Big trees of small baskets: phylogeny of the Australian genus Spyridium (Rhamnaceae: Pomaderreae), focusing on biogeographic patterns and species circumscriptions
Catherine Clowes A * , Rachael M. Fowler A , Patrick S. Fahey A , Jürgen Kellermann B C , Gillian K. Brown D and Michael J. Bayly
A * , Rachael M. Fowler A , Patrick S. Fahey A , Jürgen Kellermann B C , Gillian K. Brown D and Michael J. Bayly  A
A
A School of Biosciences, The University of Melbourne, Vic. 3010, Australia.
B State Herbarium of South Australia, Botanic Gardens and State Herbarium, Adelaide, SA 5000, Australia.
C School of Biological Sciences, The University of Adelaide, Adelaide, SA 5005, Australia.
D Department of Environment and Science, Queensland Herbarium, Toowong, Qld 4066, Australia.
Australian Systematic Botany 35(2) 95-119 https://doi.org/10.1071/SB21034
Submitted: 29 September 2021 Accepted: 10 March 2022 Published: 18 May 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Spyridium Fenzl is a genus of ~45 species endemic to south-western and south-eastern Australia. This study provides the most comprehensive phylogenies of Spyridium to date, analysing both entire chloroplast genomes and the nuclear ribosomal array (18S–5.8S–26S). There was substantial incongruence between the chloroplast and nuclear phylogenies, creating phylogenetic uncertainty, but some clear relationships and biogeographic patterns could be established. Analyses support the monophyly of Spyridium, identifying an early east–west split at the base of the nuclear phylogeny and deep divergences of New South Wales and Tasmanian endemic clades. We also found evidence of more recent dispersal events between eastern and western Australia and between Tasmania and the mainland. Eleven taxa were found to be monophyletic in the nrDNA phylogeny and two were clearly polyphyletic (S. eriocephalum Fenzl and S. phylicoides Reissek). Although the polyphyly of S. eriocephalum correlates with the two varieties, suggesting distinct taxa, further research is required on S. phylicoides.
Keywords: biogeography, chloroplast genome, molecular phylogeny, next-generation sequencing, nuclear ribosomal DNA, Rhamnaceae, species delimitation, Spyridium.
Introduction
Spyridium Fenzl is a member of the tribe Pomaderreae Reissek ex Endl., which includes 10 genera and ~230 species endemic to Australia and New Zealand (Kellermann et al. 2008). Spyridium is derived from the Greek word spyridion meaning ‘a small basket’; a reference to the flowerheads surrounded by leafy bracts (Perrin 2018). The tribe is easily distinguished within the family Rhamnaceae and is characterised by species with stellate hairs on at least some of the vegetative or floral parts.
Generic boundaries within Pomaderreae have been difficult to define (Thiele and West 2004). According to Bentham (1863, p. 410):
…most [genera], even the most natural ones, are difficult to characterize. The differences in their flowers and fruits are very trifling; they often pass into one another by the finest gradations, and habit, foliage and inflorescence must often be relied upon for fixing generic limits.
Consequently, numerous species within the tribe have been transferred from one genus to another (and in some cases back), including species from Cryptandra Sm. and Spyridium.
The distinction between Cryptandra and Spyridium had traditionally been the presence of a floral tube in the former and its absence (or close to it) in the latter. However, for some species from both genera, the floral tube grades from effectively absent to very much present, blurring these traditional generic boundaries. Recently, more distinct characters between the two genera have been described (including disc, stipule and inflorescence characters), resulting in the transfer of several species of Cryptandra to Spyridium, including S. scortechinii1 and S. buxifolium (Thiele and West 2004). In one extreme example, S. waterhousei has been moved to several genera within the tribe (including Cryptandra and Stenanthemum Reissek) before being re-instated as Spyridium waterhousei (Kellermann 2007) on the basis of its clear placement in molecular phylogenetic analyses (Kellermann et al. 2005).
The genus Spyridium currently includes ~45 species (Kellermann and Barker 2012) distributed in semi-arid to temperate regions of southern Australia (Fig. 1), including at least six potentially undescribed species identified as part of work towards a Flora of Australia treatment (Kellermann et al. 2022). The genus has two hotspots of local endemism, including ~20 species in southern South Australia (SA) and ~15 species in south-western Western Australia (WA). While the majority of species endemic to the south-west are confined to that region, two species have disjunct distributions across the Nullarbor Plain (S. tricolor and S. subochreatum). In south-eastern Australia, many of the species have narrow distributions, but several taxa (S. parvifolium, S. vexilliferum var. vexilliferum, S. eriocephalum var. eriocephalum) occur widely in this area (Coates and Kirkpatrick 1999; Kellermann and Barker 2012).

|
The monophyly of Spyridium has been confirmed by multiple molecular phylogenies (Richardson et al. 2004; Kellermann et al. 2005; Kellermann and Udovicic 2007; Hauenschild et al. 2016, 2018). The first nuclear DNA-based phylogeny (ITS) to focus on the tribe Pomaderreae was published by Kellermann et al. (2005), including 15 species of Spyridium. Spyridium was strongly supported as monophyletic, and four geographically based clades were identified, with one being endemic to the eastern mainland, one from Tasmania, one being endemic to the south-west, and one including species from south-eastern Australia more generally. In addition, two species were moved from other genera to Spyridium (S. daltonii from Trymalium and S. waterhousei from Cryptandra). A chloroplast DNA phylogeny (trnL–trnF) of the tribe, which included the same (now) 17 species of Spyridium (Kellermann and Udovicic 2007), did not resolve monophyly of the genus, instead species of Spyridium formed part of a large polytomy at the base of the tree. But, like the findings of the ITS phylogeny, species endemic to the east and Tasmania formed separate clades. Finally, Hauenschild et al. (2018) included 18 species of Spyridium in their study on Gondwanan biogeography of ziziphoid Rhamnaceae, on the basis of nuclear, plastid and mitochondrial markers, and found that eastern endemics diverged first, followed by Tasmanian endemics and, finally, those endemic to the south-west. These studies have provided valuable information about relationships in Spyridium and suggest early divergence of endemic groups from the south-west, east and Tasmania. However, more than half of the species of Spyridium have not yet been included in a molecular phylogeny.
The aim of this study is to produce a comprehensive molecular phylogeny for the genus Spyridium, by including all described species and analysing the full chloroplast genome (cpDNA) and the 18S–5.8S–26S array of nuclear rDNA (nrDNA). We use these phylogenies to investigate broad biogeographic patterns within Spyridium, to explore vicariance, dispersal and diversification in the genus and to assess the monophyly and relationships within and among currently accepted species, as well as some proposed but so far undescribed taxa.
Materials and methods
Taxon sampling
In total, 143 samples of Spyridium were analysed in this study (Table 1). All species, subspecies and varieties of Spyridium recognised in the Australian Plant Census (APC; CHAH 2020) were included with at least one sample, except for S. bifidium var. bifidum, which was excluded because of sequencing issues associated with low DNA yield and quality. Of the 56 taxa included in this study, 45 (when counted at both specific and intraspecific levels) were represented by more than one accession. For species with wide distributions, multiple samples from across the geographic range were included where possible. Samples of six proposed but so far undescribed taxa were also included (Table 2). Four samples from three other genera (Pomaderris Labill., Cryptandra and Trymalium) from the tribe Pomaderreae were included as outgroups.

|

|
For most samples, fresh leaf material was collected and dried in silica gel, along with a voucher specimen. Eight samples were obtained from existing herbarium collections at the WA Herbarium (PERTH). Herbarium voucher details are given in Table 1; many of the vouchers deposited at the University of Melbourne Herbarium (MELU) also have duplicates deposited variously at the National Herbarium of Victoria (MEL), the Tasmanian Herbarium (HO), PERTH or the State Herbarium of South Australia (AD).
DNA extraction
Total genomic DNA was extracted from ~60 mg of silica-dried leaf material, or ~30 mg of herbarium material, following a modified cetyl trimethylammonium bromide (CTAB) protocol (Shepherd and McLay 2011; McLay 2017) based on Doyle and Doyle (1987). Where possible, young leaf material from stem tips or dried floral leaves was selected for extraction. DNA quality and quantity were recorded using a Nanodrop 2000 (NanoDrop Products) and Qubit 2.0 fluorometer (Invitrogen) and used to inform library preparation.
Library preparation and DNA sequencing
Genomic DNA was prepared for multiplexed sequencing by using the library preparation and sequencing protocol of Schuster et al. (2018), with a few modifications. In total, 100 µL of each sonicated sample was transferred to a PCR plate and cleaned with solid-phase reversible immobilisation (SPRI) beads, by using a beads:sample ratio targeted to retain fragments of >300 bp (Rohland and Reich 2012). Following incubation and bead capture on a 96S super magnet plate (Alpaqua), the supernatant-free sample was washed with 100 µL of 80% ethanol. All subsequent 80% ethanol washes were also performed with 100 µL. Final q-PCRs were performed with 20 µL per reaction.
Sequence assembly
Quality filtering and base calling was conducted at Walter and Eliza Hall Institute of Medical Research (WEHI) with Illumina pipeline software (ver. 1.7) and pre-processed with custom scripts, as in Schuster et al. (2018). De-multiplexed reads were imported into Geneious (ver. 10.2, Dotmatics, see https://www.geneious.com/; Kearse et al. 2012) and trimmed using an error probability limit of 0.05. Paired-end reads were set by name and contigs were built in CLC Genomics Workbench (ver. 10.0.1, QIAGEN, see https://digitalinsights.qiagen.com/) by using default de novo settings. Contigs less than 1800 bp were discarded and remaining contigs were trimmed, removing 150 bp from each end.
Nuclear rDNA (18S–5.8S–26S, including both internal transcribed spacers and partial external and non-transcribed spacers) sequences were initially assembled by mapping contigs to the reference sequence for Helianthus annuus L. (GenBank number: KF767534), because there was no suitable extended (5′ETS–3′ETS + NTS) nrDNA reference for Spyridium or close relatives. Although Helianthus is not closely related to Spyridium, the highly conserved nature of sections of nrDNA (Jobes and Thien 1997), and success building long contigs spanning the region, resulted in successful mapping. Contigs were separated where required to assist mapping and, once completed, annotations were transferred from the reference and a draft consensus sequence was generated, with gaps preserved and the reference sequence used where data were missing. Paired reads were mapped to the draft nrDNA sequence for quality-control purposes, with a base calling threshold of 75% employed. The consensus sequence was manually adjusted where required and a final consensus sequence generated. Once the first nrDNA sequence of Spyridium was finalised (CC211; Table 1), this new sequence was used as the reference for subsequent sequence building.
The cpDNA genome for each sample was assembled using the same methodology as for the nrDNA sequences but using a different reference sequence (S. parvifolium var. parvifolium, GenBank accession MH234313; Clowes et al. 2018) and with the threshold for base calls in the final consensus sequence set at 50%. In addition, contigs and paired reads could map to multiple best-fit locations to enable the assembly of the inverted repeats.
Phylogenetic analyses
Sequences were aligned using the MAFFT (ver. 7.308, see https://mafft.cbrc.jp/alignment/software/; Katoh et al. 2002; Katoh and Standley 2013) Geneious plugin under default settings. Aligned sequences were reviewed in Geneious, base pairs were re-aligned by eye where required and ambiguous regions were excluded from the final alignment. In addition, inverted repeat A (IRA) was excluded from the cpDNA alignment at this stage.
Aligned sequences were partitioned before model testing and phylogenetic analyses. For the nrDNA, the alignment was partitioned as follows: partial external transcribed spacer (5′ETS), 18S, internal transcribed spacer 1 (ITS1), 5.8S, internal transcribed spacer 2 (ITS2), 26S, and partial non-transcribed spacer (3′ETS + NTS). The four cpDNA partitions, based on annotations from the reference sequence of S. parvifolium var. parvifolium (GenBank accession MH234313) were as follows: gene-coding sequence (CDS); transfer ribonucleic acid (tRNA); ribosomal ribonucleic acid (rRNA); and all remaining sequences, including introns and intergenic spacers (referred to in the partition as spacers).
Both nrDNA and cpDNA alignments were analysed using Bayesian inference (BI) and maximum likelihood (ML) methods. For the BI analyses, model testing was performed for each partition following Akaike’s information criterion (AIC) using MrModelltest2 (ver. 2.4, J. A. A. Nylander, see https://github.com/nylander/MrModeltest2) for nrDNA (selected models: 5′ETS GTR + G, 18S GTR + I + G, ITS1 SYM + G, ITS2 K80 + G, 26S GTR + I + G and 3′ETS + NTS HKY + G; with 5.8S alignment being excluded from analyses because all sequences were identical) and cpDNA (selected models: CDS GTR + I + G, tRNA K80 + I, rRNA HKY and spacers GTR + I + G). Bayesian inference analyses were undertaken in MrBayes XSEDE (ver. 3.2.6, C. Zhang, J. Huelsenbeck, P. van der Mark, F. Ronquist and M. Teslenko, see https://github.com/NBISweden/MrBayes/releases; Ronquist and Huelsenbeck 2003) by using the CIPRES portal (Miller et al. 2010). For the nrDNA alignment (6367 bp), two independent analyses with four chains (Markov-chain Monte Carlo) were run for 5 000 000 generations, sampling every 1000 steps, with a burnin of 25%. For the cpDNA alignment (168 343 bp), chains were run for 2 500 000 generations, sampling every 500 steps. Output files were viewed in Tracer (ver. 1.6, A. Rambaut, A. J. Drummond and M. Suchard, see https://github.com/beast-dev/tracer), checking convergence (<0.01 standard deviation of split frequencies). The 50% majority-rule consensus trees were visualised in FigTree (ver. 1.4.2, A. Rambaut and A. J. Drummond, see https://github.com/rambaut/figtree) with posterior probabilities (PP) of ≥0.95 being viewed as fully supported, and those lower considered unsupported.
Maximum likelihood analysis was performed with IQ-Tree using default settings (ver. 1.6.12, L. T. Nguyen, H. A. Schmidt, A. von Haeseler and B. Q. Minh, see https://iqtree.org/; Nguyen et al. 2015). IQ-Tree automatically employs ModelFinder (Kalyaanamoorthy et al. 2017) to select models for each partition by using Bayesian information criterion (BIC) as default. For nrDNA analysis, the models were as follows: 5′ETS TPM2 + F + G4, 18S K2P + I + G4, ITS1 TIM2e + G4, 5.8S JC, ITS2 TNe + G4, 26S TIM3 + F + I and 3′ETS + NTS TN + F + G4; for cpDNA analysis, the models were as follows: CDS TVM + F + I, tRNA K2P, rRNA HKY + F and spacers TVM + F + I + G4. In total, 261 parsimony-informative characters were reported for the nrDNA ML analysis and 3129 were reported for the cpDNA ML analyses. The bootstrap consensus trees were viewed in FigTree and exported to TreeGraph 2 (ver. 2.14.0-771 beta, Bio10, see https://treegraph.bioinfweb.info/; Stöver and Müller 2010) where branches with <50% ultrafast bootstrap (UFBS) support where collapsed to allow for easier comparison with the BI tree. UFBS values of ≥95% were viewed as supported and those below 95% were considered unsupported.
The results of the nrDNA phylogeny were mapped using QGIS (ver. 3.18.3, QGIS Development Team, see http://qgis.org/en/site/index.html). Species distributions were accessed from Atlas of Living Australia (2020) and these data filtered removing samples lacking collection dates. Outliers were reviewed and removed where errors in location or identification were suspected. Taxa were mapped by clades according to the nrDNA phylogeny.
All sequences included in this study were uploaded to GenBank (Table 1). Text files containing the alignment, partitions, BI and ML trees for each dataset (nrDNA and cpDNA) were submitted to TreeBase (accession number 28815).
Results
Nuclear rDNA phylogeny
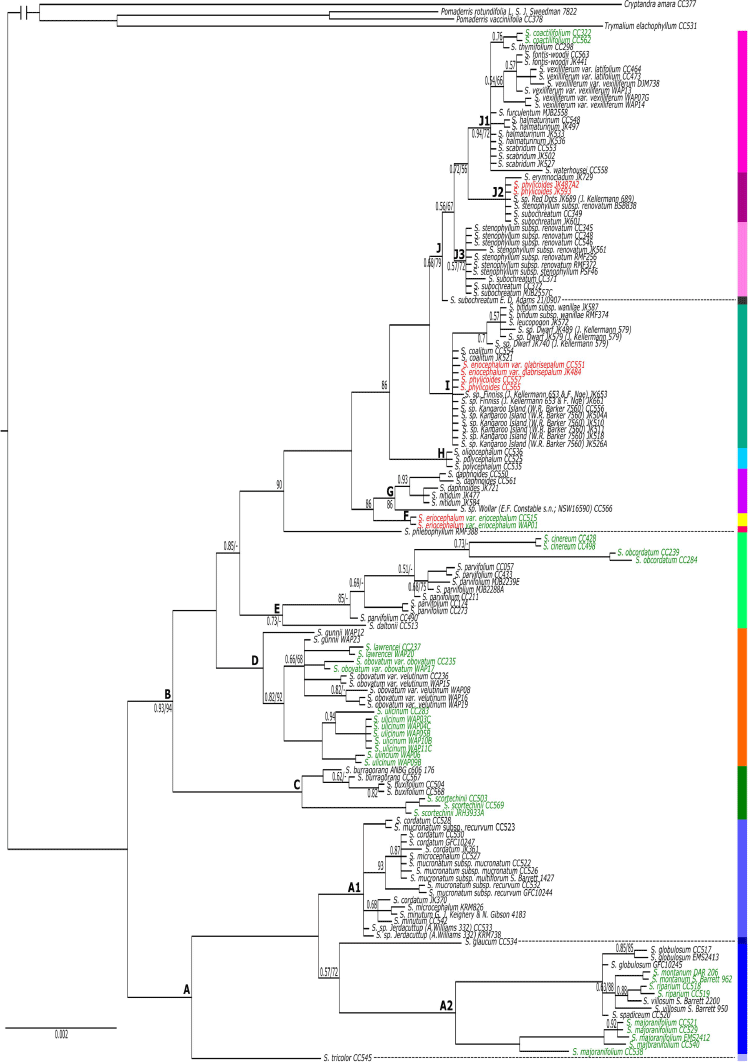
The topologies of BI and ML nrDNA trees were largely congruent, and only the BI tree is shown here, with ML bootstrap values mapped onto it (Fig. 2). A key supported difference in the ML tree was in the placement of S. daltonii as divergent from the S. parvifolium clade (UFBS = 95), with S. obcordatum and S. cinereum being unsupported as successive sisters to a clade of S. parvifolium samples (UFBS = 85 for the former and UFBS = 65 for the latter). Another supported difference was that S. burragorang formed a clade, sister to S. buxifolium (UFBS = 98).
|
Spyridium was resolved as monophyletic (PP = 1.0, UFBS = 100), with a distinct spilt at the base of the tree between largely south-western Australian endemics (Clade A) and largely south-eastern Australian species (Clade B). Clade A (PP = 1.0, UFBS = 96) grouped species endemic to south-western Australia together with one species (S. tricolor) that includes outlying populations disjunct across the Nullarbor Plain (Fig. 3a), whereas Clade B (PP = 0.93, UFBS = 94) included species from south-eastern Australia, one that occurs in both eastern and western Australia (S. subochreatum, Clade J; Fig. 2, 3j), plus two south-western endemics (S. oligocephalum and S. polycephalum, Clade H; Fig. 2, 3h) nested among the south-eastern taxa. Within the largely eastern clade (B), two early diverging lineages were a clade (PP = 1.0, UFBS = 100) of primarily New South Wales (NSW and southern Queensland2) endemics (Clade C; Fig. 2, 3b) and a clade (PP = 1.0, UFBS = 100) of Tasmanian endemics (Clade D; Fig. 2, 3b). The only other Tasmanian endemic, S. obcordatum, was placed in Clade E with S. cinereum, S. parvifolium and S. daltonii (PP = 0.73; Fig. 2, 3d).

|
Of the 45 species, subspecies or varieties represented by more than one accession, 11 (24.4%) were monophyletic and 11 (24.4%) were polyphyletic (Fig. 2, Table 3). One species polyphyletic across disparate clades was S. eriocephalum, although var. eriocephalum was monophyletic (PP = 1.0, UFBS = 100; Clade F), whereas var. glabrisepalum samples were placed in a polytomy (PP = 100, UFBS = 97; Clade I). The other example of a species polyphyletic across disparate clades was S. phylicoides, with the four accessions of this species divided among two clades (two samples in Clade I and two samples in Clade J2; Fig. 2, 4). Owing to a lack of support (BI or ML) for many branches at the tips of the nrDNA phylogeny, the remaining 23 taxa represented by more than one accession (51.1% of included taxa), were unresolved, i.e. with their monophyly neither supported nor strongly refuted.

|

|
Chloroplast genome phylogeny
The topologies of BI and ML cpDNA trees were largely congruent, and only the BI tree is shown here, with ML bootstrap values being mapped onto it (Fig. 5). One unsupported difference in the ML tree was that S. burragorang formed a clade (UFBS = 92), sister to S. buxifolium (UFBS = 100). Spyridium was resolved as monophyletic (PP = 1.0, UFBS = 100). Clades dominated by Tasmanian endemics (K; PP = 1.0, UFBS = 100) and NSW endemics (L; PP = 1.0, UFBS = 100) diverged early in the cpDNA phylogeny (Fig. 5). Spyridium phlebophyllum also diverged early on a branch between Clades K and L (PP = 1.0, UFBS = 98). Clade N predominantly contained taxa endemic to WA, as well as a SA sample of S. tricolor (PP = 1.0, UFBS = 100), a species found in WA and in SA that is disjunct on either side of the Nullarbor Plain (Fig. 3a, 5). One Tasmanian endemic species (S. obcordatum) was sister to S. parvifolium and S. daltonii in Clade O (PP = 1.0, UFBS = 100; Fig. 5). Spyridium polycephalum and S. oligocephalum, both WA endemics, were resolved in Clade M6 (PP = 1.0, UFBS = 100; Fig. 3h, 5) as sister to a WA sample of the widespread, polyphyletic S. subochreatum (Fig. 3j, 5, Table 1). These samples were found nested within the eastern Australian-dominated Clade M (PP = 1.0, UFBS = 100; Fig. 5). Of the 45 taxa that included more than one accession, nine (20.0%) were monophyletic and 30 (66.7%) were polyphyletic (Fig. 5, Table 3), two (4.4%) were paraphyletic (S. parvifolium with respect to S. daltonii in Clade O1 and S. polycephalum with respect to S. oligocephalum in Clade M6) and four (8.8%) were unresolved.

|
Incongruence between the nrDNA and cpDNA phylogenies
Although the nrDNA and cpDNA trees included some nodes in common, there was substantial incongruence between the two phylogenies (Fig. 2, 5). This included differences in the order of divergence, in species relationships (within and between clades) and between the support for species circumscriptions (e.g. monophyly).
Key clades in common between the two trees were the clade of Tasmanian endemic species (nrDNA Clade D; cpDNA Clade K), and the clade of NSW endemic species (nrDNA Clade C; cpDNA Clade L); cpDNA Clade N also included most members of the WA endemic nrDNA Clade A, but excluding S. globulosum, S. spadiceum and one sample of S. majoranifolium that were placed in Clade M4 (Fig. 5). In the clade of NSW endemics, both trees showed similar relationships among species; however, within the Tasmanian and WA clades, relationships varied substantially.
Apart from this, there were many differences in the order of divergence between the two phylogenies, with these differences being particularly apparent at the base of each tree. For example, the base of the nrDNA phylogeny showed an early east–west split (Clade A–Clade B; Fig. 2), a split not supported by the cpDNA phylogeny (Fig. 5). The cpDNA phylogeny, instead, showed the Tasmanian (Clade K) and NSW (Clade L) endemics diverging first (Fig. 5).
There were many differences in species relationships between the two phylogenies, with this incongruence being particularly apparent from Clade M to Clade Q in the cpDNA phylogeny (Fig. 5). For example, S. cinereum is found in the S. parvifolium-dominated clade in the nrDNA phylogeny (Clade E; Fig. 2), but is nested within the SA endemic Clade M1 in the cpDNA tree (Fig. 5). In another example, S. daphnoides and S. nitidum were found to be sister to Spyridium sp. Wollar (E.F.Constable s.n., NSW 16590) Kellermann and S. eriocephalum var. eriocephalum in the nrDNA phylogeny (Clades F and G), but were distantly related in the cpDNA tree (in Clades M1, M5 and P; Fig. 5).
There were also many differences relating to support for species circumscriptions between the two trees (Fig. 2, 5; Table 3). For example, S. majoranifolium was supported as monophyletic in the nrDNA phylogeny (PP = 1.0, UFBS = 100; clade A2) but found to be polyphyletic across Clades M5, N1 and N2 in the cpDNA tree. In another example, S. sp. Kangaroo Island (W.R.Barker 7560) was unresolved in the nrDNA phylogeny (Clade I; PP = 1.0, UFBS = 97; Fig. 2), but polyphyletic across Clades M2 and Q2 in the cpDNA tree (Fig. 5).
Discussion
In this study, we have presented the most comprehensive molecular phylogeny of Spyridium to date, including all recognised species, subspecies and varieties (excluding S. bifidum var. bifidum), as well as six undescribed, phrase-name taxa. These results provide an understanding of biogeographic patterns within the genus, support some species circumscriptions, call others into question, and provide support for recognition of some currently undescribed taxa. However, incongruence between the nrDNA and cpDNA phylogenies means that some relationships within the genus were unable to be resolved by these results alone.
Incongruencies: introgression or incomplete lineage sorting
Although the nrDNA and cpDNA phylogenies contained similarities, there were many differences between the two trees. These were apparent both in the relationships among major clades and when comparing implications for the monophyly, paraphyly or polyphyly of species and infraspecific taxa (Table 3). If the results of the cpDNA tree were viewed alone, as an indication of phylogenetic relationships, revising circumscriptions of half of the species in the genus might seem required. However, conversely, many of these polyphyletic taxa were found to be monophyletic or unresolved in the nrDNA tree (Table 3), providing at least some support for, or not contradicting, most of the current circumscriptions of species.
Two key processes potentially leading to incongruence between nuclear and chloroplast gene trees are introgression, including chloroplast capture (Rieseberg and Soltis 1991; Tsitrone et al. 2003), and incomplete lineage sorting (ILS; Wiley and Lieberman 2011). Distinguishing the relative influence of these processes can be difficult, a problem commonly discussed in plant phylogenetic studies (e.g. Meudt and Bayly 2008; French et al. 2016; Barrett et al. 2018; Schuster et al. 2018). Other explanations for nuclear and chloroplast incongruence could include incomplete taxon sampling, which might result in artefacts of long branch attraction in one dataset over another; however, this is unlikely, given the almost complete taxon sampling in our study.
In our analyses, we have not attempted to distinguish potential instances of introgression from ILS, or the relative importance of each, but we expect, as outlined below, that both processes have potentially had a greater influence on the cpDNA results than on the nrDNA. As such, much of the species polyphyly reported in the cpDNA phylogeny might result from either chloroplast capture or ILS, and the nrDNA tree could better match the species tree for the genus.
In angiosperms, introgression of chloroplast genomes can be more apparent than nrDNA introgression (Rieseberg and Soltis 1991; Tsitrone et al. 2003). This is because chloroplasts are (generally) maternally inherited, not recombinant or subject to concerted evolution that can obscure signals of introgression in nrDNA (Álvarez and Wendel 2003), and have lower mutation rates (Palmer 1987), meaning that, in the absence of selective pressure or stochastic events, the signal of past cpDNA introgression is preserved. Potential introgression has been inferred in other studies of Rhamnaceae, including in Pomaderris (Nge et al. 2021c), Discaria Hook. (Aagesen et al. 2005; Medan et al. 2012) and Ceanothus L. (Hardig et al. 2002; Burge et al. 2013). One possible example of introgression contributing to incongruent results in the current study may be S. sp. Kangaroo Island (W.R.Barker 7560) Kellermann, which was unresolved in the nrDNA phylogeny (Clade I; Fig. 2), but was polyphyletic across disparate clades (M2 and Q2) in the cpDNA tree (Fig. 5). Spyridium sp. Kangaroo Island (W.R.Barker 7560) has an overlapping distribution, with several species found in the same subclades in the cpDNA phylogeny (e.g. S. coalitum, clade M2; and S. thymifolium, clade Q2); therefore, chloroplast capture through introgression is a possible explanation of this incongruence.
Another potential explanation for differences between cpDNA and nrDNA phylogenies is ILS. This occurs when the coalescence for alleles at a locus present in different species pre-dates the speciation event that gave rise to these species, and it can result in a gene tree that is incongruent with the species tree (Wiley and Lieberman 2011) or with other gene trees. The usual expectation is that ILS has a greater confounding influence on phylogenetic analyses of nuclear data than of chloroplast data, because the effective population size of haploid chloroplast markers is only one quarter that of diploid nuclear markers, and this should lead to quicker coalescence of chloroplast sequences than of nuclear alleles (Birky et al. 1989). However, for nrDNA markers, as used in our study, the processes of concerted evolution (Arnheim 1983), which homogenise rDNA sequences within the genomes of organisms, can reduce the effective population size of rDNA markers relative to other nuclear sequences, by as much as 200-fold (Buckler and Holtsford 1996); this leads to substantially shorter coalescence times, and reduces the influence of ILS on nrDNA phylogenies.
For the reasons outlined above, in the following discussions of biogeography and taxonomy we place most emphasis on the nrDNA results but also incorporate additional details from cpDNA trees where relevant.
There is substantial scope to further explore the potential influence of introgression and ILS in the history of Spyridium. Multi-locus nuclear datasets, for instance, would be more amenable to statistical analyses of introgression (Joly et al. 2009; Joly 2012; García et al. 2017) than is nrDNA, and could give a stronger phylogenetic signal to assess incongruence between nuclear and plastid gene trees. Nonetheless, the data presented here highlight that nuclear and chloroplast incongruence is conspicuous in the genus.
Broad biogeographic patterns in Spyridium
This study has provided the first comprehensive (all species) phylogenetic assessment of biogeographic patterns in Spyridium. Our inferences (below) are based on the molecular phylogenies and insight from other studies of biogeography in southern Australian. We focus on broad (continent scale) patterns in general terms. The work presented here could be extended by additional analyses, such as, for example, using dated trees or probabilistic biogeographic modelling (e.g. Ree and Smith 2008; Matzke 2013), although these are not explored here.
An early split in Spyridium has been identified in the nrDNA phylogeny, between the east and west of Australia across the Nullarbor Plain (Clades A and B; Fig. 2, 3a–j). There are many examples of east–west divergences in plants distributed across southern Australia, including Phebalium Vent. (Mole et al. 2004), Eucalyptus L’Hér. subgenus Eucalyptus (Ladiges et al. 2012), Goodeniaceae R.Br. (Jabaily et al. 2014), Xanthorrhoea Sol. ex Sm. (McLay et al. 2021), Adenanthos Labill. (Nge et al. 2021a) and Pomaderris (Nge et al. 2021c). The Nullarbor Plain disjunction in a range of plant groups has been related to vicariance associated with uplifting and climatic cooling in the mid-Miocene (Crisp and Cook 2007), including in Pomaderris (Nge et al. 2021c), a close relative of Spyridium in the tribe Pomaderreae. Nge et al. (2021c) concluded that Pomaderris was widespread throughout southern and eastern Australia until c. 14 Ma, when the Nullarbor Plain uplift occurred, with subsequent rapid ‘within region’ diversification in eastern Australia from c. 10 Ma, and little movement across biomes since. Although we did not use dated trees to determine diversification dates (as per Nge et al. 2021c), a similar explanation for the early east–west divergence in Spyridium could be inferred from our results.
Assuming the deep east–west divergence in Spyridium relates to formation of the Nullarbor Plain, the nrDNA tree suggests that up to three lineages in the genus have potentially dispersed across the plain subsequent to this early east–west divergence (Fig. 2, 3a, h, j). An east-to-west dispersal of S. subochreatum is inferred from both nrDNA and cpDNA trees, because the species is nested within eastern taxa and its distribution extends to just west of the Nullarbor Plain (Fig. 3j). Conversely, for S. tricolor, a west-to-east dispersal could be inferred, because the SA sample of this species (CC545; Fig. 3a) groups with western taxa. However, because it is sister to other western taxa in Clade A in the nrDNA phylogeny (Fig. 2), a reverse scenario could not be ruled out. Like S. subochreatum, S. tricolor grows on sandy soils and limestone (FloraBase – the Western Australian Flora, see https://florabase.dpaw.wa.gov.au/, accessed 7 May 2021), suggesting it could have made use of land connections south of the Nullarbor Plain that have been exposed at times of lower sea level since the late Pliocene (Nelson 1974; Wright and Ladiges 1997). An alternative explanation, that S. tricolor was widespread across this region and became disjunct during the Nullarbor Plain uplift, would require retention of morphological resemblance, such that it is recognised as a single species, despite a considerable geographic disjunction, for a substantial period of time since the mid-Miocene. Population-level sampling of S. tricolor using variable genomic markers could provide a greater insight into geographic history of the species and be used to further test its taxonomic circumscription.
The third lineage for which dispersal over the Nullarbor Plain might possibly be inferred is that including S. polycephalum and S. oligocephalum (Fig. 2, 3h, 5). A deep east-to-west dispersal event, before the diversification of the two species, is inferred from both the nrDNA and cpDNA trees, because this clade was nested within species from east of the Nullarbor Plain. Evidence of early east–west vicariance across southern Australia, followed by subsequent dispersal events such as these have been inferred in studies of other plant groups, such as, for example, in Eucalyptus subgenus Eucalyptus (Wright and Ladiges 1997), Thelymitra J.R.Forst. & G.Forst. (Nauheimer et al. 2018), Calytrix tetragona Labill. (Nge et al. 2021b) and Pomaderris (Nge et al. 2021c). Despite this, an alternative explanation of vicariance to account for the Western Australian distribution of the S. polycephalum–S. oligocephalum clade cannot be immediately discounted on the basis of our data. Although a vicariance explanation is less parsimonious because it would infer extinction of multiple lineages in western Australia, such reasoning assumes that multiple extinctions are less probable than is a single dispersal, which might not be true (Sanmartín and Meseguer 2016), for example, in the face of substantial climatic change in Australia since the mid-Miocene. A robust time-calibrated phylogeny for Spyridium could help corroborate one of these alternative scenarios.
Within the eastern Australian branch of the nrDNA phylogeny (Clade B, Fig. 2), an early NSW divergence is inferred (Fig. 3b). This deep divergence of NSW endemics from other south-eastern Australian taxa occurs near a broadly defined area that has been termed the southern transition zone (STZ; Fig. 3b) by Milner et al. (2012). The STZ is found east of the Great Dividing Range (GDR) and north of the Victoria–NSW border and is identified as a region where genetic or distributional discontinuities are seen in a range of taxa, but with the exact position of the discontinuities being dependent on habitat requirements of individual species and potentially different timescales of divergence (Milner et al. 2012). Other plant taxa showing genetic breaks across the STZ include Hardenbergia violacea (Schneev.) Stearn (Larcombe et al. 2011), Lomatia R.Br. (Milner et al. 2012), Callitris rhomboidea R.Br. ex Rich. & A.Rich. (Worth et al. 2018) and Xanthorrhoea (McLay et al. 2021). Spyridium provides a further example of this pattern, although potential drivers of the divergence are unclear in this case.
Tasmanian endemics (excluding S. obcordatum) were found in a single, early diverging clade separate from their mainland counterparts in the nrDNA tree (Clade D, Fig. 2, 3c), suggesting early vicariance of Spyridium across Bass Strait. This early divergence and diversification of Tasmanian endemics is also supported by the cpDNA phylogeny (Clade K, Fig. 5) and the findings of Kellermann et al. (2005) and Hauenschild et al. (2018). The continued barriers to dispersal and gene-flow are likely to be the inundation of Bass Strait during interglacial periods (Galloway and Kemp 1981) and the semi-arid climate of the land-bridge exposed during glacial periods (Kirkpatrick and Fowler 1998). Major glacial and interglacial fluctuations occurred throughout the Quaternary (c. 2.2 Ma to c. 10 000 years ago; Hope 1994; Quilty 1994) and their resulting climatic extremes have been inferred to contribute to the limited distribution of narrow-range endemism in Spyridium in Tasmania (Coates and Kirkpatrick 1999).
Recent dispersal or gene flow between Victoria and Tasmania are here inferred for the lineage represented by S. obcordatum, the only endemic Tasmanian species not placed in Clades D or K (Fig. 2, 3d, 5), and several widespread taxa, including S. eriocephalum var. eriocephalum, S. vexilliferum var. vexilliferum and S. parvifolium (Fig. 2, 3d, f, j). Accessions of each of these widespread taxa collected from Tasmania (and Flinders Island for S. parvifolium) were found within the same clade as samples of the same taxa from Victoria (Table 1). Similar patterns of recent gene-flow between Victoria and Tasmania have been inferred in other plant groups, including Eucalyptus globulus Labill. (Freeman et al. 2001), Hardenbergia violacea (Larcombe et al. 2011), Correa Andrews (French et al. 2016), Zieria veronicea Sm. (Neal et al. 2019), Xanthorrhoea (McLay et al. 2021) and a range of other species (Worth et al. 2017). Evidence suggests that at least some areas of the Bassian Plain were covered in eucalypt woodland habitat (Hope 1978, 1994; Kirkpatrick and Fowler 1998) which could have been suitable for S. parvifolium, S. vexilliferum var. vexilliferum and S. eriocephalum var. eriocephalum (VicFlora 2018), i.e. potentially facilitating over-land rather than over-water dispersal between Victoria and Tasmania.
Review of circumscriptions of species
The molecular phylogenies support the circumscriptions of several Spyridium species, but raise questions about others. A quarter of the taxa represented by more than one accession were identified as monophyletic in the nrDNA phylogeny, with approximately one-third being resolved as polyphyletic, and the remainder being unresolved (Fig. 2, Table 3). Of the monophyletic taxa resolved in the nrDNA tree, several were also found to be monophyletic in the cpDNA phylogeny (e.g. S. obcordatum, S. scortechinii and S. montanum), providing additional support for these circumscriptions of species (Fig. 5, Table 3). Of the polyphyletic taxa in the nrDNA phylogeny, two of the most notable were distributed across disparate clades, namely, S. eriocephalum and S. phylicoides (Fig. 2). Given that both of these species were also resolved in separate clades in the cpDNA tree (Fig. 5), they are discussed in more detail below, along with several associated phrase-name taxa.
Spyridium eriocephalum
Spyridium eriocephalum is polyphyletic and requires taxonomic revision, because its two varieties were found in distinct clades in both nrDNA and cpDNA phylogenies (Clades F and I, Fig. 2; Clades M1 and Q2, Fig. 5). Spyridium eriocephalum var. eriocephalum is monophyletic (albeit with limited sampling, but from separated localities) and geographically distinct from other taxa in the nrDNA tree (Fig. 2, 3f). Spyridium eriocephalum var. glabrisepalum is unresolved in a polytomy in the nrDNA phylogeny (Clade I, Fig. 3) with several other taxa (Fig. 2, 3i). The two varieties of S. eriocephalum are for the most part geographically distinct, with the exception being some overlap on Kangaroo Island (J. Kellermann, unpubl. data). The typical variety is widespread in south-eastern Australia (SA, Victoria, NSW and Tasmania), whereas var. glabrisepalum is restricted to Kangaroo Island. The two varieties are also morphologically distinguished by the presence of woolly sepal hairs (var. eriocephalum) versus hairless sepals that are instead glabrous-viscid (var. glabrisepalum; Canning 1986). Although the two taxa appear distinct (from each other) in both phylogenies, given that the two samples of var. glabrisepalum are placed with some other Kangaroo Island endemic taxa (e.g. S. coalitum) in the nrDNA tree, it is possible that introgression may be influencing this placement (Fig. 2). However, placement of var. glabrisepalum in the cpDNA phylogeny is somewhat incongruent although supported, with samples being placed with accessions representing other taxa collected from a range of sites from SA to Tasmania (Fig. 5). Additional morphological or molecular work is recommended to re-assess these taxa and their relationships.
Spyridium phylicoides, S. sp. Dwarf (J.Kellermann 579) and S. sp. Red Dots (J.Kellermann 689)
Spyridium phylicoides is polyphyletic and requires taxonomic revision. In both phylogenies, samples of S. phylicoides were found in two clades (Clades I and J2, Fig. 2; Clades M1, M3 and Q1, Fig. 5). There is no biogeographic pattern to these (Fig. 4) and perhaps further unidentified forms exist within S. phylicoides, in addition to the two forms already given phrase names, namely, S. sp. Dwarf (J.Kellermann 579) Kellermann and S. sp. Red Dots (J.Kellermann 689) Kellermann (Table 2). Spyridium sp. Dwarf (J.Kellermann 579) is distinguished from S. phylicoides by generally smaller leaves and low-growing, almost prostrate habit (J. Kellermann, unpubl. data). However, the distribution of samples of S. sp. Dwarf (J.Kellermann 579; Fig. 5) overlaps with that of many species found in SA (Fig. 3f, g, i, j); therefore, it is possible that the incongruent result for this taxon (unresolved in the nrDNA phylogeny and polyphyletic in the cpDNA tree; Fig. 2, 5) may be attributed to chloroplast capture or ILS. Only one accession of S. sp. Red Dots (J.Kellermann 689) was included in this study and support for this taxon as distinct from S. phylicoides in either phylogeny is limited (Clade J2, Fig. 2; Clade M3, Fig. 5). We recommend further investigation into both phrase-name taxa, particularly S. sp. Dwarf (J.Kellermann 579), and a more detailed investigation into the circumscription of S. phylicoides.
Conclusions
Here we have presented the first comprehensive phylogenies of the genus Spyridium, representing all described species and utilising both nrDNA and whole chloroplast genomes. Most incongruencies between the two trees could relate to introgression and chloroplast capture or ILS.
We found evidence of an early east–west split at the base of the nrDNA phylogeny and early diverging clades dominated by Tasmanian and NSW endemics. Our trees provide evidence of two subsequent within-species dispersal events across the Nullarbor Plain (S. subochreatum and S. tricolor) as well as a possible dispersal and diversification of a lineage including the south-western Australian endemics S. polycephalum and S. oligocephalum (although a vicariance explanation is also plausible in that case). In Tasmania, we found S. obcordatum to be the result of a recent dispersal and subsequent diversification event, and evidence of recent gene-flow between Tasmania and Victoria in several widespread taxa (including S. vexilliferum var. vexilliferum).
Eleven taxa were supported as monophyletic in the nrDNA phylogeny and the following two were polyphyletic across disparate clades, requiring taxonomic review: S. eriocephalum (with two genetically distinct varieties) and S. phylicoides.
Data availability
The data that support this study are available in TreeBASE at https://www.treebase.org/treebase-web/home.html (Accession number 28815).
Conflicts of interest
Michael J. Bayly is an Associate Editor for Australian Systematic Botany. Despite this relationship, he did not at any stage have Associate Editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Australian Systematic Botany encourages its editors to publish in the journal and they are kept totally separate from the decision-making process for their manuscripts. The authors declare that they have no further conflicts of interest.
Declaration of funding
This project was supported by a Holsworth Wildlife Research Endowment administered by The Ecological Society of Australia, and the Hansjörg Eichler Scientific Research Fund provided by the Australasian Systematic Botany Society. Support was also provided by a Megan Klemm Postgraduate Research Award, a Sophie Ducker Postgraduate Scholarship (both administered by The University of Melbourne Botany Foundation) and a Dame Margaret Blackwood Soroptimist Scholarship (managed by Soroptimist International). The revision of the family Rhamnaceae for the Flora of Australia (Kellermann et al. 2022) has been supported by several grants from the Australian Biological Resources Study (Canberra, ACT, Australia).
Acknowledgements
We thank Matthew Dell, Rose Barrett, Mark Wapstra, Stephen Bell, Libby Sandiford, Gillian Craig, Kenneth Mills, John Hosking and Daniel Murphy for collecting silica samples and vouchers. Thanks go to Barbara Rye for advice related to WA Spyridium. We acknowledge the Western Australian Herbarium for providing destructive samples for molecular analysis. Thanks go to Todd McLay, Erin Batty and Stephen Wilcox for assistance with molecular work, and Matthew Dell and Francis Nge for general support for the project. We appreciate the assistance Joanne Birch, Will Neal and Margaret Brookes have provided while lodging samples in the University of Melbourne Herbarium. We thank Andrew Drinnan, Tanja Schuster and Peter Vesk for providing suggestions related to the project direction during committee meetings. Finally, we thank the Halls Gap Outdoor Community Facebook Group for helping retrieve a lost field book.
References
Aagesen L, Medan D, Kellermann J, Hilger HH (2005) Phylogeny of the tribe Colletieae (Rhamnaceae) – a sensitivity analysis of the plastid region trnL-trnF combined with morphology. Plant Systematics and Evolution 250, 197–214.| Phylogeny of the tribe Colletieae (Rhamnaceae) – a sensitivity analysis of the plastid region trnL-trnF combined with morphology.Crossref | GoogleScholarGoogle Scholar |
Álvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution 29, 417–434.
| Ribosomal ITS sequences and plant phylogenetic inference.Crossref | GoogleScholarGoogle Scholar | 14615184PubMed |
Arnheim N (1983) Concerted evolution of multigene families. In ‘Evolution of genes and proteins’. (Eds M Nei, R Koehn) pp. 38–61. (Sinauer: Sunderland, MA, USA)
Atlas of Living Australia (2020) Occurrence records download on 2020-06-26: taxon_name: ‘Spyridium’. Available at
| Crossref | [Verified 26 June 2020]
Barker WR (2005) Standardising informal names in Australian publications. Australian Systematic Botany Society Newsletter 122, 11–12.
Barrett RA, Bayly MJ, Duretto MF, Forster PI, Ladiges PY, Cantrill DJ (2018) Phylogenetic analysis of Zieria (Rutaceae) in Australia and New Caledonia based on nuclear ribosomal DNA shows species polyphyly, divergent paralogues and incongruence with chloroplast DNA. Australian Systematic Botany 31, 16–47.
| Phylogenetic analysis of Zieria (Rutaceae) in Australia and New Caledonia based on nuclear ribosomal DNA shows species polyphyly, divergent paralogues and incongruence with chloroplast DNA.Crossref | GoogleScholarGoogle Scholar |
Bentham G (1863) Rhamneae. In ‘Flora Australiensis, a description of the plants of the Australian territory. Vol. 1: Ranunculaceae to Anacardiaceae’. pp. 409–445. (Lovell Reeve and Co: London, UK)
Birky CW, Fuerst P, Maruyama T (1989) Organelle gene diversity under migration, mutation, and drift: equilibrium expectations, approach to equilibrium, effects of heteroplasmic cells, and comparison to nuclear genes. Genetics 121, 613–627.
| Organelle gene diversity under migration, mutation, and drift: equilibrium expectations, approach to equilibrium, effects of heteroplasmic cells, and comparison to nuclear genes.Crossref | GoogleScholarGoogle Scholar | 2714640PubMed |
Buckler ES, Holtsford TP (1996) Zea systematics: ribosomal ITS evidence. Molecular Biology and Evolution 13, 612–622.
| Zea systematics: ribosomal ITS evidence.Crossref | GoogleScholarGoogle Scholar | 8882504PubMed |
Burge DO, Hopkins R, Tsai Y-HE, Manos PS (2013) Limited hybridization across an edaphic disjunction between the gabbro-endemic shrub Ceanothus roderickii (Rhamnaceae) and the soil-generalist Ceanothus cuneatus. American Journal of Botany 100, 1883–1895.
| Limited hybridization across an edaphic disjunction between the gabbro-endemic shrub Ceanothus roderickii (Rhamnaceae) and the soil-generalist Ceanothus cuneatus.Crossref | GoogleScholarGoogle Scholar | 24018856PubMed |
Canning EM (1986) Spyridium. In ‘Flora of South Australia’, Vol. 2. (Eds JP Jessop, HR Toelken) (Government Printer: Adelaide, SA, Australia)
CHAH (2020) Spyridium Fenzl. In ‘Australian Plant Census’. (Council of Heads of Australian Herbaria) Available at https://biodiversity.org.au/nsl/services/search?product=APNI&tree.id=&name=Spyridium&inc._scientific=&inc._cultivar=&inc._other=&max=100&display=&search=true [Verified 17 November 2020]
Clowes C, Fowler RM, Brown GK, Bayly MJ (2018) The complete chloroplast genome sequence of Spyridium parvifolium var. parvifolium (family Rhamnaceae; tribe Pomaderreae). Mitochondrial DNA. Part B, Resources 3, 807–809.
| The complete chloroplast genome sequence of Spyridium parvifolium var. parvifolium (family Rhamnaceae; tribe Pomaderreae).Crossref | GoogleScholarGoogle Scholar | 33474330PubMed |
Coates F, Kirkpatrick JB (1999) Is geographic range correlated with climatic range in Australian Spyridium taxa? Australian Journal of Botany 47, 755–767.
| Is geographic range correlated with climatic range in Australian Spyridium taxa?Crossref | GoogleScholarGoogle Scholar |
Crisp MD, Cook LG (2007) A congruent molecular signature of vicariance across multiple plant lineages. Molecular Phylogenetics and Evolution 43, 1106–1117.
| A congruent molecular signature of vicariance across multiple plant lineages.Crossref | GoogleScholarGoogle Scholar | 17434758PubMed |
Department of Agriculture Water and the Environment (2020) Interim Biogeographic Regionalisation for Australia (Regions – States and Territories) v. 7 (IBRA). Available at https://www.environment.gov.au/fed/catalog/search/resource/details.page?uuid=%7B1273FBE2-F266-4F3F-895D-C1E45D77CAF5%7D [Verified 26 July 2020]
Doyle J, Doyle JL (1987) Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochemical Bulletin 19, 11–15.
Freeman JS, Jackson HD, Steane DA, McKinnon GE, Dutkowski GW, Potts BM, Vaillancourt RE (2001) Chloroplast DNA phylogeography of Eucalyptus globulus. Australian Journal of Botany 49, 585–596.
| Chloroplast DNA phylogeography of Eucalyptus globulus.Crossref | GoogleScholarGoogle Scholar |
French PA, Brown GK, Bayly MJ (2016) Incongruent patterns of nuclear and chloroplast variation in Correa (Rutaceae): introgression and biogeography in south-eastern Australia. Plant Systematics and Evolution 302, 447–468.
| Incongruent patterns of nuclear and chloroplast variation in Correa (Rutaceae): introgression and biogeography in south-eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Galloway RW, Kemp EM (1981) Late Cainozoic environments of Australia. In ‘Ecological Biogeography of Australia’, Vol. 1. (Ed. A Keast) pp. 53–80. (Junk: The Hague, Netherlands)
García N, Folk RA, Meerow AW, Chamala S, Gitzendanner MA, de Oliveira RS, Soltis DE, Soltis PS (2017) Deep reticulation and incomplete lineage sorting obscure the diploid phylogeny of rain-lilies and allies (Amaryllidaceae tribe Hippeastreae). Molecular Phylogenetics and Evolution 111, 231–247.
| Deep reticulation and incomplete lineage sorting obscure the diploid phylogeny of rain-lilies and allies (Amaryllidaceae tribe Hippeastreae).Crossref | GoogleScholarGoogle Scholar | 28390909PubMed |
Hardig TM, Soltis PS, Soltis DE, Hudson RB (2002) Morphological and molecular analysis of putative hybrid speciation in Ceanothus (Rhamnaceae). Systematic Botany 27, 734–746.
Hauenschild F, Matuszak S, Muellner-Riehl AN, Favre A (2016) Phylogenetic relationships within the cosmopolitan buckthorn family (Rhamnaceae) support the resurrection of Sarcomphalus and the description of Pseudoziziphus gen. nov. Taxon 65, 47–64.
| Phylogenetic relationships within the cosmopolitan buckthorn family (Rhamnaceae) support the resurrection of Sarcomphalus and the description of Pseudoziziphus gen. nov.Crossref | GoogleScholarGoogle Scholar |
Hauenschild F, Favre A, Michalak I, Muellner‐Riehl AN (2018) The influence of the Gondwanan breakup on the biogeographic history of the ziziphoids (Rhamnaceae). Journal of Biogeography 45, 2669–2677.
| The influence of the Gondwanan breakup on the biogeographic history of the ziziphoids (Rhamnaceae).Crossref | GoogleScholarGoogle Scholar |
Hope GS (1978) The late Pleistocene and Holocene vegetational history of Hunter Island, north-western Tasmania. Australian Journal of Botany 26, 493–514.
| The late Pleistocene and Holocene vegetational history of Hunter Island, north-western Tasmania.Crossref | GoogleScholarGoogle Scholar |
Hope GS (1994) Quaternary vegetation. In ‘History of the Australian vegetation: Cretaceous to recent’. (Ed. GS Hope) pp. 368–389. (University of Adelaide Press: Adelaide, SA, Australia)
Jabaily RS, Shepherd KA, Gardner AG, Gustafsson MHG, Howarth DG, Motley TJ (2014) Historical biogeography of the predominantly Australian plant family Goodeniaceae. Journal of Biogeography 41, 2057–2067.
| Historical biogeography of the predominantly Australian plant family Goodeniaceae.Crossref | GoogleScholarGoogle Scholar |
Jobes DV, Thien LB (1997) A conserved motif in the 5.8 S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) sequences. Plant Molecular Biology Reporter 15, 326–334.
| A conserved motif in the 5.8 S ribosomal RNA (rRNA) gene is a useful diagnostic marker for plant internal transcribed spacer (ITS) sequences.Crossref | GoogleScholarGoogle Scholar |
Joly S (2012) JML: testing hybridization from species trees. Molecular Ecology Resources 12, 179–184.
| JML: testing hybridization from species trees.Crossref | GoogleScholarGoogle Scholar | 21899723PubMed |
Joly S, McLenachan PA, Lockhart PJ (2009) A statistical approach for distinguishing hybridization and incomplete lineage sorting. American Naturalist 174, E54–E70.
| A statistical approach for distinguishing hybridization and incomplete lineage sorting.Crossref | GoogleScholarGoogle Scholar | 19519219PubMed |
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587–589.
| ModelFinder: fast model selection for accurate phylogenetic estimates.Crossref | GoogleScholarGoogle Scholar | 28481363PubMed |
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30, 3059–3066.
| MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform.Crossref | GoogleScholarGoogle Scholar | 12136088PubMed |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772–780.
| MAFFT multiple sequence alignment software version 7: improvements in performance and usability.Crossref | GoogleScholarGoogle Scholar | 23329690PubMed |
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649.
| Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data.Crossref | GoogleScholarGoogle Scholar | 22543367PubMed |
Kellermann J (2007) Re-instatement of the name Spyridium waterhousei from Kangaroo Island, South Australia, with a short history of the tribe Pomaderreae (Rhamnaceae). Journal of the Adelaide Botanic Gardens 21, 55–62.
Kellermann J (2021) The importance of the ‘h’ – parahomonymy in Trymalium (Rhamnaceae: Pomaderreae). Swainsona 35, 23–29.
Kellermann J, Barker WR (2012) Revision of the Spyridium bifidum–S. halmaturinum complex (Rhamnaceae: Pomaderreae) from South Australia and Victoria. Muelleria 30, 26–58.
Kellermann J, Udovicic F (2007) Large indels obscure phylogeny in analysis of chloroplast DNA (trnL–F) sequence data: Pomaderreae (Rhamnaceae) revisited. Telopea 12, 1–22.
Kellermann J, Udovicic F, Ladiges PY (2005) Phylogenetic analysis and generic limits of the tribe Pomaderreae (Rhamnaceae) using internal transcribed spacer DNA sequences. Taxon 54, 619–631.
| Phylogenetic analysis and generic limits of the tribe Pomaderreae (Rhamnaceae) using internal transcribed spacer DNA sequences.Crossref | GoogleScholarGoogle Scholar |
Kellermann J, Rye BL, Thiele KR (2008) Nomenclatural notes, typifications and name changes in Trymalium (Rhamnaceae: Pomaderreae). Transactions of the Royal Society of South Australia 132, 18–28.
| Nomenclatural notes, typifications and name changes in Trymalium (Rhamnaceae: Pomaderreae).Crossref | GoogleScholarGoogle Scholar |
Kellermann J, Thiele KR, Udovicic F, Walsh NG (2022) Rhamnaceae. In ‘Flora of Australia’. (Ed. PG Kodela) (Australian Biological Resources Study: Canberra, ACT, Australia) Available at https://profiles.ala.org.au/opus/foa/profile/Rhamnaceae [Verified 10 May 2022]
Kirkpatrick JB, Fowler M (1998) Locating likely glacial forest refugia in Tasmania using palynological and ecological information to test alternative climatic models. Biological Conservation 85, 171–182.
| Locating likely glacial forest refugia in Tasmania using palynological and ecological information to test alternative climatic models.Crossref | GoogleScholarGoogle Scholar |
Ladiges PY, Bayly MJ, Nelson G (2012) Searching for ancestral areas and artifactual centers of origin in biogeography: with comment on east–west patterns across southern Australia. Systematic Biology 61, 703–708.
| Searching for ancestral areas and artifactual centers of origin in biogeography: with comment on east–west patterns across southern Australia.Crossref | GoogleScholarGoogle Scholar | 22234419PubMed |
Larcombe MJ, McKinnon GE, Vaillancourt RE (2011) Genetic evidence for the origins of range disjunctions in the Australian dry sclerophyll plant Hardenbergia violacea. Journal of Biogeography 38, 125–136.
| Genetic evidence for the origins of range disjunctions in the Australian dry sclerophyll plant Hardenbergia violacea.Crossref | GoogleScholarGoogle Scholar |
Matzke NJ (2013) Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing. Frontiers of Biogeography 5, 242–247..
| Probabilistic historical biogeography: new models for founder-event speciation, imperfect detection, and fossils allow improved accuracy and model-testing.Crossref | GoogleScholarGoogle Scholar |
McLay TG (2017) High quality DNA extraction protocol from recalcitrant plant tissue. Available at https://www.protocols.io/view/high-quality-dna-extraction-protocol-from-recalcit-i8jchun [Verified 12 July 2018]
McLay TGB, Ladiges PY, Doyle SR, Bayly MJ (2021) Phylogeographic patterns of the Australian grass trees (Xanthorrhoea Asphodelaceae) shown using targeted amplicon sequencing. Australian Systematic Botany 34, 206–225.
| Phylogeographic patterns of the Australian grass trees (Xanthorrhoea Asphodelaceae) shown using targeted amplicon sequencing.Crossref | GoogleScholarGoogle Scholar |
Medan D, Arbetman M, Chaia EE, Premoli AC (2012) Interspecific and intergeneric hybridization in south American Rhamnaceae-Colletieae. Plant Systematics and Evolution 298, 1425–1435.
| Interspecific and intergeneric hybridization in south American Rhamnaceae-Colletieae.Crossref | GoogleScholarGoogle Scholar |
Meudt HM, Bayly MJ (2008) Phylogeographic patterns in the Australasian genus Chionohebe (Veronica s.l., Plantaginaceae) based on AFLP and chloroplast DNA sequences. Molecular Phylogenetics and Evolution 47, 319–338.
| Phylogeographic patterns in the Australasian genus Chionohebe (Veronica s.l., Plantaginaceae) based on AFLP and chloroplast DNA sequences.Crossref | GoogleScholarGoogle Scholar | 18299210PubMed |
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In ‘Proceedings of the Gateway Computing Environments Workshop (GCE)’, 14 November 2010, New Orleans, LA, USA. INSPEC Accession Number 11705685. (IEEE)
| Crossref |
Milner ML, Rossetto M, Crisp MD, Weston PH (2012) The impact of multiple biogeographic barriers and hybridization on species‐level differentiation. American Journal of Botany 99, 2045–2057.
| The impact of multiple biogeographic barriers and hybridization on species‐level differentiation.Crossref | GoogleScholarGoogle Scholar | 23221499PubMed |
Mole BJ, Udovicic F, Ladiges PY, Duretto MF (2004) Molecular phylogeny of Phebalium (Rutaceae: Boronieae) and related genera based on the nrDNA regions ITS 1+ 2. Plant Systematics and Evolution 249, 197–212.
| Molecular phylogeny of Phebalium (Rutaceae: Boronieae) and related genera based on the nrDNA regions ITS 1+ 2.Crossref | GoogleScholarGoogle Scholar |
Nauheimer L, Schley RJ, Clements MA, Micheneau C, Nargar K (2018) Australasian orchid biogeography at continental scale: molecular phylogenetic insights from the sun orchids (Thelymitra, Orchidaceae). Molecular Phylogenetics and Evolution 127, 304–319.
| Australasian orchid biogeography at continental scale: molecular phylogenetic insights from the sun orchids (Thelymitra, Orchidaceae).Crossref | GoogleScholarGoogle Scholar | 29870858PubMed |
Neal WC, James EA, Bayly MJ (2019) Phylogeography, classification and conservation of pink zieria (Zieria veronicea; Rutaceae): influence of changes in climate, geology and sea level in south-eastern Australia. Plant Systematics and Evolution 305, 503–520.
| Phylogeography, classification and conservation of pink zieria (Zieria veronicea; Rutaceae): influence of changes in climate, geology and sea level in south-eastern Australia.Crossref | GoogleScholarGoogle Scholar |
Nelson EC (1974) Disjunct plant distributions on the south-western Nullarbor Plain, Western Australia. Journal of the Royal Society of Western Australia 57, 105–117.
Nge FJ, Biffin E, Thiele KR, Waycott M (2021a) Reticulate evolution, ancient chloroplast haplotypes, and rapid radiation of the Australian plant genus Adenanthos (Proteaceae). Frontiers in Ecology and Evolution 8, 616741
Nge FJ, Biffin E, Waycott M, Thiele KR (2021b) Phylogenomics and continental biogeographic disjunctions: insight from the Australian starflowers (Calytrix). American Journal of Botany 109, 291–308.
| Phylogenomics and continental biogeographic disjunctions: insight from the Australian starflowers (Calytrix).Crossref | GoogleScholarGoogle Scholar |
Nge FJ, Kellermann J, Biffin E, Waycott M, Thiele KR (2021c) Historical biogeography of Pomaderris (Rhamnaceae): continental vicariance in Australia and repeated independent dispersals to New Zealand. Molecular Phylogenetics and Evolution 158, 107085
| Historical biogeography of Pomaderris (Rhamnaceae): continental vicariance in Australia and repeated independent dispersals to New Zealand.Crossref | GoogleScholarGoogle Scholar | 33540078PubMed |
Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32, 268–274.
| IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies.Crossref | GoogleScholarGoogle Scholar | 25371430PubMed |
Palmer JD (1987) Chloroplast DNA evolution and biosystematic uses of chloroplast DNA variation. American Naturalist 130, S6–S29.
| Chloroplast DNA evolution and biosystematic uses of chloroplast DNA variation.Crossref | GoogleScholarGoogle Scholar |
Perrin D (2018) ‘Dictionary of botanical names: Australian plant names, meanings, derivation and application.’ (JT Press)
Quilty PG (1994) The background: 144 million years of Australian palaeoclimate and palaeogeography. In ‘History of the Australian vegetation: Cretaceous to recent’. (Ed. RS Hill) Vol. 14, pp. 14–43. (University of Adelaide Press: Adelaide, SA, Australia)
Ree RH, Smith SA (2008) Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology 57, 4–14.
| Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis.Crossref | GoogleScholarGoogle Scholar | 18253896PubMed |
Richardson JE, Chatrou LW, Mols JB, Erkens RHJ, Pirie MD (2004) Historical biogeography of two cosmopolitan families of flowering plants: annonaceae and Rhamnaceae. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 359, 1495–1508.
| Historical biogeography of two cosmopolitan families of flowering plants: annonaceae and Rhamnaceae.Crossref | GoogleScholarGoogle Scholar | 15519968PubMed |
Rieseberg LH, Soltis DE (1991) Phylogenetic consequences of cytoplasmic gene flow in plants. Evolutionary Trends in Plants 5, 65–84.
Rohland N, Reich D (2012) Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Research 22, 939–946.
| Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture.Crossref | GoogleScholarGoogle Scholar | 22267522PubMed |
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574.
| MrBayes 3: Bayesian phylogenetic inference under mixed models.Crossref | GoogleScholarGoogle Scholar | 12912839PubMed |
Sanmartín I, Meseguer AS (2016) Extinction in phylogenetics and biogeography: from timetrees to patterns of biotic assemblage. Frontiers in Genetics 7, 35
| Extinction in phylogenetics and biogeography: from timetrees to patterns of biotic assemblage.Crossref | GoogleScholarGoogle Scholar | 27047538PubMed |
Schuster TM, Setaro SD, Tibbits JFG, Batty EL, Fowler RM, McLay TGB, Wilcox S, Ades PK, Bayly MJ (2018) Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae). PLoS One 13, e0195034
| Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae).Crossref | GoogleScholarGoogle Scholar | 29668710PubMed |
Shepherd LD, McLay TGB (2011) Two micro-scale protocols for the isolation of DNA from polysaccharide-rich plant tissue. Journal of Plant Research 124, 311–314.
| Two micro-scale protocols for the isolation of DNA from polysaccharide-rich plant tissue.Crossref | GoogleScholarGoogle Scholar | 20927638PubMed |
Stöver BC, Müller KF (2010) TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11, 7
| TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar | 20051126PubMed |
Thiele KR, West JG (2004) Spyridium burragorang (Rhamnaceae), a new species from New South Wales, with new combinations for Spyridium buxifolium and Spyridium scortechinii. Telopea 10, 823–829.
Tsitrone A, Kirkpatrick M, Levin DA (2003) A model for chloroplast capture. Evolution 57, 1776–1782.
| A model for chloroplast capture.Crossref | GoogleScholarGoogle Scholar | 14503619PubMed |
VicFlora (2018) Spyridium. In ‘Flora of Victoria’. (Royal Botanic Gardens Foundation Victoria) Available at https://vicflora.rbg.vic.gov.au/flora/taxon/829f1202-25c7-429c-97f7-fc7a9ba62a2d [Verified 23 April 2018]
Wiley EO, Lieberman BS (2011) ‘Phylogenetics: theory and practice of phylogenetic systematics.’ (Wiley: Hoboken, NJ, USA)
Wilkins C, Kern S, Rathborne D, Markey A (2011). ‘Rare and poorly known flora of the Raventhorpe Range and Bandalup Hill.’ (Government of Western Australia: Perth, WA, Australia)
Worth JRP, Holland BR, Beeton NJ, Schönfeld B, Rossetto M, Vaillancourt RE, Jordan GJ (2017) Habitat type and dispersal mode underlie the capacity for plant migration across an intermittent seaway. Annals of Botany 120, 539–549.
| Habitat type and dispersal mode underlie the capacity for plant migration across an intermittent seaway.Crossref | GoogleScholarGoogle Scholar | 28961707PubMed |
Worth JRP, Sakaguchi S, Harrison PA, Brüniche-Olsen A, Janes JK, Crisp MD, Bowman DMJS (2018) Pleistocene divergence of two disjunct conifers in the eastern Australian temperate zone. Biological Journal of the Linnean Society 125, 459–474.
| Pleistocene divergence of two disjunct conifers in the eastern Australian temperate zone.Crossref | GoogleScholarGoogle Scholar |
Wright IJ, Ladiges PY (1997) Geographic variation in Eucalyptus diversifolia (Myrtaceae) and the recognition of new subspecies E. diversifolia subsp. hesperia and E. diversifolia subsp. megacarpa. Australian Systematic Botany 10, 651–680.
| Geographic variation in Eucalyptus diversifolia (Myrtaceae) and the recognition of new subspecies E. diversifolia subsp. hesperia and E. diversifolia subsp. megacarpa.Crossref | GoogleScholarGoogle Scholar |
1 Author names for all taxa at species level and lower are shown in Table 1; vouchers and authors of phrase-name taxa are in Table 2.
2 Spyridium scortechinii has a distribution that extends approximately 20 km into southern Queensland. However, this species is largely restricted to NSW. For ease of reading, for the remainder of the text, we will refer to this clade (and the equivalent clade in the cpDNA phylogeny) as NSW endemics.


