Individual identification via remote video verified by DNA analysis: a case study of the American black bear
Alan B. Ramsey A C , Michael A. Sawaya B , Lorinda S. Bullington A and Philip W. Ramsey A
A C , Michael A. Sawaya B , Lorinda S. Bullington A and Philip W. Ramsey A
A MPG Ranch, 1001 South Higgins Avenue Suite A3, Missoula, MT 59801, USA.
B Sinopah Wildlife Research Associates, 127 North Higgins Avenue Suite 310, Missoula, MT 59802, USA.
C Corresponding author. Email: aramsey@mpgranch.com
Wildlife Research 46(4) 326-333 https://doi.org/10.1071/WR18049
Submitted: 14 March 2018 Accepted: 8 March 2019 Published: 27 May 2019
Journal Compilation © CSIRO 2019 Open Access CC BY-NC-ND
Abstract
Context: Researchers and managers often use DNA analysis and remote photography to identify cryptic animals and estimate abundance. Remote video cameras are used less often but offer an increased ability to distinguish similar-looking individuals as well as to observe behavioural patterns that cannot be adequately captured with still photography. However, the use of this approach in species with minimally distinguishing marks has not been tested.
Aims: To determine the utility and accuracy of distinguishing characteristics of American black bears, Ursus americanus, observed on remote video for identifying individuals in an open population.
Methods: We compared individuals identified on video with individuals and their sex identified by DNA analysis of hairs collected from hair traps visited by the bears.
Key results: We found that remote video could be used to determine the number of male and female black bears sampled by the video cameras. Specifically, we matched 13 individual bear genotypes with 13 video identifications, one genotype for each individual. We correctly matched ~82% of video identifications with all 38 genotypes collected from hair traps.
Conclusions: We demonstrated that distinguishing characteristics of a cryptic animal in remote video can be used to accurately identify individuals. Remote video complements genetic analysis by providing information about habitat use and behaviour.
Implications: When remote video cameras can be used to identify individuals, a wealth of other information will subsequently be obtained. Multi-year video-based studies can show sex ratios, and relative physical condition; shed light on fine-scale habitat use, such as when and where animals feed and what they eat; and display social interactions and rare behaviours.
Additional keywords: behaviour, camera trap, cryptic animal, hair trap, human–bear conflict, individual identity, Montana, population demographics, rub object, Ursus americanus, wildlife management.
Introduction
Identification of individual animals is important for monitoring the demographics of populations and learning about behaviour (Sloane et al. 2000; Merrick and Koprowski 2017). Many tools are available to identify individuals, with methods to retrieve genetic samples and remote photography among those that are being used the most often (Kays et al. 2015). Remote photography using camera traps triggered by motion or heat is a non-invasive way to study wildlife, and over 100 (and rising) still-camera trap studies are being published each year (Meek et al. 2014; Swinnen et al. 2014; Burton et al. 2015). Remote photographs contain enough information to identify animals with highly distinctive body markings, such as stripes on tigers (Panthera tigris; Karanth and Nichols 1998), rosettes on jaguars (Panthera onca; Sollmann et al. 2011) and spots on bobcats (Lynx rufus; Heilbrun et al. 2003). Species with less distinctive markings, such as bears and mustelids, nonetheless, have traits and body markings that can be used to identify individuals and provide insight for management (Sollmann et al. 2013; Burton et al. 2015).
Animals with less distinctive markings may be more easily identified as individuals by using video than by using still images. Reyes et al. (2017) found videos more effective than still images for individual identification of Andean bears (Tremarctos ornatus). Remote video is continuous, allows seamless viewing from moment to moment, and requires no re-triggering of the camera once the initial trigger occurs. Even though many remote photography cameras have video capability, it is rarely used in wildlife research studies, because it consumes more batteries, uses more storage space and requires more processing time. However, because video may contain enough information to identify animals with minimally distinctive visual markings, whereas still photography may not, the extra cost and time required may be justified. Minimally distinguishing visual markings include permanent characteristics, such as blazes, and temporary characteristics, such as coat-shedding patterns. Such markings may appear in frames of a video, but be absent in photographs. Sex characteristics, as described by Marks and Erickson (1966), are also identifiable on video.
Genetic data and still-camera data have been used in conjunction to estimate abundance by using an occupancy framework (Fisher and Bradbury 2014). Non-invasive genetic sampling is an excellent method to identify individuals, estimate population parameters, explore relatedness and determine sex without handling the animal (Taberlet et al. 1999; Carmichael et al. 2005; Long et al. 2008). Non-invasive genetic sampling used in conjunction with remote video offers the possibility of confirming the identity of cryptic animals and using the identifications to learn about demographics and behaviour. We explored this possibility with a case study of the American black bear, Ursus americanus.
We assessed the reliability of identifying black bear individuals with remote video cameras, using 2 years of video data collected in the Sapphire Mountains, Montana, USA. We compared identifications of bears in video with those from non-invasive genetic data collected concurrently. The aim was to determine whether reliable identification of individual bears from video (as confirmed by hair-DNA analysis) could be achieved and, thus, provide information on demographics, habitat use and behaviour. We discuss several benefits of remote video compared with still images for learning about the behaviour of individual animals.
Study site
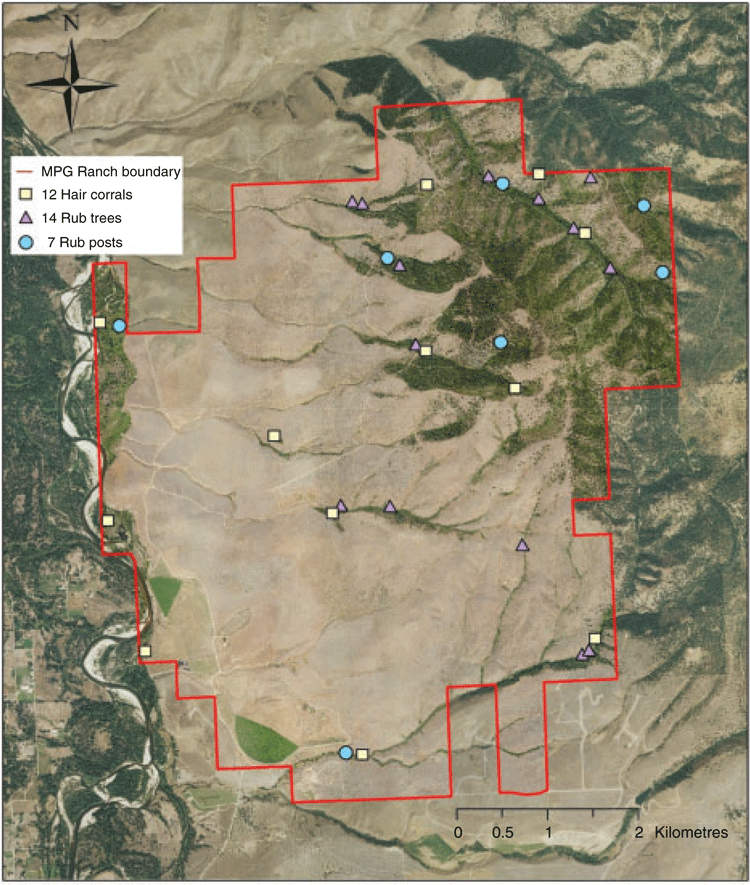
The study was located in the northern Sapphire Mountains in western Montana, USA. Cameras and hair traps were placed on MPG Ranch, a 38.4-km2 private nature preserve and science centre near Missoula (population 70 000; Fig. 1).
Bears freely moved in and out of the study area, so by ‘population’ we referred to bears that used the study area during any time of the year. Black bear research activities were concentrated in the South and Middle Forks of Davis Creek (~1123 ha). The area was uninhabited by people and consisted of land managed for timber production. All of the drainage had been logged at various times between the 1960s and 2007. The site supported most mammals native to western Montana forests, with exceptions such as grizzly bears (Ursus arctos), which were not observed.
Materials and methods
Remote-video collection
We collected videos from remote cameras to determine whether we could identify individual black bears. During the study period, 2013–2014, we deployed 91 video cameras at hair-collection stations (hair corrals, rub trees and rub posts). We placed 58 additional cameras in areas with high bear traffic, such as known travel routes and foraging areas, to maximise detection. We used Stealth Cam Model STC-DVIRHD, Stealth Cam model STC-G42NG (Grand Prairie, Texas, USA), and Bushnell Model 1197678 (Overland Park, Kansas, USA). We placed cameras 0.3–1.0 m off the ground to view distinguishing characteristics such as blazes, genitals and facial features. Cameras recorded for 1-min intervals at up to 30 frames a second for the entire study period unless the camera malfunctioned (see Video S1, available as Supplementary material to this paper). Camera detection of bears was highly likely, because they are large-bodied animals at rub objects or scent lures. Animal body mass and time spent within detection range correlate with camera-trap detection success (Lyra-Jorge et al. 2008). If multiple bears appeared in a single video, we treated each bear on video as a single bear event.
Individual identifications
To start video analysis, we separated videos of bears from other wildlife video footage. We assigned individual identifiers to each bear in each video. We first identified bears on video ‘blind’ (i.e. without knowledge from genetic data) and then evaluated data ‘post-blind’ (i.e. with knowledge from genetic data) to determine the achievable accuracy of using both methods simultaneously. To create the 2013/14 blind dataset, we used a set of bear video identifications with a ‘date modified’ (the last time the computer file was modified) before the date we received 2013/14 genetic results. Next, the videos from this dataset were sorted chronologically. Chronological sorting helped us spot misidentified bears that could not be in two different locations at the same time. One researcher identified 2013/14 bears on video as data became available. In 2014, a technician helped retrieve data from the field and process videos.
We evaluated and defined identifiable traits, including a combination of temporary characteristics, permanent characteristics and individual behaviours. Some characteristics were temporary and only selectively used in conjunction with other more permanent features (File S1, available as Supplementary material to this paper). Temporary characteristics included bear coat-shedding patterns during 1 year, coat shedding across both years, coat-shedding appearance in night videos, coat colour in sunlight, wet coats, burr presence, nipple visibility, general weight gain and uneven weight gain. Permanent characteristics used were body size, blazes, bare spots, ear notches, relative ear size, eyebrow colour, forehead hair pattern, head muscle (temporalis and masseter) size, profile snout shape, snout and head shape, snout moustaches, snout colour, snout scars and lines, female genitals, male genitals and shoulder humps (trapezius area). Some permanent characteristics such as body size and scarring change over time, but not fast enough to be included in temporary characteristics. Individual behaviours involved back floating, head scratching, leg elevating, bear water-entrance technique and berry technique (see File S1 for extensive images and descriptions of characteristics). We, generally, used multiple characteristics to add more certainty to each identification. While we identified bears in videos, we recorded unusual behaviours, repeated behaviours and social behaviours. Many identifying characteristics were often evident even at night, such as, for example, blazes, shed patterns and body sizes. We identified sex on video by observing male and female genitalia (File S1).
DNA collection and analysis
We collected bear hair by using non-invasive sampling methods to obtain genetic-based individual and sex identifications. Methods included 33 total hair-collection stations, including 12 hair corrals consisting of barbed-wire enclosures encircling woody debris covered in scent lure (Woods et al. 1999), seven rub posts consisting of railroad-ties with barbed-wire wrapped around them (Mulders et al. 2007) and 14 rub trees, which were trees with barbed-wire on which bears rub (Kendall et al. 2009). We distributed the hair-collection stations over 38.4 km2 to maximise the number of individuals sampled (Fig. 1). We rebaited hair corrals with scent lure, checked their corresponding video cameras, and collected bear hair every 2–3 weeks from 6 June 2013 to 7 September 2013 and from 30 June 2014 to 30 August 2014. We checked the rub posts and rub trees for bear hair and checked corresponding cameras once every 4 weeks, on average, from 1 April to 1 November each year. Hair-trap barbs were flame-sterilised between collections to limit cross-contamination of samples. We dried hair samples in paper envelopes and stored them with silica desiccant to minimise DNA degradation.
The National Genomics Center for Wildlife and Fish Conservation Laboratory at USFS Rocky Mountain Research Station (RMRS) and MPG Ranch extracted DNA from each hair sample individually, using a DNeasy® Blood and Tissue kit (QIAGEN, Valencia, CA, USA). The RMRS laboratory conducted the individual identity analysis of the bear hair by using a panel of nine microsatellite loci, including G1A, G10D, G10B (Paetkau and Strobeck 1994), G10H, G10J, G10 L, G10P, G10X and UarMu59 (Paetkau and Strobeck 1998), plus one sex identification locus, SRY (Carmichael et al. 2005). G10J was used to distinguish between black and grizzly bears (Kendall et al. 2009), and all samples were found consistent with black bears. To minimise genotyping errors, all samples were genotyped in duplicate or triplicate. Samples that were inconsistent or mixed (producing 3–4 alleles at a given locus, Roon et al. 2005) were re-extracted and amplified until they produced a high-quality genotype or were otherwise removed from further analyses. To identify potential errors such as allelic drop-out and false alleles, the RMRS laboratory ran resulting genotypes through two programs, namely, DROPOUT (McKelvey and Schwartz 2005) and Micro-checker (Van Oosterhout et al. 2004). The results of these tests indicated that the final dataset was free of any genotyping errors. The probability of identity, that is, the probability that two individuals drawn at random from the population would have the same genotype at these loci, as reported by RMRS, was <1 × 10−10. The probability of identity for siblings was 1.8 × 10−4. Both of these statistics were calculated using GenAlEx (Peakall and Smouse 2006).
Video versus genetic identifications
To assess the accuracy of video identifications, we assembled genetic ID–video ID match sets. A match set was one genetic identification and all blind video identifications from the same location within a timeframe. The timeframe was from the time we collected the genetic sample to the previous time we collected or checked for samples. Within each match set, we identified an individual in a video to make a genetic ID–video ID match. Because some bears left multiple (i.e. redundant) samples at the same time and location, we included only one sample (i.e. non-redundant) in each genetic ID match set (Table 1; Table S1, available as Supplementary material to this paper). We narrowed the video ID sets involving the genetic ID–video ID matches to include only bears that rubbed against a rub object or contacted a hair-corral wire.
Results
Remote-video captures and hair collections
The video dataset of all cameras included 2627 bear videos. Some videos contained multiple bears, resulting in a total of 3228 recorded bear events. We recorded 983 bear videos in 2013, with 1074 bear events, and 1644 bear videos in 2014, with 2154 bear events. We designated 107 videos in 2013 and 118 in 2014 as unidentified. Post-blind (i.e. after consulting the genetic data), we changed identifications on 52 (1.6%) bear events of 11 individual bears, including 17 events of eight bears in 2013 and 35 events of nine bears in 2014. Comparing the video and genetic analysis showed that identifications agreed 98.4% of the time.
We collected 239 total hair samples over two seasons for subsequent DNA analysis. Rocky Mountain Research Station (RMRS) provided high-quality genotypes for 75 of those samples, (39 from 2013 and 36 from 2014). As bears often left multiple samples while rubbing, we further narrowed the dataset to include only the 47 non-redundant genotyped samples, (21 from 2013 and 26 from 2014). We then eliminated nine non-redundant genotyped samples because the corresponding camera was not functional for the sample period. We were left with 38 samples, 18 for 2013 and 20 for 2014, which provided the genotypes used for comparison to images for All Matches (Table 1). For this matched set, 28 samples were collected at 14 rub trees, six samples at seven rub posts, and four samples at 12 hair corrals.
Individual identifications
In 2013/14, we blind identified 22 bears on video (8 males, 7 females and 7 unidentified cubs) and post-blind identified 21 bears (8 adult males, 6 adult females, 1 male cub, 1 female cub and 5 unidentified cubs). Fourteen individuals (7 males and 7 females) were present in both 2013 and 2014. In 2013, we blind identified 19 bears on video, including eight males, seven females and four cubs of unknown sex. Post-blind, we added one bear and removed two bears for a new total of 18 individuals, including eight males, six females and four cubs of unknown sex. In 2014, we blind identified 17 bears on video, including six males, six females and five cubs of unknown sex. Post-blind and blind identifications were the same, but we identified the sex of two cubs, namely one male and one female. We post-blind identified, with video, six cubs, one adult male and one adult female whose genotype we did not determine. We identified four putative family groups of mothers with cubs with video. Three groups had two cubs, and one group had one cub.
Seventy-five bear events during which hair could have been left occurred in the dataset All Matches. Sex was apparent in 67% of these 75 events. By identifying the bears of known sex throughout the set, we accounted for the sex of all events except one, namely an unidentified event at night. Also, for one cub event, we learned his sex post-blind from genetics. We did not change the sex of any individuals post-blind to the opposite sex. We recorded an average of 1.7 bears on video of the genetically identified hair sample sex and 2.3 bears on video of both sexes rubbing or contacting hair-corral wire at each sample location per sample period. In all, 70% of these 75 events took place during the day and 30% at night. We misidentified four daytime and three night-time bear events.
Video versus genetic identifications
Rocky Mountain Research Station (RMRS) identified 13 genetically unique individuals and assigned them genetic ID numbers; we identified 22 unique individuals from video and assigned them Video ID letters. We matched all 13 genetically identified individuals with correct Video IDs (Table S2). Nine matches were blind matches. Four matches were post-blind matches with both genetic and video data available. In six matches, only one individual was present. Here, it was highly unlikely we mismatched genetic IDs and video IDs. We included repeat visits by the same individual as one individual present. In three matches, only one individual of the genetic ID sex was present but individual(s) of the opposite sex were also present; here, it was very unlikely that we mismatched genetic IDs and video IDs, but we were dependent on knowing the sex of each bear. In one match, we erred and had to change one misidentified bear to a new bear. In one match we changed one bear identified as ‘unidentified’ to a known bear on the basis of genetic data and other relevant videos. We made two matches by process of elimination. In one, we had identified the two bears present in other matches. In the other, we distinguished between one of two cubs who were identical except for sex. We clearly saw the cub leave hair on video. We identified correctly (defined as when genetic and video identifications aligned) blind that the genetic sample was one of two cubs, but could not tell which one without genetic information on one cub’s sex and video information on the other’s sex.
Of the Best Matches (Table S2), we identified correctly 9 of the 13 matches blind and four post-blind. We made three errors in identifying the 13 matches, namely, we misidentified one bear, changed one bear labelled ‘unidentified’ to a known bear, and could not differentiate between one of two cubs. If we count these three identifications as incorrect, we had a 77% success rate (Table S2). Genetic analysis identified bears successfully 93% of the time. Fourteen bears were originally identified through genetic analysis; however, a mixed sample was detected, and the total was adjusted to 13 bears. When both video and genetic data were collected at rub objects and hair corrals, we estimated that the two methods returned a 98.4% success rate at correctly identifying individuals, because both datasets were compared with evaluate accuracy.
Of All Matches, we correctly identified 31 of the 38 observations blind and changed seven for a blind success rate of 82% (Tables 1, S1). For 20 of these observations, multiple members of the genetic ID sex were present, so we relied on the process of elimination to confirm identifications after identifying the 13 genetic ID–video ID matches. We changed two misidentified bears to known bears, changed two unidentified bears to known bears, changed one unidentified bear to a new bear, distinguished the sex of one of two identical cubs, and found that one bear was absent during the sampling period; we suspected this hair sample was left over from a previous sampling period.
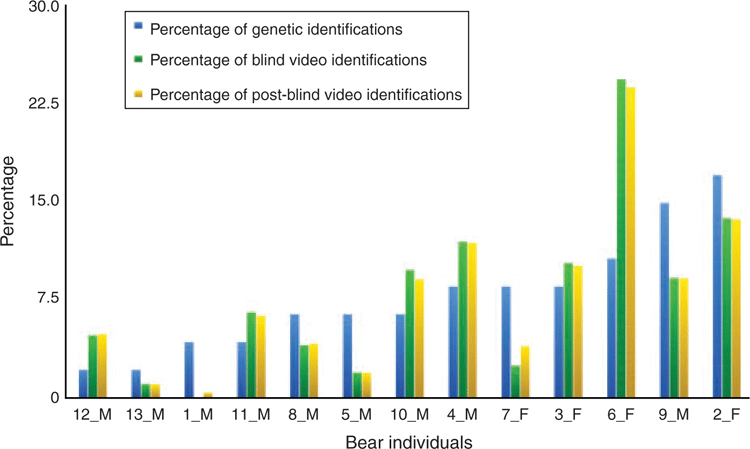
We compared all genotyped samples, excluding multiple samples of the same bear at the same time and location, versus all video identifications as percentages (Fig. 2). In general, individuals with more genetic IDs had more video IDs. Important exceptions were evident. For example, 6F had the third-most genetic IDs (10.6%), but the most video IDs (23.7%). We documented 6F on 62 of a possible 158 days at 21 locations in the study area, from 15 May 2014 to 19 October 2014.
|
Discussion
We used video and genetic analysis to identify individuals of a species, the American black bear, with marginally distinguishable body markings. We matched 13 bear genotypes with 13 bears we identified on video. When we labelled all recorded bear videos at rub objects and hair corrals without genetic information, we achieved a success rate of ~82%. Genetic analysis identified bears successfully the majority (93%) of the time. With access to both genetic and video information, we estimate a 98.4% success rate was achieved when matching individuals on video at rub objects and hair corrals with their genetic identification. Confirmed identifications allowed the remote video to be used to learn about behavioural and reproductive characteristics of individual bears over months to years. These variables provided more insight into the ecology of bears in the study area than would have been possible through the use of video or genetics alone. In a future publication, we plan to publish ecological and behavioural results from the present study.
Individual identification of animals by video can aid management and research by providing insights into population demographics, physical condition, fine-scale habitat use and behaviour. Regarding demographics, the approach used here allowed sex to be identified with video and putative family-group identity to be inferred with video (4 putative family groups of mothers with cubs identified with video). We gained additional demographic information about the number of individuals, namely, we identified eight more bears on video, mostly cubs, than we did with genetics. In addition, where multiple bears use the same rub, genetic samples will often become ‘mixed’ and uninformative. In these cases, video data may still allow identification of multiple individuals where genetic work does not. Video will also often be better than still photos as the lag time between still photos can confuse identification. Genetic data, still images, and identifying individuals with remote video can all lead to misidentification and inaccurate estimation of population size. Because of this, multiple methods of identification should be used wherever possible, to improve accuracy of population estimates and observations (Vine et al. 2009).
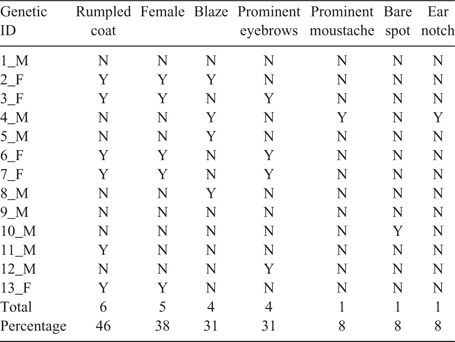
The use of photography, both still images and video, is limited by how distinguishing an animal’s body markings are. Here, coat colour (e.g. Fig. 3) was the most easily determined characteristic during daylight of the 25 distinguishing physical characteristics (File S1) used to identify individual bears. In the Rocky Mountains, Oregon and Washington, 30–90% of bears are a colour phase other than black, with non-black bears more common in less forested areas (Rounds 1987). In the present study, 81% of the bears were a colour other than black. The present study would have been much more difficult in eastern North America where close to 100% of the bears are of the black colour phase (Rounds 1987). However, there is a report of visual identification of individual released black-phased American black bears in New Hampshire, USA (Kilham 2013).

|
In the present study, we were close to the maximum number of individuals that could be reliably distinguished using characteristics such as those described in Table 2. Here, in a population of 20–30 bears with a variety of coat colours, most bears were distinguishable; in a population of 20–30 bears with little coat-colour variety, many bears would still be distinguishable through characteristics elaborated in Table 2. In a video study conducted over a larger area or over a longer period, we would eventually lose the ability to distinguish individuals. Larger-scale applications might need to focus on putative family groups only, marked animals, or animals with unusual physical characteristics. We found that for video and genetic identification, rub trees were worth the effort to locate. We were more successful at collecting hair at rub trees than at rub posts, and at rub posts more than from hair corrals. Unbaited rub trees and posts caused fewer human visits to bear habitat than did hair corrals, which required rebaiting. At hair corrals, it was more difficult to see blazes and sex bears. We recommend a priority order of rub trees, rub posts, and then hair corrals. Where bear rub trees are scarce, rub posts, and then hair corrals, may work.
|
In addition to population parameters, the videos generated from the present study allowed for many observations that could not be obtained from still images or genetics alone. For example, assessments of physical condition can be useful for black bear management because bears in poor condition are more likely to come into conflict with people (Black et al. 2004). The video footage allowed us to assess the physical condition of identified animals on multiple occasions, including in different seasons, noting spring sluggishness, fall weight gain, frost bite on ear tips and lacerations to ankles, presumably from foot traps. Video was also useful for showing which species of plants, and even which plant parts, bears ate as well as when and where bears ate them. Concerning behaviour, with video and accompanying audio, we determined whether one bear or many bears repeated certain behaviours, such as swimming in a pool in Davis Creek. In this pool, we documented multiple adult bears at the same time plus individual thermoregulation, swim times, day versus night swimming, and stick and mud holding (Sawaya et al. 2016). We documented social behaviour between individuals such as mating, including a male and female together on three consecutive days. We documented non-mating social behaviour among unrelated bears. Although unrelated to population estimates and demographics, ancillary observations from video can help develop a fuller view of an animal’s behaviour and are lacking for bears and many other species (Harding 2004).
Conclusions
Species that live secretively in dense cover, mostly alone, or at a low density in a landscape challenge managers seeking to make informed decisions (Piggott and Taylor 2003; McCall et al. 2013). Although remote photography has some advantages over video in terms of battery consumption, storage-space use and processing time, video offers a richer dataset for learning about individuals and behaviour. The principal advantages of video are (1) fewer individuals being missed moving in and out of frame between image captures and (2) more opportunity to capture unique characteristics and behaviours. As such, more use of video in wildlife research will advance efforts to document and manage cryptic species.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
We thank MPG Ranch ownership for generously funding this research and thank Jeffrey Stetz and Jessica Jenne of Sinopah Wildlife Research Associates; Thain Bertin, Tor Bertin, and Lauren Shreading of MPG Operations; and USFS Rocky Mountain Research Station staff Michael Schwartz (Ph.D.), Kristine Pilgrim (M.Sc.) and Cory Engkjer. No wildlife/ethics permits were necessary in the study area to remotely collect black bear hair.
References
Black, H., Auger, J., and Smith, R. V. (2004). Long-term trapping of black bears on the East Tavaputs Plateau. Black Bears of Utah’s East Tavaputs Plateau 1, 5–19.Burton, A.C., Neilson, E., Moreira, D., Ladle, A., Steenweg, R., Fisher, J. T., Bayne, E., and Boutin, S. (2015). Wildlife camera trapping: a review and recommendations for linking surveys to ecological processes. Journal of Applied Ecology 52, 675–685.
| Wildlife camera trapping: a review and recommendations for linking surveys to ecological processes.Crossref | GoogleScholarGoogle Scholar |
Carmichael, L. E., Krizan, P., Blum, S. P., and Strobeck, C. (2005). Genotyping of pseudohermaphrodite polar bears in Nunavut and advances in DNA sexing techniques. Journal of Mammalogy 86, 160–169.
| Genotyping of pseudohermaphrodite polar bears in Nunavut and advances in DNA sexing techniques.Crossref | GoogleScholarGoogle Scholar |
Fisher, J. T., and Bradbury, S. (2014). A multi-method hierarchical modeling approach to quantifying bias in occupancy from noninvasive genetic tagging studies. The Journal of Wildlife Management 78, 1087–1095.
| A multi-method hierarchical modeling approach to quantifying bias in occupancy from noninvasive genetic tagging studies.Crossref | GoogleScholarGoogle Scholar |
Harding, L. E. (2004). Habitat use, behaviors, and movements of black bears on the East Tavaputs Plateau Utah Black Bears of Utah’s East Tavaputs Plateau 1, 55–64.
Heilbrun, R. D., Silvy, N. J., Tewes, M. E., and Peterson, M. J. (2003). Using automatically triggered cameras to individually identify bobcats. Wildlife Society Bulletin 31, 748–755.
Karanth, K. U., and Nichols, J. D. (1998). Estimation of tiger densities in India using photographic captures and recaptures. Ecology 79, 2852–2862.
| Estimation of tiger densities in India using photographic captures and recaptures.Crossref | GoogleScholarGoogle Scholar |
Kays, R., Crofoot, M.C., Jetz, W., and Wikelski, M. (2015). Terrestrial animal tracking as an eye on life and planet. Science 348, aaa2478-1–aaa2478-9.
| Terrestrial animal tracking as an eye on life and planet.Crossref | GoogleScholarGoogle Scholar |
Kendall, K. C., Stetz, J. B., Boulanger, J., Macleod, A. C., Paetkau, D., and White, G. C. (2009). Demography and genetic structure of a recovering grizzly bear population. The Journal of Wildlife Management 73, 3–16.
| Demography and genetic structure of a recovering grizzly bear population.Crossref | GoogleScholarGoogle Scholar |
Kilham, B. (2013). ‘Out on a Limb: Bear Society.’ (Chelsea Green Publishing: White River Junction, VT, USA.)
Long, R. A., MacKay, P., Zielinksi, W., and Ray, J. (2008). ‘Noninvasive Survey Methods for Carnivores.’ (Island Press: Washington DC, USA.)
Lyra-Jorge, M. C., Ciocheti, G., Pivello, V. R., and Meirelles, S. T. (2008). Comparing methods for sampling large-and medium-sized mammals: camera traps and track plots. European Journal of Wildlife Research 54, 739.
| Comparing methods for sampling large-and medium-sized mammals: camera traps and track plots.Crossref | GoogleScholarGoogle Scholar |
Marks, S. A., and Erickson, A. W. (1966). Age determination in the black bear. The Journal of Wildlife Management 30, 389–410.
| Age determination in the black bear.Crossref | GoogleScholarGoogle Scholar |
McCall, B. S., Mitchell, M. S., Schwartz, M. K., Hayden, J., Cushman, S. A., Zager, P., and Kasworm, W. F. (2013). Combined use of mark–recapture and genetic analyses reveals response of a black bear population to changes in food productivity. The Journal of Wildlife Management 77, 1572–1582.
| Combined use of mark–recapture and genetic analyses reveals response of a black bear population to changes in food productivity.Crossref | GoogleScholarGoogle Scholar |
McKelvey, K. S., and Schwartz, M. K. (2005). DROPOUT: a program to identify problem loci and samples for noninvasive genetic samples in a capture-mark-recapture framework. Molecular Ecology Notes 5, 716–718.
| DROPOUT: a program to identify problem loci and samples for noninvasive genetic samples in a capture-mark-recapture framework.Crossref | GoogleScholarGoogle Scholar |
Meek, P. D., Ballard, G., Claridge, A., Kays, R., Moseby, K., O’Brien, T., O’Connell, A., Sanderson, J., Swann, D. E., Tobler, M., and Townsend, S. (2014). Recommended guiding principles for reporting on camera trapping research. Biodiversity and Conservation 23, 2321–2343.
| Recommended guiding principles for reporting on camera trapping research.Crossref | GoogleScholarGoogle Scholar |
Merrick, M. J., and Koprowski, J. L. (2017). Should we consider individual behavior differences in applied wildlife conservation studies? Biological Conservation 209, 34–44.
| Should we consider individual behavior differences in applied wildlife conservation studies?Crossref | GoogleScholarGoogle Scholar |
Mulders, R., Boulanger, J., and Paetkau, D. (2007). Estimation of population size for wolverines Gulo gulo at Daring Lake, Northwest Territories, using DNA based mark-recapture methods. Wildlife Biology 13, 38–51.
| Estimation of population size for wolverines Gulo gulo at Daring Lake, Northwest Territories, using DNA based mark-recapture methods.Crossref | GoogleScholarGoogle Scholar |
Paetkau, D., and Strobeck, C. (1994). Microsatellite analysis of genetic variation in black bear populations. Molecular Ecology 3, 489–495.
| Microsatellite analysis of genetic variation in black bear populations.Crossref | GoogleScholarGoogle Scholar | 7952329PubMed |
Paetkau, D., and Strobeck, C. (1998). Ecological genetic studies of bears using microsatellite analysis. Ursus 10, 299–306.
Peakall, R., and Smouse, P. E. (2006). genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes 6, 288–295.
| genalex 6: genetic analysis in Excel. Population genetic software for teaching and research.Crossref | GoogleScholarGoogle Scholar |
Piggott, M. P., and Taylor, A. C. (2003). Remote collection of animal DNA and its applications in conservation management and understanding the population biology of rare and cryptic species. Wildlife Research 30, 1–13.
| Remote collection of animal DNA and its applications in conservation management and understanding the population biology of rare and cryptic species.Crossref | GoogleScholarGoogle Scholar |
Reyes, A., Rodríguez, D., Reyes-Amaya, N., Rodríguez-Castro, D., Restrepo, H., and Urquijo, M. (2017). Comparative efficiency of photographs and videos for individual identification of the Andean bear (Tremarctos ornatus) in camera trapping Therya 8, 83–87.
Roon, D. A., Thomas, M. E., Kendall, K. C., and Waits, L. P. (2005). Evaluating mixed samples as a source of error in non‐invasive genetic studies using microsatellites. Molecular Ecology 14, 195–201.
| Evaluating mixed samples as a source of error in non‐invasive genetic studies using microsatellites.Crossref | GoogleScholarGoogle Scholar | 15643963PubMed |
Rounds, R. C. (1987). Distribution and analysis of colourmorphs of the black bear (Ursus americanus). Journal of Biogeography 14, 521–538.
| Distribution and analysis of colourmorphs of the black bear (Ursus americanus).Crossref | GoogleScholarGoogle Scholar |
Sawaya, M. A., Ramsey, A. B., and Ramsey, P. W. (2016). American black bear thermoregulation at natural and artificial water sources. Ursus 27, 1–7.
Sloane, M. A., Sunnucks, P., Alpers, D., Beheregaray, B., and Taylor, A. C. (2000). Highly reliable genetic identification of individual northern hairy-nosed wombats from single remotely collected hairs: a feasible censusing method. Molecular Ecology 9, 1233–1240.
| Highly reliable genetic identification of individual northern hairy-nosed wombats from single remotely collected hairs: a feasible censusing method.Crossref | GoogleScholarGoogle Scholar | 10972763PubMed |
Sollmann, R., Furtado, M. M., Gardner, B., Hofer, H., Jácomo, A. T., Tôrres, N. M., and Silveira, L. (2011). Improving density estimates for elusive carnivores: accounting for sex-specific detection and movements using spatial capture–recapture models for jaguars in central Brazil. Biological Conservation 144, 1017–1024.
| Improving density estimates for elusive carnivores: accounting for sex-specific detection and movements using spatial capture–recapture models for jaguars in central Brazil.Crossref | GoogleScholarGoogle Scholar |
Sollmann, R., Gardner, B., Chandler, R. B., Shindle, D. B., Onorato, D. P., Royle, J. A., and O’Connell, A. F. (2013). Using multiple data sources provides density estimates for endangered Florida panther. Journal of Applied Ecology 50, 961–968.
| Using multiple data sources provides density estimates for endangered Florida panther.Crossref | GoogleScholarGoogle Scholar |
Swinnen, K. R., Reijniers, J., Breno, M., and Leirs, H. (2014). A novel method to reduce time investment when processing videos from camera trap studies. PLoS One 9, e98881.
| A novel method to reduce time investment when processing videos from camera trap studies.Crossref | GoogleScholarGoogle Scholar | 24918777PubMed |
Taberlet, P., Waits, L. P., and Luikart, G. (1999). Noninvasive genetic sampling: look before you leap. Trends in Ecology & Evolution 14, 323–327.
| Noninvasive genetic sampling: look before you leap.Crossref | GoogleScholarGoogle Scholar |
Van Oosterhout, C., Hutchinson, W. F., Wills, D. P. M., and Shipley, P. (2004). MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4, 535–538.
| MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data.Crossref | GoogleScholarGoogle Scholar |
Vine, S. J., Crowther, M. S., Lapidge, S. J., Dickman, C. R., Mooney, N., Piggott, M. P., and English, A. W. (2009). Comparison of methods to detect rare and cryptic species: a case study using the red fox (Vulpes vulpes). Wildlife Research 36, 436–446.
| Comparison of methods to detect rare and cryptic species: a case study using the red fox (Vulpes vulpes).Crossref | GoogleScholarGoogle Scholar |
Woods, J. G., Paetkau, D., Lewis, D., McLellan, B. N., Proctor, M., and Strobeck, C. (1999). Genetic tagging of free-ranging black and brown bears. Wildlife Society Bulletin 27, 616–627.