Bollinger RR, Parker W (2009) Comparative anatomy and phylogenetic distribution of the mammalian cecal appendix. Journal of Evolutionary Biology 22, 1984–1999.
| Comparative anatomy and phylogenetic distribution of the mammalian cecal appendix.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DC%2BD1MnjslehtA%3D%3D&md5=4ab8258b9da2b489accfe3d9ac006a7aCAS |
Sommer F, Bäckhed F (2013) The gut microbiota-masters of host development and physiology. Nature Reviews. Microbiology 11, 227–238.
| The gut microbiota-masters of host development and physiology.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXivFOrtbY%3D&md5=4ca3a813a7d00ae8ece299c501b5a9a7CAS |
Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D (2011) CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity 34, 122–134.
| CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1Wlu7w%3D&md5=a285d6e14f1b2fb00adbc235765059b2CAS |
Stoodley P, Sauer K, Davies DG, Costerton JW (2002) Biofilms as complex differentiated communities. Annual Review of Microbiology 56, 187–209.
| Biofilms as complex differentiated communities.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xos1Gis7o%3D&md5=a7aec4230331d8666335c204c6023ee5CAS |
Tyndale-Biscoe H (2005) ‘Life of marsupials.’ (CSIRO Publishing: Melbourne)
von Engelhardt W, Wolter S, Lawrenz H, Hemsley JA (1978) Production of methane in two non-ruminant herbivores. Comparative Biochemistry and Physiology. Part A. Physiology 60, 309–311.
| Production of methane in two non-ruminant herbivores.Crossref | GoogleScholarGoogle Scholar |
Walker JA, Barlow JL, McKenzie ANJ (2013) Innate lymphoid cells: how did we miss them? Nature Reviews. Immunology 13, 75–87.
| Innate lymphoid cells: how did we miss them?Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXjslGjsQ%3D%3D&md5=1bb0f86eead58b22ea7ecbf302a2faa0CAS |
Wang ZW, Chen S (2009) Potential of biofilm-based biofuel production. Applied and Environmental Microbiology 83, 1–18.
Wang J, Wong ESW, Whitely JC, Li J, Stringer JM, Short KR, Renfree MB, Belov K, Cocks B (2011) Ancient AMPS kill antibiotic-resistant pathogens: Australian mammals provide new options. PLoS One 6, e24030
| Ancient AMPS kill antibiotic-resistant pathogens: Australian mammals provide new options.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXht1Wrt77N&md5=a5100031472b7329b9413b2efd48aee5CAS |
Weimer PJ, Russell JB, Muck RE (2009) Lessons from the cow: what the ruminant animal can teach us about consolidated bioprocessing of cellulosic biomass. Bioresource Technology 100, 5323–5331.
| Lessons from the cow: what the ruminant animal can teach us about consolidated bioprocessing of cellulosic biomass.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXoslGgsrY%3D&md5=712091419e50d293fcaaf57d987365cbCAS |
Williams AG (2007) Rumen holotrich ciliate protozoa. Microbiological Reviews 50, 25–49.
Zinder SH, Koch M (1984) Non-aceticlastic methanogenesis from acetate: acetate oxidation by a thermophilic syntrophic coculture. Archives of Microbiology 138, 263–272.
| Non-aceticlastic methanogenesis from acetate: acetate oxidation by a thermophilic syntrophic coculture.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL2cXkvVOiu70%3D&md5=6d6eadee168d6f284ab7f37cafe28753CAS |

Ronald Alfred Leng is Professor Emeritus of Nutritional Biochemistry at the University of New England (UNE). Ron studied Agricultural Chemistry at the University of Nottingham before coming to Australia in 1959 to pursue a PhD in the Faculty of Rural Science at UNE. On successful completion of his PhD he was appointed Lecturer in Nutrition in 1963 and was progressively promoted over the next 10 years to a Personal Chair in Nutritional Biochemistry. He was awarded a DRurSc in 1972. Ron was made an Officer of the Order of Australia in 1991 for his contribution to the development of systems of using poor quality feeds for ruminant meat and milk production in Australia and in developing countries. Ron has been a Distinguished Visiting Professor at Iowa State University (USA) and Nihon University (Japan). The Australian Society of Animal Production made him a Fellow in 1996. In 2002, he received the Han Award from the Asian-Australasian Association of Animal Production Societies in recognition of his outstanding contribution to animal production of international significance. He has been a consultant to the governments of more than 40 countries through United Nations Development Programs. He retired from the University to undertake a private consultancy and to continue his research on a global scale. Ron has published in excess of 500 papers in peer-reviewed journals and has published eight books in areas related to animal production science. He has a Hirsch index of 56 and is ranked by Google citations profile in the top 1000 scientists in Australian institutions.