Genomic data show little geographical structure across the naturally fragmented range of the purple-gaped honeyeater
Leo Joseph A C , Catriona D. Campbell A , Lynn Pedler B and Alex Drew A
A C , Catriona D. Campbell A , Lynn Pedler B and Alex Drew A
A Australian National Wildlife Collection, CSIRO National Research Collections Australia, GPO Box 1700, Canberra, ACT 2601, Australia.
B PO Box 58, Koolunga, SA 5464, Australia.
C Corresponding author. Email: leo.joseph@csiro.au
Australian Journal of Zoology 67(4) 226-230 https://doi.org/10.1071/ZO20074
Submitted: 13 August 2020 Accepted: 11 December 2020 Published: 23 December 2020
Journal compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
Using single nucleotide polymorphisms and mitochondrial DNA sequences we find some evidence of genetic structure within a widespread and naturally fragmented species, the purple-gaped honeyeater (Lichenostomus cratitius), of southern Australian mallee shrublands. The very earliest stages of differentiation either side of the Nullarbor Barrier may already have been arrested by gene flow, some of which may have been anthropogenically induced.
Keywords: purple-gaped honeyeater, honeyeater, phylogeography, Nullarbor Barrier, Eyrean Barrier, southern Australia, Lichenostomus.
Introduction
Interest in the roles of biogeographical barriers in speciation and differentiation arises from several sources. These include whether present-day distributions of species and recognised subspecies correlate with and can be explained by biogeographical barriers. One approach to address these interests is to test for molecular phylogeographic structure within a species and link it to barriers. These approaches underpin the ever-burgeoning phylogeographic literature on the evolution of terrestrial and marine species. Examples from across southern Australia, this paper’s area of interest, are typical.
Two biogeographical barriers of interest in many southern Australian phylogeographic studies are the Nullarbor and Eyrean Barriers (Fig. 1). They have affected different species at different times (Crisp and Cook 2007) and according to different ecologies (e.g. diet: granivory, insectivory, nectarivory; habitat: mesic or xeric: Dolman and Joseph 2012). Some species with geographical ranges naturally fragmented by these two barriers have no subspecies and no discernible phylogeographic structure (e.g. tawny-crowned honeyeater, Gliciphila melanops: Dolman and Joseph 2012). Others are more continuously distributed across all of southern Australia. They show contrasting taxonomic and phylogeographic patterns, indicating a range of temporal and spatial effects of these two barriers (cf. Alpers et al. 2016 in wombats; Donnellan et al. 2009 in birds). More examples from the growing body of literature relevant to these questions in southern Australia now include work on reptiles (Edwards et al. 2015; Ansari et al. 2019), mammals (Cooper et al. 2000, 2018; Pestell et al. 2008; Neaves et al. 2012), fish (Donnellan et al. 2015), invertebrates (Pons et al. 2006; Rix et al. 2017) and plants (Crisp and Cook 2007); birds have been especially studied (Joseph and Wilke 2006; Dolman and Joseph 2015; McElroy et al. 2020; Norman et al. 2014).
Here we contribute a phylogeographic study of another southern Australian bird species, the purple-gaped honeyeater (Lichenostomus cratitius) (Fig. 1). The species occurs in semiarid habitats across southern Australia, specifically mallee woodlands, tall heath and associated low eucalypt woodlands (Menkhorst et al. 2017). Its range is naturally fragmented by the Nullarbor and Eyrean Barriers. The Kangaroo Island population (Fig. 1) is isolated by sea. The species is almost phenotypically invariant. Two weakly differentiated subspecies are currently recognised (Schodde and Mason 1999): L. c. cratitius on Kangaroo Island and L. c. occidentalis for all other populations. L. c. occidentalis is thus isolated into three groups of populations by the Nullarbor and Eyrean Barriers. Isolation by the Nullarbor Barrier may well have been partial. Pockets of suitable habitat do occur along otherwise unsuitable coastal cliffs where the Nullarbor Plain reaches the sea. Their connectivity has likely been augmented by anthropogenically provided habitats such as gardens around Eucla (Fig. 1). Here, we ask whether there is cryptic genetic differentiation in L. cratitius and, if so, can it be related to the Nullarbor and Eyrean Barriers, or indeed a sea barrier, and to currently recognised subspecies? To address these questions, we have gathered mitochondrial DNA (mtDNA) and single nucleotide polymorphism (SNP) data from the nuclear genome.
Methods
Study design, specimens
We wished to sample the purple-gaped honeyeater (Lichenostomus cratitius) evenly across its range. Fig. 1 and Table S1 (Supplementary Material) show locations and other details of the specimens studied. We lacked samples from what may be an isolated population at the species’ easternmost extremity well east of the Eyrean Barrier (Fig. 1) and east of the Bassian Volcanic Barrier (sensu Schodde and Mason 1999). That population aside, the largest gap from which no tissue samples were available before our study was ~1050 km, comprising 710 and 340 km west and east, respectively of Eucla, Western Australia (Fig. 1). We collected 16 specimens from the former region west of Eucla through field work in 2017. We were unable to obtain any further samples from east of Eucla. Nonetheless, we are confident that Fig. 1 and Table S1 show reasonably even sampling of the species across its range.
Genotype-by-sequencing
DNA was extracted from liver samples of L. cratitius (n = 34) and its sister species, the yellow-tufted honeyeater (L. melanops) (n = 3) (Nyári and Joseph 2011; Marki et al. 2017; specimens listed in Table S1) using the Qiagen Puregene® Tissue Kit following the manufacturer’s protocols. DNA was provided to Diversity Arrays Technology (DArT) with a concentration of 30 ng µL–1. The DArTseq™ genotype-by-sequencing approach uses a combination of DArT complexity reduction and high-throughput sequencing (Kilian et al. 2012; Courtois et al. 2013; Cruz et al. 2013) to simultaneously identify and genotype SNP markers in the absence of a reference genome. A combination of two enzymes were used for complexity reduction (PstI–HpaII), before addition of custom adapters (Kilian et al. 2012) to restriction site overhangs. Fragments were amplified using primers complementary to the adapters. These also incorporated molecular identifier barcode tags, to allow multiplexing of up to 96 samples per sequencing run. PCR conditions consisted of denaturation at 94°C for 1 min; 30 cycles of 94°C for 20 s, 58°C for 30 s and 72°C for 45 s; and a final extension period of 72C for 7 min. PCR products were pooled for sequencing on an Illumina HiSeq 2000 platform using 77 cycles of single end sequencing. Raw sequence reads were filtered and processed using a proprietary DArT analytical pipeline. Poor quality sequences were removed, with more stringent criteria being placed upon the barcode region than the rest of the sequence. Approximately 2 000 000 sequences per barcode were identified and used in marker calling, during which identical sequences were collapsed and filtered before screening to identify variable markers using DArT proprietary SNP and SilicoDArT algorithms (DArTsoft14).
The R package dartR (Gruber et al. 2018) was used for population genomic analyses of the DArTseq SNP data, including filtering data and principal coordinates analysis (PCoA). SNPs were filtered using the callrate function (99.5%) and the RepAvg function (1). No monomorphic loci and no duplicate loci per sequence tag were identified. The proportion of genetic variation that is attributable to between-population differentiation, FST, was calculated using the R package StAMPP (Pembleton et al. 2013).
Mitochondrial DNA
We obtained mtDNA sequences for the ND2 gene of mitochondrial DNA from the 31 L. cratitius (including most of those analysed for SNP data) and three L. melanops samples. We used protocols described in Dolman and Joseph (2015) for DNA extraction and sequencing. Forward and reverse raw sequences were aligned in Geneious 10.2 (Biomatters) and subsequently checked and edited manually. Consensus sequences for all individuals including those of the outgroup L. melanops were aligned in MUSCLE (Edgar 2004). Gamma distribution, percentage of invariant sites and model of sequence evolution were chosen using AIC calculations in MEGA 7.0 (Kumar et al. 2018). Forward and reverse raw sequences were aligned in Geneious 10.2 (Biomatters) and subsequently checked and edited manually. The consensus sequence for all individuals were aligned in MUSCLE (Edgar 2004). The Tamura–Nei (TN93+G) (Tamura and Nei 1993) model was chosen using AIC calculations in MEGA X 10.0.5 (Kumar et al. 2018). Phylogenetic trees were prepared using Garli 2.01 (Zwickl 2006) and visualised in SumTrees (Sukumaran and Holder 2015).
Nodal support under maximum likelihood was assessed with 100 bootstrap pseudoreplicates. We used DnaSP 6.11.01 (Rozas et al. 2017) to calculate Da (net nucleotide divergence), which corrects for shared diversity among the individuals within groups being compared.
Results
SNP data
We generated 170 551 SNPs. After filtering to remove loci with missing data, we retained 40 437 SNPs for further analyses (data lodged at doi: 10.6084/m9.figshare.12798446). FST between L. cratitius and L. melanops were high, as expected (0.49–0.51). Within the focal species, L. cratitius, FST was low, at 0.01. A PCoA plot including both species (Fig. 1a) shows that 19.8% of the variation in the data was explained by the first two axes of variation, PCo1 and PCo2, most of this reflecting the separation, as expected, between the species L. cratitius and L. melanops. Still with L. melanops included, very narrow but sharp separation of L. cratitius samples east and west of the Nullarbor Barrier was suggested on PCo1 (Fig. 1a). This particular effect disappeared when the analysis was confined to L. cratitius (Fig. 1b) and a much smaller proportion of the variation within the species, 9.6%, was explained by PCo1 (5.2%) and PCo2 (4.4%). The only structure then apparent on PCo1 was that of all three Kangaroo Island samples being apart from all others. PCo2 similarly showed almost all samples clustered together and mostly with no geographical structure. Two pairs of samples, however – a pair from Eyre Peninsula and a pair from south of Madura at the western edge of the Nullarbor Plain – were each well separated on PCo2. Other samples from both localities fell in the main cluster. Further filtering the data to remove loci where rare alleles have frequencies less than 0.25 yielded essentially identical results (not shown).
MtDNA
Thirty-two ND2 sequences newly acquired in this study have been lodged in GenBank (accession numbers MW310592–MW310623). Of these, 30 were from L. cratitius and three were from L. melanops. Net divergence, Da, between the two species L. cratitius and L. melanops was substantial at 3.31%. Within L. cratitius itself nucleotide diversity was low at 0.82%. So too, net divergence, Da, between eastern and western groups was low at 0.06%. Da between Kangaroo Island and all mainland samples was an order of magnitude higher but still low at 0.2%. Phylogenetic analysis showed two haplogroups, each comprising samples from across the range (Fig. 2). Only one of these haplogroups was strongly supported. It comprised five and two samples from the eastern and western parts of the species’ range, respectively. It was robustly supported by 99% bootstrap support. The other haplogroup was weakly supported at 79%. Only one specimen, ANWC B46696 from near Cummins, Eyre Peninsula, was in both this mtDNA clade and among weakly differentiated samples on PCo2 of the SNP analysis (Fig. 1b).
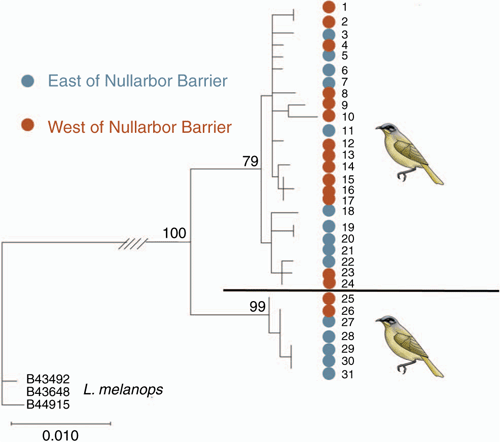
|
Discussion
We asked whether the Nullarbor and Eyrean Barriers and a sea barrier have generated genetic differentiation among populations of the purple-gaped honeyeater (Lichenostomus cratitius). We were interested in linking any structure to the species’ very low phenotypic diversity. Based on single nucleotide polymorphism (SNP) and mitochondrial DNA sequences, we conclude that the species has very low genetic diversity and appears almost panmictic. The most strongly supported genetic structure in the SNP data parallels the currently recognised subspecies, L. c. cratitius (Kangaroo Island) and L. c. occidentalis (mainland). Two aspects of our results warrant further comment.
First, closer inspection of the PCoA analysis of SNP data including both species L. melanops and L. cratitius (Fig. 1a) suggested a pattern arguably consistent with early stages of differentiation in the latter. Importantly, this pattern disappeared in the analysis using only data for L. cratitius, the species of interest (Fig. 1b). Still at this geographical scale, robust support for one mtDNA haplogroup is notable (Fig. 2). That haplogroup comprises seven samples (samples 25–31: Table S1), of which five (samples 27–31) are from east of the Eyrean Barrier. The other two samples (samples 25 and 26) are from west of the Nullarbor Barrier but they are among the easternmost such samples. If the Nullarbor Barrier initiated differentiation in mtDNA in this species, that differentiation has probably long since been arrested by gene flow. We note here that the species occurs in domestic gardens at Eucla (Fig. 1; authors’ obs.). Eucla is itself located in what is otherwise a gap in the species’ range (Atlas of Living Australia, http://www.ala.org.au, accessed 4 August 2020). This would at least facilitate recent bidirectional gene flow across the Nullarbor Barrier. Second, the differentiation of the Kangaroo Island population should be interpreted with care. That is, genetic drift in an island population may explain this as well as, if not better than, longer term biogeographical separation implied by treating the population as a separate subspecies.
We conclude that some differentiation may have occurred historically during more arid glacial cycles, which presumably fragmented the distribution across the Nullarbor Barrier. We infer that this has since been largely obliterated, however, by gene flow. Gene flow may also explain the diversity within L. cratitius at 0.82%. It is approximately equal to or potentially lower than what might be expected, given that L. cratitius and L. melanops diverged from a common ancestor ~2.5 million years ago (see Joseph et al. 2014; Marki et al. 2017). The resulting genetic patterns nonetheless correlate with currently recognised subspecies in L. cratitius. Overall, this is very similar to patterns seen in the western pygmy possum (Cercartetus concinnus), the distribution and habitat of which closely mirror those of L. cratitius (Pestell et al. 2008). No effect of the Eyrean Barrier is evident, unlike in many other bird species, some of which have even less of a contemporary geographical separation across that barrier than L. cratitius (examples cited in Introduction). If phenotypic variation recognised by the currently recognised subspecies is itself valid, then it may also include an effect of drift in the Kangaroo Island population. The species seems poised at an interesting stage in its evolutionary history.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We are very grateful for the enormously helpful critiques of reviewers and the editor. Jo Sumner (Museum Victoria) kindly arranged a tissue loan of the westernmost sample of L. cratitius currently available from South Australia. Specimens we collected in 2017 were collected under Permit 08-000827-1 from the Department of Parks and Wildlife, Western Australia, and CSIRO Animal Ethics Permits 2015–16. Mark Robbins helped with field work. Kevin Winker helpfully commented on an early draft. Julian Teh kindly prepared the figures. This research did not receive any specific funding.
References
Alpers, D. L., Walker, F. M., Taylor, A. C., Sunnucks, P., Bellman, S., Hansen, B. D., and Sherwin, W. B. (2016). Evidence of subdivisions on evolutionary timescales in a large, declining marsupial distributed across a phylogeographic barrier. PLoS One 11, e0162789.| Evidence of subdivisions on evolutionary timescales in a large, declining marsupial distributed across a phylogeographic barrier.Crossref | GoogleScholarGoogle Scholar | 27732594PubMed |
Ansari, M. H., Cooper, S. J. B., Schwarz, M. P., Ebrahimie, M., Dolman, G., Reinberger, L., Saint, K. M., Donnellan, S. C., Bull, C. M., and Gardner, M. G. (2019). Plio-Pleistocene diversification and biogeographic barriers in southern Australia reflected in the phylogeography of a widespread and common lizard species. Molecular Phylogenetics and Evolution 133, 107–119.
| Plio-Pleistocene diversification and biogeographic barriers in southern Australia reflected in the phylogeography of a widespread and common lizard species.Crossref | GoogleScholarGoogle Scholar | 30553880PubMed |
Cooper, S. J. B., Adams, M., and Labrinidis, A. (2000). Phylogeography of the Australian dunnart Sminthopsis crassicaudata (Marsupialia: Dasyuridae). Australian Journal of Zoology 48, 461–473.
| Phylogeography of the Australian dunnart Sminthopsis crassicaudata (Marsupialia: Dasyuridae).Crossref | GoogleScholarGoogle Scholar |
Cooper, S. J. B., Ottewell, K., MacDonald, A. J., Adams, M., Byrne, M., Carthew, S. M., Eldridge, M. D. B., Li, Y., Pope, L. C., Saint, K. M., and Westerman, M. (2018). Phylogeography of southern brown and golden bandicoots: implications for the taxonomy and distribution of endangered subspecies and species. Australian Journal of Zoology 66, 379–393.
| Phylogeography of southern brown and golden bandicoots: implications for the taxonomy and distribution of endangered subspecies and species.Crossref | GoogleScholarGoogle Scholar |
Courtois, B., Audebert, A., Dardou, A., Roques, S., Ghneim-Herrera, T, Droc, G, Frouin, J, Rouan, L, Gozé, E, Kilian, A, Ahmadi, N, and Dingkuhn, M (2013). Genome-wide association mapping of root traits in a japonica rice panel. PLoS One 8, e78037.
| Genome-wide association mapping of root traits in a japonica rice panel.Crossref | GoogleScholarGoogle Scholar | 24223758PubMed |
Crisp, M. D., and Cook, L. G. (2007). A congruent molecular signature of vicariance across multiple plant lineages. Molecular Phylogenetics and Evolution 43, 1106–1117.
| A congruent molecular signature of vicariance across multiple plant lineages.Crossref | GoogleScholarGoogle Scholar | 17434758PubMed |
Cruz, V. M. V., Kilian, A., and Dierig, D. A. (2013). Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop lesquerella and related species. PLoS One 8, e64062.
| Development of DArT marker platforms and genetic diversity assessment of the US collection of the new oilseed crop lesquerella and related species.Crossref | GoogleScholarGoogle Scholar |
Dolman, G., and Joseph, L. (2012). A species assemblage approach to comparative phylogeography of birds in southern Australia. Ecology and Evolution 2, 354–369.
| A species assemblage approach to comparative phylogeography of birds in southern Australia.Crossref | GoogleScholarGoogle Scholar | 22423329PubMed |
Dolman, G., and Joseph, L. (2015). Evolutionary history of birds across southern Australia: structure, history and taxonomic implications of mitochondrial DNA diversity in an ecologically diverse suite of species. Emu 115, 35–48.
| Evolutionary history of birds across southern Australia: structure, history and taxonomic implications of mitochondrial DNA diversity in an ecologically diverse suite of species.Crossref | GoogleScholarGoogle Scholar |
Donnellan, S. C., Armstrong, J., Pickett, M., Milne, T., Baulderstone, J., Holfelder, T., and Bertozzi, T. (2009). Systematic and conservation implications of mitochondrial DNA diversity in emu-wrens, Stipiturus (Aves: Maluridae). Emu 109, 143–152.
| Systematic and conservation implications of mitochondrial DNA diversity in emu-wrens, Stipiturus (Aves: Maluridae).Crossref | GoogleScholarGoogle Scholar |
Donnellan, S. C., Foster, R., Junge, C., Huveneers, C., Rogers, P., Kilian, A., and Bertozzi, T. (2015). Fiddling with the proof: the magpie fiddler ray is a colour pattern variant of the common southern fiddler ray (Rhinobatidae: Trygonorrhina). Zootaxa 3981, 367–384.
| Fiddling with the proof: the magpie fiddler ray is a colour pattern variant of the common southern fiddler ray (Rhinobatidae: Trygonorrhina).Crossref | GoogleScholarGoogle Scholar | 26250000PubMed |
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797.
| MUSCLE: multiple sequence alignment with high accuracy and high throughput.Crossref | GoogleScholarGoogle Scholar | 15034147PubMed |
Edwards, D. L., Melville, J., Joseph, L., and Keogh, J. S. (2015). Ecological divergence, adaptive radiation and the evolution of sexual signaling traits in a complex of Australian agamid lizards. American Naturalist 186, E144–E161.
| Ecological divergence, adaptive radiation and the evolution of sexual signaling traits in a complex of Australian agamid lizards.Crossref | GoogleScholarGoogle Scholar |
Gruber, B., Unmack, P. J., Berry, O. F., and Georges, A. (2018). dartR: an R package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Molecular Ecology Resources 18, 691–699.
| dartR: an R package to facilitate analysis of SNP data generated from reduced representation genome sequencing.Crossref | GoogleScholarGoogle Scholar | 29266847PubMed |
Joseph, L., and Wilke, T. (2006). Molecular resolution of population history, systematics and historical biogeography of the Australian ringneck parrots Barnardius: are we there yet? Emu 106, 49–62.
| Molecular resolution of population history, systematics and historical biogeography of the Australian ringneck parrots Barnardius: are we there yet?Crossref | GoogleScholarGoogle Scholar |
Joseph, L., Toon, A., Nyári, Á. S., Longmore, N. W., Rowe, K. M. C., Haryoko, T., Trueman, J., and Gardner, J. (2014). A new synthesis of the molecular systematics and biogeography of honeyeaters (Passeriformes: Meliphagidae) highlights biogeographical complexity of a spectacular avian radiation. Zoologica Scripta 43, 235–248.
| A new synthesis of the molecular systematics and biogeography of honeyeaters (Passeriformes: Meliphagidae) highlights biogeographical complexity of a spectacular avian radiation.Crossref | GoogleScholarGoogle Scholar |
Kilian, A., Wenzl, P., Huttner, E., Carling, J., Xia, L., Blois, H., Caig, V., Heller-Uszynska, K., Jaccoud, D., Hopper, C., Aschenbrenner-Kilian, M., Evers, M., Peng, K., Cayla, C., Hok, P., and Uszynski, G. (2012). Diversity arrays technology: a generic genome profiling technology on open platforms. Methods in Molecular Biology 888, 67–89.
| Diversity arrays technology: a generic genome profiling technology on open platforms.Crossref | GoogleScholarGoogle Scholar | 22665276PubMed |
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35, 1547–1549.
| MEGA X: Molecular evolutionary genetics analysis across computing platforms.Crossref | GoogleScholarGoogle Scholar | 29722887PubMed |
Marki, P. Z., Jønsson, K. A., Irestedt, M., Nguyen, J., Rahbek, C., and Fjeldså, J. (2017). Supermatrix phylogeny and biogeography of the Australasian Meliphagides radiation (Aves: Passeriformes). Molecular Phylogenetics and Evolution 107, 516–529.
| Supermatrix phylogeny and biogeography of the Australasian Meliphagides radiation (Aves: Passeriformes).Crossref | GoogleScholarGoogle Scholar | 28017855PubMed |
McElroy, K., Dolman, G., Horton, P., Black, A., Pedler, L., Drew, A., and Joseph, L. (2020). Robbery in progress: historical museum collections bring to light a mitochondrial capture within a bird species widespread across southern Australia, the copperback quail-thrush Cinclosoma clarum. Ecology and Evolution 10, 6785–6793.
| Robbery in progress: historical museum collections bring to light a mitochondrial capture within a bird species widespread across southern Australia, the copperback quail-thrush Cinclosoma clarum.Crossref | GoogleScholarGoogle Scholar | 32724551PubMed |
Menkhorst, P., Rogers, D., Clarke, R., Davies, J., Marsack, P., and Franklin, K. (2017). ‘The Australian Bird Guide.’ (CSIRO Publishing: Melbourne.)
Neaves, L. E., Zenger, K. R., Prince, R. I. T., and Eldridge, M. D. B. (2012). Impact of Pleistocene aridity oscillations on the population history of a widespread, vagile Australian mammal, Macropus fuliginosus. Journal of Biogeography 39, 1545–1563.
| Impact of Pleistocene aridity oscillations on the population history of a widespread, vagile Australian mammal, Macropus fuliginosus.Crossref | GoogleScholarGoogle Scholar |
Norman, J. A., Blackmore, C. J., Rourke, M., and Christidis, L. (2014). Effects of mitochondrial DNA rate variation on reconstruction of Pleistocene demographic history in a social avian species, Pomatostomus superciliosus. PLoS One 9, e106267.
| Effects of mitochondrial DNA rate variation on reconstruction of Pleistocene demographic history in a social avian species, Pomatostomus superciliosus.Crossref | GoogleScholarGoogle Scholar | 25181547PubMed |
Nyári, A., and Joseph, L. (2011). Systematic dismantlement of Lichenostomus improves the basis for understanding relationships within the honeyeaters (Meliphagidae) and historical development of Australo-Papuan bird communities. Emu 111, 202–211.
| Systematic dismantlement of Lichenostomus improves the basis for understanding relationships within the honeyeaters (Meliphagidae) and historical development of Australo-Papuan bird communities.Crossref | GoogleScholarGoogle Scholar |
Pembleton, L. W., Cogan, N. O. I., and Forster, J. W. (2013). StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Molecular Ecology Resources 13, 946–952.
| StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations.Crossref | GoogleScholarGoogle Scholar | 23738873PubMed |
Pestell, A. J. L., Cooper, S. J. B., Saint, K., and Petit, S. (2008). Genetic structure of the western pygmy possum, Cercartetus concinnus Gould (Marsupialia: Burramyidae) based on mitochondrial DNA. Australian Mammalogy 29, 191–200.
| Genetic structure of the western pygmy possum, Cercartetus concinnus Gould (Marsupialia: Burramyidae) based on mitochondrial DNA.Crossref | GoogleScholarGoogle Scholar |
Pons, J., Barraclough, T. G., Gomez-Zurita, J., Cardoso, A., Duran, D. P., Hazell, S., Kamoun, S., Sumlin, W. D., and Vogl, A. P. (2006). Sequence-based species delimitation for the DNA taxonomy of undescribed insects Systematic Biology 55, 595–609.
| Sequence-based species delimitation for the DNA taxonomy of undescribed insectsCrossref | GoogleScholarGoogle Scholar | 16967577PubMed |
Rix, M. G., Cooper, S. J. B., Meusemann, K., Klopfstein, S., Harrison, S. E., Harvey, M. S., and Austin, A. D. (2017). Post-Eocene climate change across continental Australia and the diversification of Australasian spiny trapdoor spiders (Idiopidae: Arbanitinae). Molecular Phylogenetics and Evolution 109, 302–320.
| Post-Eocene climate change across continental Australia and the diversification of Australasian spiny trapdoor spiders (Idiopidae: Arbanitinae).Crossref | GoogleScholarGoogle Scholar | 28126515PubMed |
Rozas, J, Ferrer-Mata, A, Sanchez-Del Barrio, J. C., Guirao-Rico, S, Librado, P, Ramos-Onsins, S. E., and Sánchez-Gracia, A (2017). DnaSP v6: DNA sequence polymorphism analysis of large datasets. Molecular Biology and Evolution 34, 3299–3302.
| DnaSP v6: DNA sequence polymorphism analysis of large datasets.Crossref | GoogleScholarGoogle Scholar | 29029172PubMed |
Schodde, R., and Mason, I. J. (1999). ‘The Directory of Australian Birds: Passerines.’ (CSIRO Publishing: Melbourne.)
Sukumaran, J., and Holder, M. T. (2015). SumTrees: phylogenetic tree summarization. 4.0.0. Available at: https://github.com/jeetsukumaran/DendroPy [accessed 14 December 2020].
Tamura, K., and Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10, 512–526.
| Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees.Crossref | GoogleScholarGoogle Scholar | 8336541PubMed |
Zwickl, D. J. (2006). Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion, Ph.D. Dissertation, University of Texas.



