Wedged between two congeners: will the real Gompholobium nitidum (Fabaceae: Mirbelieae) please stand up?
C. L. Simmons A B * , T. G. B. McLay A C and M. T. Mathieson B
A B * , T. G. B. McLay A C and M. T. Mathieson B
A
B
C
Abstract
The most recent review of the genus Gompholobium Sm. (Fabaceae: Mirbelieae), published in 2008, considered Gompholobium nitidum Sol. ex Benth. to exhibit a high degree of variation, ranging from very compact shrubs with linear leaflets, to open shrubs of a taller stature with lanceolate leaflets, through to dense shrubs with oblong leaflets with obtuse-emarginate apices. We use morphological and ddRAD sequencing data to recognise three species from this broad complex, linked with topographic and edaphic complexity in the study area (Laura south to the Atherton Tablelands, north-east Queensland): (1) G. nitidum sens. strict. is a narrowly distributed taxon limited to the white sands of coastal dunes north of Cooktown, with a single disjunct population south of Cooktown; (2) a novel species described here, G. cinctum M.T.Mathieson & C.L.Simmons, has a slightly broader distribution across the skeletal sandy loams of sandstone formations in the Cooktown region; and (3) G. papuanum Merr. & L.M.Perry, previously subsumed into G. nitidum, is reinstated and occurs broadly across north-east Queensland in woodlands from west of Townsville to Cape York, the Torres Strait Islands and into Papua New Guinea.
Keywords: Australian flora, conservation status, ddRAD, Fabaceae, Gompholobium, Queensland flora, species complex, taxonomy.
Introduction
The Australasian endemic wedge-pea genus Gompholobium Sm. (Fabaceae: Mirbelieae) comprises 46 prostrate to medium-sized erect shrub species distributed across all Australian states, with one species found in the Lesser Sunda Islands and one in New Guinea (Crisp et al. 2005; Chappill et al. 2008; Council of Heads of Australasian Herbaria 2011). The species occurring in Queensland are found in coastal and subcoastal heaths and woodlands, and are easily recognisable by pinnate leaves, the production of conspicuous yellow or yellow–orange flowers in spring and short, cylindrical fruit following fertilisation. The genus name is derived from the inflated shape of the seed pods. No specific pollination studies have been conducted on any Australian Gompholobium species but that Fabaceae species tend to be insect pollinated, primarily by Hymenoptera (Aronne et al. 2012; Scaccabarozzi et al. 2020), is widely accepted and seeds are likely dispersed locally through gravity. Although widespread, long-term land clearing has occurred throughout the range and habitat of the currently described species, most Queensland Gompholobium species are afforded some protection within conservation reserves and none are considered threatened (Nature Conservation Act 1992 (Qld)).
Phylogenetic studies of Australian papilionoid legumes have advanced our understanding of the relationships between genera (Crisp et al. 2000; Crisp and Cook 2003a, 2003b; Chappill et al. 2008; Cardoso et al. 2012), especially with the continuing development of phylogenomic datasets (Renner et al. 2022). ALthough there are some complexities in large genera such as Pultenaea (Barrett et al. 2021, 2024), the generic identity of Gompholobium has always been well established, and forms part of the ‘giant antipodal egg and bacon pea’ group with Bossiaea (Crisp and Cook 2003b; Barrett et al. 2021). A taxonomic revision of Gompholobium recognised 44 species distributed through all Australian states, and New Guinea and Wetar Island in the Lesser Sunda Islands, Indonesia; with eight of these species found in Queensland (Chappill et al. 2008). Since the revision, two new species from Western Australia have been described (Wilkins and Trudgen 2012; Wilkins and Sandiford 2020).
However, despite the taxonomic classification of the genus being reasonably well understood, the general acceptance of extreme variation in leaf and leaflet morphology within the north-east Queensland species Gompholobium nitidum Sol. ex Benth. sens. lat. (Chappill et al. 2008) is curious and has not been systematically investigated. Under G. nitidum sens. strict., Chappill et al. (2008) incorporated G. papuanum Merr. & L.M.Perry and a phrase-named taxon ‘G. sp. Q1 – aff. nitidum Cooktown’, also known by the phrase name Gompholobium sp. (Point Archer J.Wrigley + NQ1301) (Brown and Halford 2024) (Table 1). A review of specimens within the Queensland Herbarium (BRI) broadly divided these into three groups based on leaflet shape: (1) plants that match Solander’s type specimen of G. nitidum with oblong–obovate leaflets with rounded, obtuse or emarginate apices found in near-coastal aeolian dune formations near Cooktown; (2) plants matching the formerly recognised G. papuanum with lanceolate to narrowly lanceolate leaflets bearing acuminate apices, occurring widely in the eastern Cape York Peninsula and New Guinea; and (3) plants matching Gompholobium sp. (Point Archer J.Wrigley + NQ1301) that has narrow, linear leaflets, and is found in coastal and subcoastal sandstone hills and ranges in the Cooktown area. These three morphotypes are referred to according to general leaflet shape for clarity, namely (1) G. nitidum ‘oblong’; (2) G. nitidum ‘lanceolate’; and (3) G. nitidum ‘linear’ respectively (Table 1).
| Phrase names and synonyms | Formal names recognised by this study | |
|---|---|---|
| Gompholobium nitidum ‘oblong’ | Gompholobium nitidum | |
| Gompholobium nitidum ‘lanceolate’ | Gompholobium papuanum | |
| Gompholobium sp. (Tozers Gap C.H.Gittins 1030) | ||
| Gompholobium nitidum ‘linear’ | Gompholobium cinctum | |
| Gompholobium sp. (Point Archer J.Wrigley + NQ1301) | ||
| Gompholobium sp. Q1 ─ aff. nitidum ‘Cooktown’ |
We aimed to clarify the circumscription, distribution, ecological characteristicsy and conservation status of the G. nitidum species complex from north-east Queensland by establishing patterns of morphological and molecular variation among individuals. Morphological variation was investigated to determine if this was continuous among populations or partitioned into discrete clusters aligning with the three informally identified forms. The broad circumscription of G. nitidum sens. lat. was tested using a ddRADseq molecular dataset designed to capture thousands of genome-wide single-nucleotide polymorphisms (SNPs). Resolving the narrow-ranged taxon G. nitidum ‘linear’ formed the basis of this study that was partially funded by Bush Blitz in 2020–21 (see bushblitz.org.au/taxonomy-research-projects/, accessed 2 May 2024). If the three morphotypes G. nitidum ‘oblong’, G. nitidum ‘lanceolate’ and G. nitidum ‘linear’ are proven to be both genetically and morphologically distinct entities within the Gompholobium nitidum species complex, formal description will be justified and conservation assessments may be warranted.
Materials and methods
Specimen collection
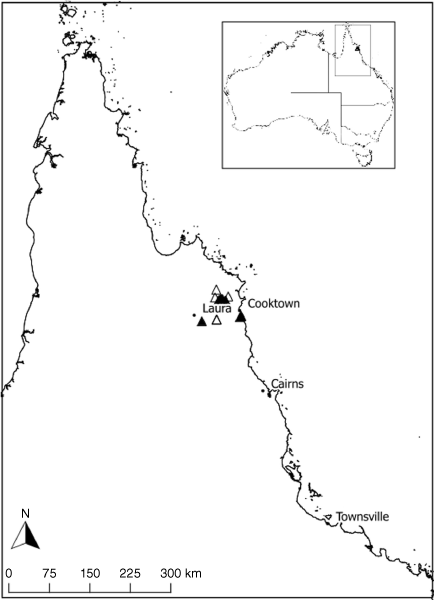
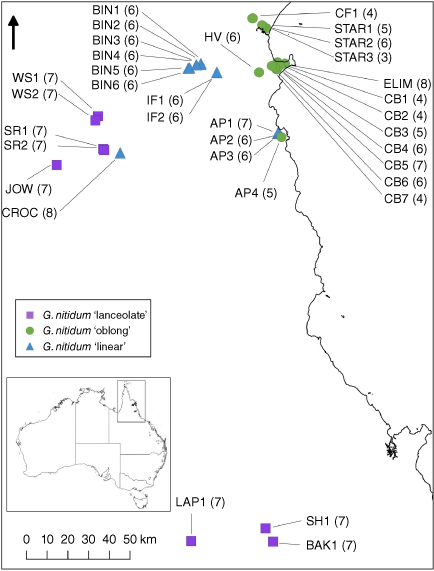
Herbarium specimens at the Queensland Herbarium (BRI) and Bush Blitz data were used to establish locations to inform field surveys. Populations were sampled across the range of each entity within the species complex (Fig. 1), except for G. nitidum ‘lanceolate’. This morphotype is more widespread than the other two taxa and sampling the full distribution (i.e. northern Cape York Peninsula and outside of Australia) was beyond the scope of this study. Therefore, sampling of G. nitidum ‘lanceolate’ was limited to the area bounded by Starcke and Laura in the north and the Atherton Tablelands in the south to focus on the delimitation of the narrow-ranged G. nitidum ‘linear’. Populations were often composed of groupings of plants patchily distributed across a broad geographic area. We sampled 1–10 individuals from multiple patches of plants within the broader geographic range of the population (Fig. 1, Supplementary Table S1). Samples were collected from plants ~5 m apart to minimise chances of sampling clones or close relatives. Newest leaves showing minimal signs of disease or damage were placed in labelled coffee filters and placed in airtight zip-lock bags containing silica desiccating beads and at least one voucher herbarium specimen was taken per sampling location (Supplementary Table S1). Flowers were placed in 70% ethanol for morphological examination. Five samples of the geographically and morphologically distinct taxon G. pinnatum were collected from locations in south-east Queensland following the same methods to provide an outgroup for genomic analysis.
Map of north-east Queensland showing collecting locations for the three Gompholobium nitidum species complex morphotypes. Circles represent G. nitidum ‘oblong’, squares represent G. nitidum ‘lanceolate’ and triangles represent G. nitidum ‘linear’. Inset map shows location within Australia, on which grey dots indicate herbarium records for G. pinnatum.
Morphological data collection
Morphological characters were measured from a representative selection of all dried herbarium specimens held at AD, BRI, DNA, MEL, NE and NSW, including vouchers collected in 2021 for the molecular analysis below (see Results and Taxonomic treatment sections). Floral measurements were made from specimens preserved in 70% ethanol or from reconstituted flowers removed from BRI specimens. Description format follows that of Chappill et al. (2008).
Type material of Gompholobium papuanum at BRI and G. nitidum at MEL was examined directly. Scanned images of all other type material held in other institutions were viewed in JSTOR Global Plants (see http://plants.jstor.org/, accessed 10 March 2024). Geological associations are described based primarily on the State surface geology – Queensland (see https://www.business.qld.gov.au/industries/mining-energy-water/resources/geoscience-information/gsq, accessed 17 October 2023).
Molecular data collection
Leaf tissue from living plants was placed onto silica gel. DNA extraction was undertaken by the Australian Genome Research Facility (AGRF, Adelaide, SA, Australia) from 30 mg of dry leaf material from 332 samples using a modified CTAB protocol (Doyle and Doyle 1987). Extractions were quantified using QuantiFluor (Promega) and visualised for quality on a Genomic DNA ScreenTape (Agilent) at AGRF (Melbourne). In total, 285 samples passed quality control thresholds. Subsequent ddRAD libraries were prepared at AGRF following the methods of Peterson et al. (2012) using the PstI and MseI restriction enzymes to digest DNA. The library was sequenced on an Illumina NovaSeq. 6000 using a 2 × 150-bp single-end reads kit at AGRF (Melbourne).
Molecular data analysis
Raw sequence reads were put through the ipyrad pipeline (ver. 0.9.53, see https://ipyrad.readthedocs.io/en/latest/; Eaton and Overcast 2020) to demultiplex, trim and assemble loci on the Volvox HPC (Royal Botanic Gardens Victoria). Data were assembled using the ‘denovo’ option. We applied a range of filters on subsequent datasets to ensure that downstream analyses were conducted on high-quality SNP data and reduce missing data. Multiple values for several important ipyrad parameters (Orel et al. 2023) were tested to determine the impact on the resulting dataset and subsequent analysis. The following settings were used for each dataset described below, to retain as much data as possible while allowing for confidence in the base calls and analyses not being computationally limited: a 0.85 clustering threshold, six read minimal coverage and a minimum of 95% of samples represented at each locus. We also removed secondary loci by reducing the dataset to a single SNP per locus to avoid linkage.
Molecular phylogenetic analyses
A refined ‘phylogenetics’ dataset was created to reduce computational time but still interrogate the phylogenetic relationships between taxa, using one representative from each sampled location of Gompholobium nitidum ‘oblong’, ‘lanceolate’ and ‘linear’, with all samples of G. pinnatum included as an outgroup (Supplementary Table S1). Filtering steps were applied through ipyrad as described above to produce the high-quality ‘phylogenetic’ dataset composed of 6012 SNP loci across 42 individuals.
Coalescent‐based phylogenetic trees were constructed to test species hypotheses and infer evolutionary relationships with the SVDquartets package (ver. 1, see https://evomics.org/learning/phylogenetics/svdquartets/; Chifman and Kubatko 2014) implemented in PAUP* (ver. 4.0a169, see http://phylosolutions.com/paup-test/; Swofford 2002). This program is designed to accept SNP data and produces robust phylogenetic results. All analyses used at least three runs with the multispecies coalescent tree model selected, evaluating 100,000 quartets and 1000 bootstrap replicates. Trees were evaluated using the taxon groups as tip labels.
A phylogeny by maximum likelihood was also estimated from the SNP alignment using IQ-TREE (ver. 2.2.0, see http://www.iqtree.org/; Nguyen et al. 2015); with ModelFinder (see http://www.iqtree.org/ModelFinder/; Kalyaanamoorthy et al. 2017) used to select the optimal substitution model, the ascertainment bias flag was used to account for the SNP-only alignment and the best model was determined using the Bayesian information criterion (BIC). The best fitting model selected and used in the tree reconstruction was TVMe + G4. We used the Shimodaira–Hasegawa-like approximate likelihood-ratio test (SH-aLRT; Guindon et al. 2010) to assess consistency and node support, and 1000 ultrafast bootstrap replicates (UFBoot in IQ-TREE, see http://iqtree.cibiv.univie.ac.at; Minh et al. 2013; Hoang et al. 2018) were used to assess consistency and support for nodes within the phylogeny. G. pinnatum was originally intended to be used as an outgroup, on the assumption that this species was related to, but not part of, the G. nitidum complex. However, long branch lengths between the ingroup and outgroup (indicative of substantial phylogenetic distance), limited visual interpretation of the ingroup relationships. Instead, a midpoint root was applied and deemed appropriate given the geographic distance between the south-east and north-east taxa.
Species complex genetic structure analyses
Phylogenetic analyses supported the Gompholobium nitidum morphotypes belonging to a well-defined clade (see results below) and therefore the outgroup G. pinnatum samples were removed from further analyses. Removing outgroup taxa eliminates the influence of orthogonal variation in other taxa, thus more clearly displaying variation within the focal species complex. The filtering steps described above produced a high-quality ‘species complex’ dataset composed of 8314 SNP loci across 204 individuals from the G. nitidum complex. The structure and complexity in the species complex SNP dataset were visualised in an abstract phylogenetic network computed by SplitsTree (ver. 4.16.2, see https://github.com/husonlab/splitstree4; Huson and Bryant 2006) using the Neighbour-Net distance transformation on uncorrected characters with ambiguous states ignored. Neighbour-Net provides a visual summary of conflicting signals within the dataset, owing to reticulation and other processes, to guide further investigation of the data.
Genetic structure within the samples was assessed using principal component analysis (PCA) that provides a model-free representation of similarities and differences among individuals through a low-dimensional projection of the data that preserves the variance–covariance structure. The PCA was conducted with gl.pcoa that performs a Gower ordination using Euclidean distances, in the packages dartR (ver. 1.9.9.1, see https://cran.r-project.org/package=dartR; Gruber et al. 2018) and adegenet (ver. 2.1.7, see https://cran.r-project.org/package=adegenet; Jombart 2008) in R (R Foundation for Statistical Computing, Vienna, Austria, see https://www.R-project.org/).
We used model-based clustering analysis implemented by STRUCTURE (ver. 2.3, see https://web.stanford.edu/group/pritchardlab/structure.html; Pritchard et al. 2000), under an admixture model because each of the populations under examination had been considered part of a single species and are or were therefore presumably in recent genetic contact. STRUCTURE assigns samples to K distinct genetic groups to minimise deviations from Hardy–Weinberg equilibrium and linkage disequilibrium within each genetic group, using Bayesian inference. We ran MCMC chains for 200,000 iterations after a burnin of 50,000 iterations for each value of K from 1 to 6 and completed 10 runs for each value of K to assess and confirm run convergence. STRUCTURE HARVESTER (ver. 0.6.94, see http://taylor0.biology.ucla.edu/structureHarvester/) was utilised to determine the most likely number of genetic clusters based on the Evanno method and reported using the delta-K statistic (Evanno et al. 2005).
Fine-scale genetic structure
The two putative novel taxa, Gompholobium nitidum ‘lanceolate’ and G. nitidum ‘linear’ had a closer genetic affinity (see results below), with two populations showing some levels of admixture. A final subset of data, ‘linear–lanceolate’, was created using the filtering steps described above to further interrogate the relationship between populations of G. nitidum ‘lanceolate’ and G. nitidum ‘linear’, and this produced a high-quality species complex dataset composed of 9065 SNP loci across 131 individuals of G. nitidum ‘lanceolate’ and G. nitidum ‘linear’. A PCA was performed to display the variation more clearly in this subgroup of taxa and identify any intergrades between the putative taxa.
Results
Morphological analyses
The examination of specimens from AD, BRI, DNA, MEL, NE and NSW confirmed the presence of three consistent morphotypes within the broader circumscription of Gompholobium nitidum (Chappill et al. 2008). Complete descriptions, line drawings and distribution maps are in the taxonomic treatment and summarised below.
The first, Gompholobium nitidum ‘oblong’, is a small to medium-sized dense shrub 0.75–1.75 m tall with branches that are largely glabrous with occasional white hairs to ~0.5 mm, leaflets that are strongly discolourous, oblong–obovate with obtuse to emarginate apices and weakly to moderately decurved, thickened margins (Fig. 2a). The flowers are bright lemon–yellow and relatively larger than those of the other morphotypes, with the standard petal ~25 × 15 mm (width × length). This morphotype is currently known only from the white sands of consolidated and unconsolidated aeolian dunefields over a reasonably small geographic area.
Photographs of plants of the three morphotypes within the broader circumscription of Gompholobium nitidum. (a) G. nitidum ‘oblong’ = G. nitidum sens. strict., Elim Rd, just behind Elim Beach, east of Hopevale, 18 June 2021, M.T.Mathieson MTM3346 & R.Booth (BRI AQ 1023568); (b) G. nitidum ‘lanceolate’ = G. papuanum, roadside north-east of Laura, towards ‘Welcome’ Station, 11 Apr. 2021, M.T.Mathieson MTM3292 & S.L.Thompson (BRI AQ 1021927) and (c) G. nitidum ‘linear’ = G. cinctum, sp. nov. Battlecamp Rd, Biniirr NP, west of Isabella Falls, 8 Apr. 2021, M.T.Mathieson et al. MTM3260 (BRI AQ 1021877). Photo credits: M. T. Mathieson.
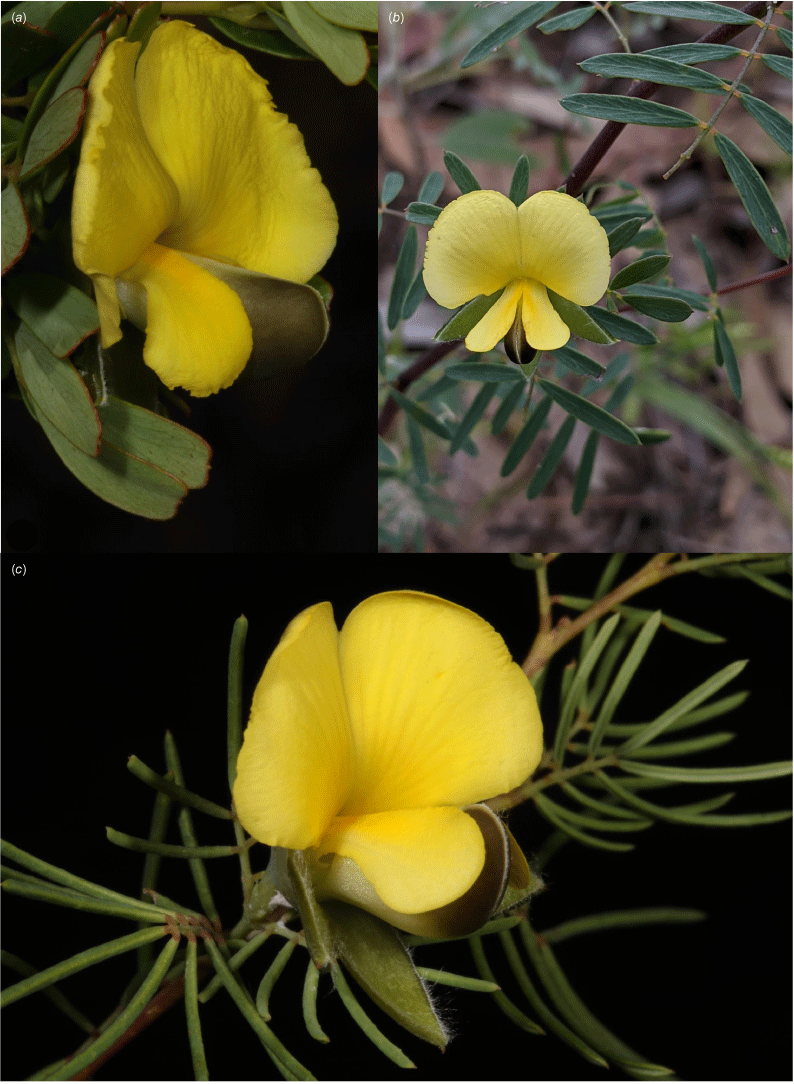
The second morphotype, Gompholobium nitidum ‘lanceolate’, is a small open shrub 0.75–1.2 m tall, with glabrous branches with occasional small white hairs to ~0.3 mm long, leaflets that are moderately–strongly discolourous, lanceolate–narrowly elliptical with acute–apiculate apices and weakly to moderately decurved, thickened margins (Fig. 2b). The flowers are commonly a dull yellow with the standard petal measuring up to 11 × 15 mm. This morphotype is widespread across north-east Queensland from west of Townsville to Cape York and into New Guinea, occurring on a wide variety of geological formations including alluvial plains, sandstone slopes and plateaux, metamorphics and volcanic formations of rhyolite and granites.
The third morphotype, Gompholobium nitidum ‘linear’, is a compact, erect, somewhat open shrub 0.2–0.5 m tall with moderately discolourous, narrow, linear leaflets (the strongly revolute margins giving a terete appearance) with apiculate–mucronate apices (Fig. 2c). The flowers are generally bright yellow with the standard petal measuring up to 10 × 16 mm, very similar to flowers of G. nitidum ‘lanceolate’. This morphotype is known to occur on sandstone slopes and ranges, and at one location, on sandsheets atop coastal metamorphic formations.
Currently there are no known locations in which any of these morphotypes are sympatric, and no populations have been found with consistently intermediate features between morphotypes.
Molecular analyses
The initial ipyrad assembly pipeline generated 190,328 RAD loci. After filtering, the final SNP matrix for the phylogenetic dataset contained 6012 SNP loci across 42 individuals (95.2% average call rate, 4.28% overall missing data rate); the final SNP matrix for the species complex dataset contained 8314 SNP loci across 204 individuals (95.2% average call rate, 4.8% overall missing data rate); and the linear–lanceolate dataset was composed of 9065 SNP loci across 131 individuals (95.2% average call rate, 4.77% overall missing data rate).
Phylogenetic analysis of northern Queensland Gompholobium
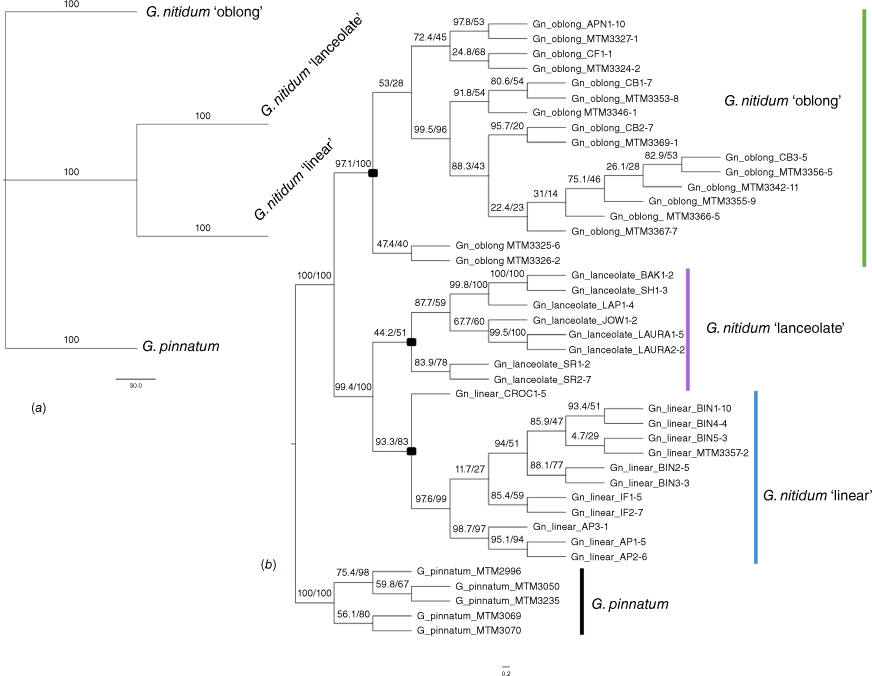
The IQ-TREE and SVDquartets analyses on the phylogeny dataset provided the same well-supported topology with three clades recovered in the phylogeny rooted at the midpoint based on the phylogenetic dataset (Fig. 3). There was strong bootstrap support (>95%) separating the south-east taxon Gompholobium pinnatum from the north-east G. nitidum species complex. Two clades were resolved within the G. nitidum species complex: one composed of G. nitidum ‘oblong’, and a second composed of G. nitidum ‘lanceolate’ and G. nitidum ‘linear’. The latter clade suggests that G. nitidum ‘lanceolate’, and G. nitidum ‘linear’ are monophyletic sister species that are more closely related to each other than to G. nitidum ‘oblong’. Bootstrap support is lower beyond the node separating G. nitidum ‘lanceolate’ and G. nitidum ‘linear’, suggesting differences driven by population divergence. For example, the G. nitidum ‘linear’ population Crocodile Station (CROC) is located further inland than other collecting locations, and consequently occurs in a separate branch to the remaining populations of this morphotype. The phylogenetic analyses supported the north-east Queensland taxa belonging to a well-defined clade with further analyses focusing on the species complex dataset.
Phylogenomic reconstruction of relationships among the Gompholobium nitidum taxa based on the reduced phylogeny dataset (6012 SNPs across 42 individuals), transformed to equal lengths. (a) SVDquartets coalescent species tree. Bootstrap support values are shown above branches with total incompatible quartets = 0 (0%). (b) IQ-TREE maximum likelihood tree. Support values are SH-aLRT support/ultrafast bootstrap support, both expressed as percentages. The composition of each clade by the informal taxa tested in this study is indicated, with species accepted in our revision demarcated by black squares on the corresponding ancestral node.
Interrogating the Gompholobium nitidum species complex
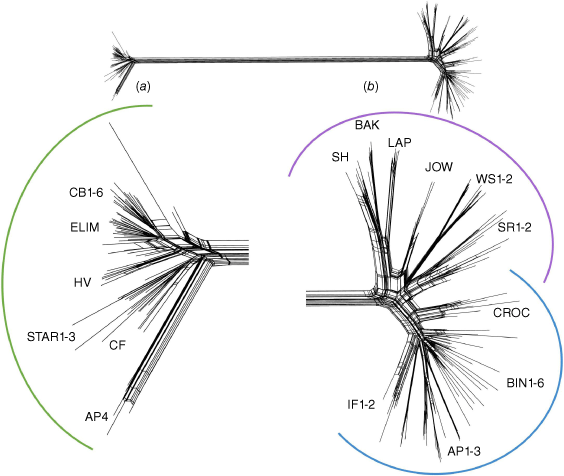
The Neighbour-Net, PCA and STRUCTURE analyses of the species complex dataset showed similar patterns. The network of the Gompholobium nitidum complex recovered two largely separated main clusters (Fig. 4), one consisting of all samples of G. nitidum ‘oblong’, and the second consisting of G. nitidum ‘lanceolate’ and G. nitidum ‘linear’. The G. nitidum ‘oblong’ cluster had a group of samples from the southern part of the species range at Archer Point (AP4) at the termini of long narrow rectangular cycles (Fig. 4a). Despite populations of G. nitidum ‘linear’ being located in geographic proximity (within ~2 km) to populations of G. nitidum ‘oblong’ at Archer Point (AP1-4, Fig. 1), no connections were drawn between these populations, indicating that no contemporary gene flow has occurred between these taxa at this location. Geographically clustered G. nitidum ‘oblong’ populations formed regions of complex localised cycling indicative of relationships between locally occurring populations (CB1-6, ELIM, HV). Groups of samples corresponding to populations were clustered at the termini of rectangular cycles of the G. nitidum ‘lanceolate’ and G. nitidum ‘linear’ cluster, with cycles present throughout the entire network (Fig. 4b). Populations of G. nitidum ‘lanceolate’ formed groups of populations on the left while G. nitidum ‘linear’ formed groups on the right (Fig. 4b). Some populations, such as WS1-2 in the northern sampling range of G. nitidum ‘lanceolate’ were at the ends of long, narrow rectangular cycles, whereas others, such as LAP, SH and BAK in the southern sampling range of G. nitidum ‘lanceolate’ and BIN1-6 in the northern range of G. nitidum ‘linear’, formed regions of complex localised cycling indicative of relationships between locally occurring populations. Individuals from the G. nitidum ‘linear’ population CROC formed a group towards the centre of the two taxa, linked at the base to the geographically close SR populations of G. nitidum ‘lanceolate’ (Fig. 4b).
SplitsTree Neighbour-Net network analysis of the Gompholobium nitidum species complex dataset (8314 SNPs, 204 individuals). Two largely separated clusters are illustrated in the whole network above, separating (a) G. nitidum ‘oblong’ (green) from (b) G. nitidum ‘lanceolate’ (blue) and G. nitidum ‘linear’ (purple). Close-up images of the splits show the cycles within (a) G. nitidum ‘oblong’ (green); and (b) G. nitidum ‘lanceolate’ (blue) and G. nitidum ‘linear’ (purple).
The first and second principal components of the species complex SNP matrix plotted the individuals into three broad clusters corresponding to morphotype (Fig. 5a). Reflecting the splits network, the first principal component captured 48.68% of the molecular variation and separated G. nitidum ‘oblong’ from G. nitidum ‘linear’ and G. nitidum ‘lanceolate’. All samples of G. nitidum ‘oblong’ were tightly clustered in the positive space of the first component. The second principal component captured 3.88% of the molecular variation and separated G. nitidum ‘lanceolate’ in the positive space from G. nitidum ‘linear’ in the negative space. Most populations of both morphotypes were clustered in respective single groups. The exceptions were G. nitidum ‘linear’ population CROC that formed a cluster separate from the main group, and G. nitidum ‘lanceolate’ populations SR1-2 that were separate from the respective main cluster, toward the midpoint of the axis.
Graphical representations of genetic similarity among individuals within the Gompholobium nitidum species complex dataset (8314 SNPs, 204 individuals): (a) Principal component analysis (PCA) for the species complex dataset. (b) STRUCTURE results, where each individual is represented by a vertical bar, coloured according to the proportional assignment to each of three genetic clusters. (c) STRUCTURE results for each population mapped on collecting locations, colours matching those in (a). Circles represent individuals of G. nitidum ‘oblong’, squares represent G. nitidum ‘lanceolate’ and triangles represent G. nitidum ‘linear’. Labelled ellipses indicate populations with admixture detected in the STRUCTURE analysis.
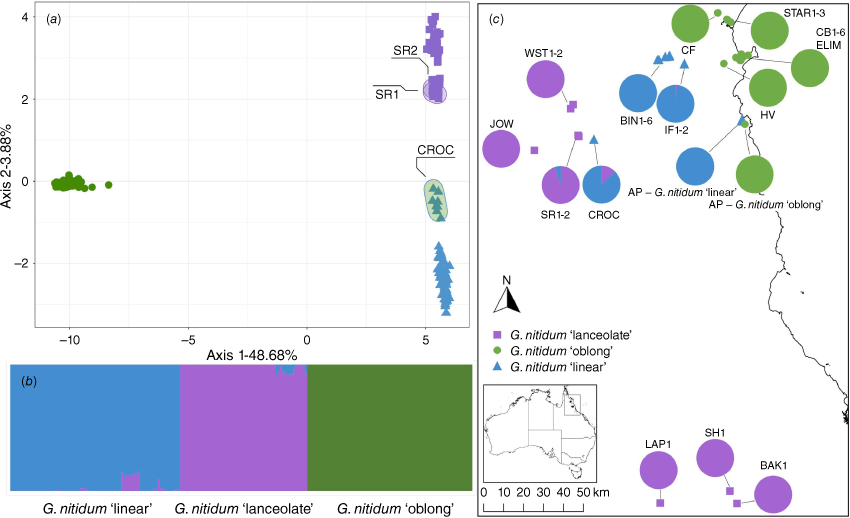
The Evanno method of delta K selection inferred three genetic clusters in the STRUCTURE results, with a broad pattern of genetic ancestry representing the three morphotypes, echoing the patterns of the NeighbourNet and PCA (Fig. 5b, c). Gompholobium nitidum ‘oblong’ formed a distinct group with no admixture with the other morphotypes. Populations CROC, IF2 and BIN6 of G. nitidum ‘linear’ had admixture with the G. nitidum ‘lanceolate’ cluster and G. nitidum ‘lanceolate’ populations SR1-2 had admixture with the G. nitidum ‘linear’ cluster. The remaining G. nitidum ‘linear’ and G. nitidum ‘lanceolate’ populations had no admixture outside the primary cluster. Populations CROC and SR1-2 are in proximity to each other, whereas IF2 and BIN6 are at the northern range of G. nitidum ‘linear’ that may be in proximity to G. nitidum ‘lanceolate’ populations that were not included in this study.
Fine-scale genetic structure of ‘linear–lanceolate’ morphotypes
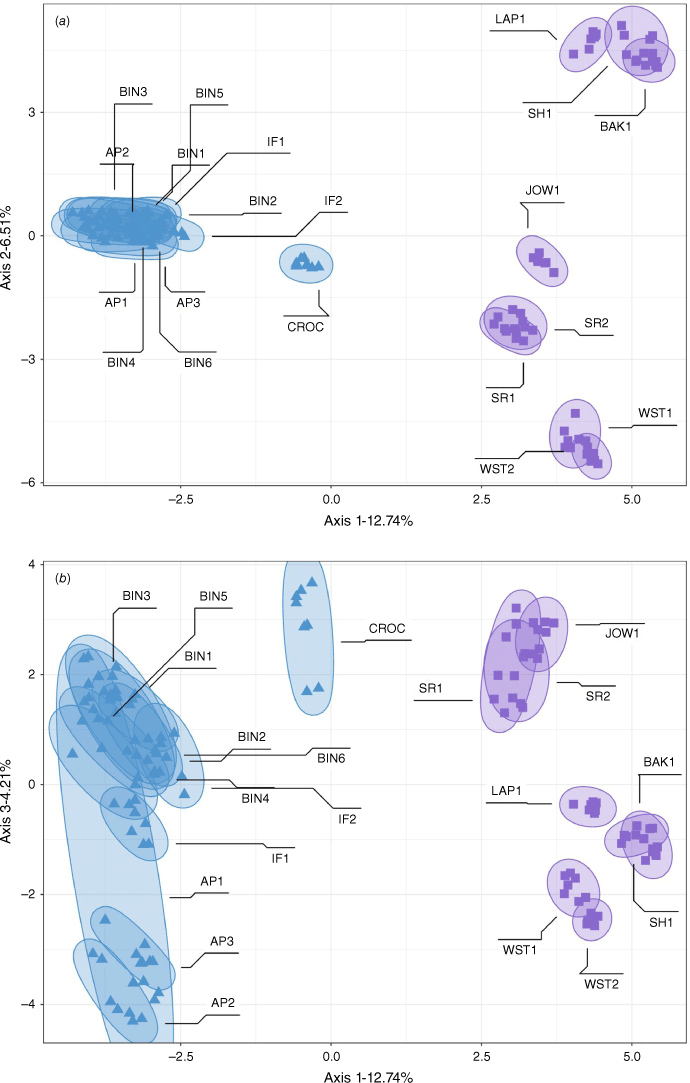
The PCA for the linear–lanceolate dataset focusing on the sister taxa Gompholobium nitidum ‘lanceolate’ and G. nitidum ‘linear’ shows separation of the two morphotypes across the first axis (12.74%, Fig. 6a). The G. nitidum ‘linear’ population CROC is located in an intermediate position between G. nitidum ‘lanceolate’ and the remaining G. nitidum ‘linear’ populations on the first axis, reflecting the patterns of admixture observed in the STRUCTURE results. The second principal component (6.51%) visualises the genetic differentiation of geographic clusters of G. nitidum ‘lanceolate’ populations (Fig. 1, 6a). The third principal component (4.21%) separates the populations of G. nitidum ‘linear’ with CROC clearly separated from the geographically closest populations BIN1-6, IF1-2 (Fig. 6b). The Archer Point populations AP2-3 are separated from the northern populations (BIN1-6, IF1-2) and there is more variability in AP1 with some individuals grouped more closely to AP2-3 whereas others fall within the northern grouping.
Principal component plot of genetic similarity among individuals within the Gompholobium nitidum ‘Point Archer’ dataset consisting of G. nitidum ‘linear’ and G. nitidum ‘lanceolate’ (9065 SNPs, 131 individuals). (a) Axes 1 and 2. (b) Axes 1 and 3. Squares represent individuals of G. nitidum ‘lanceolate’ and triangles represent G. nitidum ‘linear’. Ellipses and labels represent populations of each morphotype.
Discussion
This study has demonstrated the utility of coupling genomic data with morphological studies to resolve species complexes in plants. The patterns of genetic differentiation among populations present compelling evidence against Gompholobium nitidum being a single variable species. Each of the three morphotypes are both morphologically discrete and genetically distinct, rather than tending towards a continuum of either morphological or genetic variation. Population sampling of ddRAD genomic data enabled the identification of entities that maintain distinctiveness, despite growing in proximity to each other. Together, these results indicate three distinct morphotypes that warrant formal description.
Species delimitation
Delimitation in plant species complexes is challenging as species boundaries are often semipermeable and difficulties arise when determining the transition from population-level differentiation to independent evolutionary lineages that can be recognised as species (de Queiroz 2005; Harrison and Larson 2014). Sequencing of restriction site-associated DNA through methods such as ddRAD or DArTseq has proven useful for resolution of species complexes at both the population genetic and phylogenetic levels in several Australian plant genera (e.g. Geleznowia, Anderson et al. 2023; Pultenaea, Renner et al. 2022; Seringia, Binks et al. 2020; Zieria, Orel et al. 2023) and highlighted areas of reticulate evolution in others (e.g. Eucalyptus, Fahey et al. 2022).
Investigating the complexities within the Tribe Mirbelieae has been the subject of many recent studies (e.g. Crisp and Cook 2003b; Orthia et al. 2005; summarised in Barrett et al. 2021), including work to resolve Pultenaea species complexes within the Mirbelia clade (Renner et al. 2022; Barrett et al. 2024). The genus Gompholobium is part of the ‘giant antipodal egg and bacon pea’ group and recognised as a natural morphological and monophyletic group within Tribe Mirbelieae, with a full taxonomic revision undertaken within the last two decades (Chappill et al. 2008). The work presented here represents the first genetic species delimitation study to be undertaken on any Gompholobium species, though this is not comprehensive across the genus. Initially focused on delimitation of the very narrowly distributed G. nitidum ‘linear’, the study was expanded to provide insight into the two other morphotypes that share the surrounding geographic area within a radius of ~120 km that also encompassed the entire known range of G. nitidum ‘oblong’. Sampling across the entire range of the widely distributed G. nitidum ‘lanceolate’ was beyond the scope of this study. Each of the three genomic datasets analysed in this work established separation of the three morphotypes of G. nitidum, with limited evidence of ongoing gene flow among the three entities, with greater phylogenetic distance between G. nitidum ‘oblong’ and the other taxa.
No evidence of gene flow was detected between G. nitidum ‘oblong’ and G. nitidum ‘linear’ despite these morphotypes occurring in proximity, found within ~2 km of each other near Archer Point (Fig. 1). Larger pollinators would be capable of moving this distance, even if only occasionally, suggesting the presence of specific breeding barriers. At this location, the taxa occur on different substrates and within dissimilar habitat. Additionally, no gene flow was detected between G. nitidum ‘oblong’ and G. nitidum ‘lanceolate’, though this is not entirely surprising given that the populations are more geographically distant, with ~40 km separating the closest populations. However, evidence of historical gene flow was detected in both directions between the broadly distributed G. nitidum ‘lanceolate’ and at the very western limit of G. nitidum ‘linear’ near the township of Laura (CROC, SR1-2; Fig. 1). A low level of historical gene flow between two species is not uncommon and does not preclude these from being considered different species (e.g. Anderson et al. 2023 found similar evidence of admixture between two groups in Geleznowia currently recognised at species rank). The presence of admixture suggests that reproductive isolation between G. nitidum ‘linear’ and G. nitidum ‘lanceolate’ is not complete, highlighting the complexities involved in species delimitation and the recognition of entities at the rank of subspecies or species (de Queiroz 2005, 2020; Wiens 2007).
Evidence from the species complex-level genomic analyses consistently identifies three genetic groupings, with G. nitidum ‘oblong’ having a large degree of genomic divergence from the other two taxa. When the orthogonal variation exerted by G. nitidum ‘oblong’ is removed from the dataset, there is clear genetic differentiation between the G. nitidum ‘linear’ and G. nitidum ‘lanceolate’ entities, with the G. nitidum ‘linear’ population CROC falling in a somewhat intermediate position but not overlapping with any populations of the other taxon. Similarly, there are one or more qualitative or quantitative morphological characters that show no overlap between these closely related taxa. Within populations CROC and SR1-2 there was some genetic evidence of hybridisation but morphological consistency is maintained between of each of the morphotypes. Where the genotype of each of the two morphotypes dominates, the morphological features (e.g. leaf shape) also do. Re-examination of plants and populations with mixed ancestry yielded no evidence of mixed morphology in these specimens, with both taxa holding the morphological form true to that of the genetic majority. The leaflet shape of G. nitidum ‘linear’ and G. nitidum ‘lanceolate’ appears to be diagnostic, in that these are either linear or lanceolate; however floral structure is more similar between these taxa than that of either taxon to G. nitidum ‘oblong’.
Pollination of the papilionoid Fabaceae is primarily attributed to bees (Leppik 1966; Westerkamp and Weber 1999). Opportunistic hybridisation (Bello et al. 2018; Heenan et al. 2018) and pollination mimicry (Uluer et al. 2022) are also associated with the papilionoid Fabaceae, with multiple evolutionary origins of keel flower pollinator syndrome being shown to be consistent with but not proof of mimicry (Uluer et al. 2022). The potential for mimicry among pea-flowered Fabaceae (and non-Fabaceae mimics) is an indication of a non-pollinator-specific pollination strategy. Hence, cross pollination between species with similar flower characteristics would be expected at zones of overlap, resulting in gene mixing through time. Although the ~8 km between population CROC of G. nitidum ‘linear’ and populations Split Rock (SR1-2) of G. nitidum ‘lanceolate’ is most likely beyond the foraging distance of bees and smaller insects, there is a high likelihood that the distribution of the latter taxon continues on the elevated sandstone plateaus edges adjacent to Crocodile Station and further gene mixing may have occurred there, or these populations may have been more extensive and in greater proximity in the recent past. To date, no populations have been found with leaflet shapes that are intermediate between G. nitidum ‘linear’ and G. nitidum ‘lanceolate’. Finally, although the three morphotypes of the G. nitidum species complex occur across a small range, there are landscape complexities and each morphotype appears to occur on unique substrates. The widespread G. nitidum ‘lanceolate’ occurs on Carboniferous through to middle Jurassic (c. 360–160 Ma) conglomerate and granodiorite derived soils in Far North Queensland and Torres Strait Islands, whereas the very narrowly distributed G. nitidum ‘linear’ is primarily restricted to younger late Jurassic, early Cretaceous (c. 100–160 Ma) sandstones and siltstones. Despite admixture between the two entities identified at some locations, we consider G. nitidum ‘linear’ and G. nitidum ‘lanceolate’ to be separate species owing to the genetic, morphological and habitat differences.
Consistently, G. nitidum ‘oblong’ was the most genetically differentiated of the three morphotypes, occurring as sister to the two other species in the phylogenetic analyses, at the termini of well-defined neighbour networks, and with no evidence of admixture between the other entities in the PCA or STRUCTURE analysis. In contrast to the older substrates that G. nitidum ‘lanceolate’ and G. nitidum ‘linear’ occupy, G. nitidum ‘oblong’ occurs on younger Quaternary coastal and subcoastal sand dunes and, much more rarely, alluvial deposits. There are also more morphological distinctions between G. nitidum ‘oblong’ and the other taxa. This morphotype is a dense, medium to large shrub, with bright golden yellow flowers almost twice the diameter of those of G. nitidum ‘lanceolate’ and G. nitidum ‘linear’, whereas G. nitidum ‘lanceolate’ and G. nitidum ‘linear’ are small, open shrubs with smaller, somewhat dull yellow flowers. The contrasting lack of evidence for hybridisation between G. nitidum ‘linear’ and G. nitidum ‘oblong’ at Archer Point, despite populations AP1-4 occurring in proximity could be attributed to longer lineage divergence as a result of greater reproductive isolation due to differences in floral structure.
Similar species delimitation studies have recently been undertaken on other complexes within the Mirbelieae. Renner et al. (2022) segregated six new species from Pultenaea glabra based on the fine geographic structuring of both genetic and morphological diversity in the Greater Blue Mountains World Heritage Area, New South Wales. Many of the newly described species in this study were almost wholly allopatric, often associated with different microsites or vegetation ecosystems, similar to our findings within the G. nitidum species complex. The correlated differences in distribution, morphology and genetic identity, similarly support the recognition of the three morphological and genetic entities within the G. nitidum species complex at species level. A broader understanding of the phylogenetic relationships of the genera and species within the Mirbelieae is still needed, particularly for the ‘giant antipodal egg and bacon pea’ group that includes Gompholobium and Bossiaea. Therefore further opportunities exist for placing the Gompholobium nitidum species complex within a more widely sampled study at the tribe level. Additional surveys between populations CROC for G. nitidum ‘linear’ and SR1-2 for G. nitidum ‘lanceolate’ may reveal mixed morphology populations that could be included. Such future phylogenetic work could include the representatives of all three focal taxa from north-east Australia and population-level sampling of the variable G. subulatum from Western Australia and the Northern Territory, and G. papuanum specimens from New Guinea. Our finding of three entities encompassed within a single species is likely only scratching the surface of the undescribed diversity within Gompholobium in north-eastern Australia.
Conservation implications
An objective of this study was to determine well-supported species delimitations of potentially narrowly ranged taxa to guide conservation assessments. Field observations provided opportunities to assess population size and the proposed delimitation of three taxa within the broadly circumscribed G. nitidum. We have refined the determinations of specimens housed at BRI and on loan from other institutions, enabling an accurate assessment of the distribution of the three taxa we recognise as distinct species. Despite G. nitidum ‘linear’ having a narrower distribution than the other morphotypes within the species complex, as with the other morphotypes, this has an abundance of suitable habitat, is present in numerous protected areas and lacks immediate threatening processes. Although Common Assessment Method (CAM) evaluations have not been formally undertaken to evaluate the risk of extinction of the proposed taxa within the G. nitidum species complex against the International Union for Conservation of Nature (IUCN) Red List Categories, all three proposed taxa meet the IUCN category of Least Concern (Threatened Species Scientific Committee 2021; IUCN Standards and Petitions Committee 2022). The well-researched species delimitation study presented here has had the additional effect of minimising the time and effort spent on species conservation assessments, reducing the cost of species protection (Wiens 2007).
Taxonomic treatment
Two of the three taxa recognised in this study as comprising Gompholobium nitidum sens. lat. (as defined by Chappill et al. 2008) have existing names, viz.: 1. G. nitidum Sol. ex Benth., currently deemed to represent the narrowly distributed species first described from the Endeavour River area near Cooktown, has been referred to in this manuscript as G. nitidum ‘oblong’; and 2. G. papuanum Merr. & L.M.Perry has been referred to in this manuscript as G. nitidum ‘lanceolate’, and is a widespread species recorded through north-east Queensland and eastern New Guinea. The third taxon, referred to as G. nitidum ‘linear’, also known by the phrase name Gompholobium sp. (Point Archer J.Wrigley + NQ1301), is formally named Gompholobium cinctum M.T.Mathieson & C.L.Simmons. The traditional use of an exclamation mark (!) has been applied where type specimens have been examined directly and an asterisk (*) used to indicate type specimens viewed as online images from JSTOR Global Plants (see https://plants.jstor.org/).
Two-dimensional measurements in the descriptions below are provided as length × width unless otherwise stated.
| 1. | Leaflets narrow, 0.4–0.9 mm wide with strongly revolute margins giving a terete appearance; low, compact yet open shrubs to 0.2–0.5 m tall...3. G. cinctum Leaflets >1 mm wide, with weakly to moderately recurved margins, not terete in appearance; small to medium-sized open or dense shrubs, 0.50–1.75 m tall...2 |
| 2. | Leaflet apices acute to apiculate; width of standard petal <20 mm...2. G. papuanum Leaflet apices obtuse or emarginate, oroccasionally rounded, with a minute apiculum; width of standard petal 22–25 mm...1. G. nitidum sens. strict. |
Species descriptions
1. Gompholobium nitidum Sol. ex Benth., Fl. austral. 2: 48 (1864)
Type citation: ‘Queensland. Endeavour River, Banks and Solander, R. Brown (Herb. Banks and R.Br.).’ Type: Endeavour River, Banks & Solander s.n. (lecto fide J.A.Chappill, C.F.Wilkins & M.D.Crisp, Austral. Syst. Bot. 21: 125 (2008): BM 000550705* [sheet labelled photo 205]; isolecto: BM 000550706* [sheet labelled photo 206)], MEL!, P*, W); Endeavour River, R.Brown s.n. (residual syn: BM 000550704*).
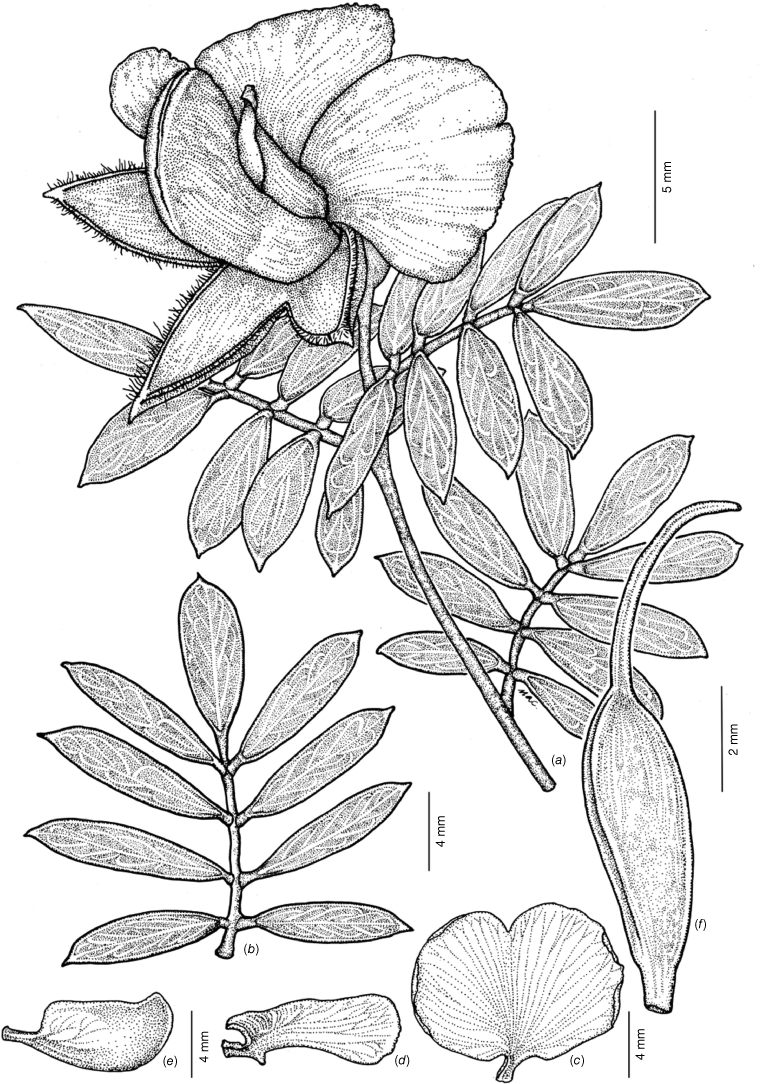
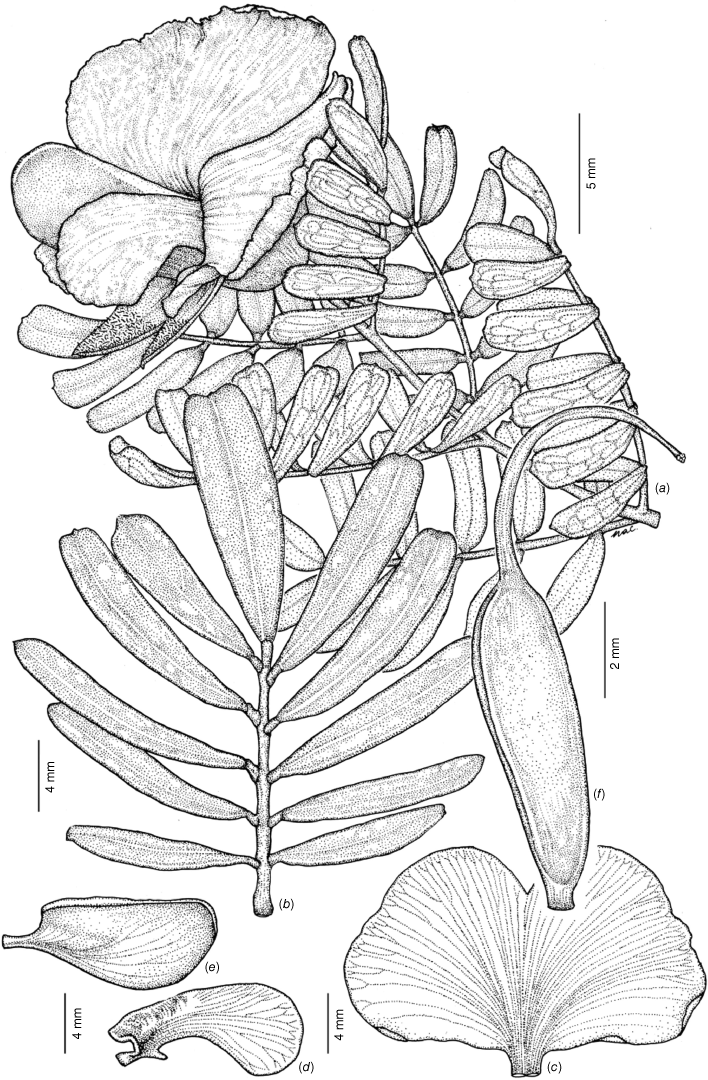
Erect shrubs 0.75–1.75 m tall, not viscid. Branchlets straight, terete, with faint ribs, not tuberculate, glabrous or with occasional white hairs to 0.2–0.5 mm long. Stipules subulate, erect, 0.4–0.7 × 0.1–0.2 mm. Leaves imparipinnate; petiole 0.7–1.7 mm long; petiolules 0.6–0.8 mm long. Leaflets glabrous, 7–11(–15), oblong to oblanceolate, (10–)12–18(–20) × 3–5.5(–7) mm, not tuberculate, strongly discolourous, dark glossy green adaxially, grey–green and papillate abaxially; bases cuneate, rarely attenuate, margin entire, thickened abaxially and slightly–moderately revolute; apices generally emarginate, rarely obtuse or rounded, apiculum 0–0.2 mm long. Flowers solitary at apex of branchlets, subtending leaves usually longer than inflorescence. Peduncles 0.7–1.0 mm long, glabrous. Pedicels 1.5–4 mm long, glabrous. Bracts subulate, 1.2–1.4 × 0.25–0.3 mm, glabrous. Bracteoles persistent on centre of the pedicel, subulate, 0.9–1.0 × 0.2 mm, glabrous, margin entire. Buds ellipsoid, ecostate, green, 12–15 × 6.5–7.5 mm, glabrous, apiculum straight, 0.2–0.4 mm, tips of calyx fused. Hypanthium 1.0–1.2 mm long. Calyx with adaxial lobes fused at base for 2.5–4 mm, symmetrical, 7.5–12 × 2.5–4 mm in total length, abaxial lobes fused at base for 2–3.2 mm, symmetrical, 7.5–11.5 × 2.5–4.2 mm in total length. Petals: standard claw ~2 × 2 mm, lamina transversely obovate, 22–25 × 13–16 mm (width × height), without auricles, bright lemon yellow on the front and grey–green on the back, eye marking absent, deeply emarginate with notch 3–4 mm deep; wings straight to curved slightly downwards, oblanceolate, 12–14 × 3–6 mm including claw, auricle present, pale yellow with grey–brown on back, apex obtuse; keel 14–16 × 6–8 mm including claw, dark green–brown from centre to the apex, pale green–yellow proximally, apex obtuse with margins minutely serrulate. Stamens with filaments orange–red, 9.2–11 × 0.5–0.6 mm; anthers sub-basifixed, narrowly ovoid, uniform in size, 1.8–2 × 0.7–1.1 mm, yellowish-cream with a narrow red connective. Gynoecium with stipe 1.3–1.5 mm long; ovary 5.5–6.2 × 1.7–2.1 mm, glabrous; style 8.0–8.5 × 0.25–0.3 mm, glabrous; ovules 2–6. Fruit ovoid, 8.5–8.8 × 6.0–6.9 mm, glabrous, funicle 2.5–3 mm long. Seeds brown, glabrous, 3–3.7 × 2–2.3 mm, cuticular wrinkles present, rim aril absent. Fig. 7.
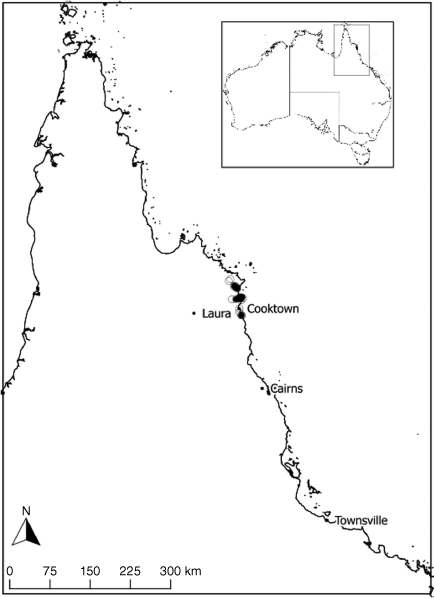
This species predominantly occurs in low, open or closed heathlands and occasionally in low shrubby woodlands, growing on Quaternary coastal and sub-coastal sand dune fields, stabilised dunes and plains or very rarely, alluvium sand silt and gravel, south of Cooktown to Cape Flattery (Fig. 8). Flowering occurs from the late wet season through the drier months, March–August, although apparently this is never profuse.
Although narrowly distributed, this species would qualify as being of Least Concern under IUCN criteria (IUCN Standards and Petitions Committee 2022).
Queensland [Cook District]: East of Hopevale, 20 June 2021, M.T.Mathieson MTM3369 & R.Booth (BRI AQ1023579); North of Archer Point Rd, south of Cooktown, 15 June 2021, M.T.Mathieson MTM3316 & R.Booth (BRI AQ1023538); Cape Flattery Rd, east of Starcke–Hopevale Rd, north of Cooktown, 17 June 2021, M.T.Mathieson MTM3325, J.Harrigan, R.Booth & E.Harrigan (BRI AQ1023559); 10 km SE of Starcke Station on Starcke–Cape Flattery Rd, Starcke Station is 82 km N of Cooktown, 22 Sep. 1990, P.C.Jobson 1206 (BRI AQ 519125, MEL); 6.7 km from the Cooktown–Lakeland Downs Road, on the road to Archer Point, 21 June 1984, J.R.Clarkson 5449 (DNA, L, MEL, QRS, BRI AQ 399394); S of Cape Bedford, ~25 km NNE of Cooktown, 30 July 1980, J.R.Clarkson 3299 (BRI AQ 345204); ~1 km S of Annan River mouth, 26 Aug. 1974, J.G.Tracey 14736 (BRI AQ 385687); 4 km E of the Hopevale–Starke Rd on track to the mouth of the McIvor River, 13 Aug. 1984, J.R.Clarkson 5470 (BRI AQ 397170); between Nob Point and Cape Bedford, ~2 miles [~3.2 km] back from beach, 1 Sep. 1986, V.Scarth-Johnson 1669A (BRI AQ 379852); Hopevale mission, On Elim Creek Road to Bert’s camp on the beach, 1 June 2005, M.A.Semple 852 (BRI AQ 870556); Elim Rd, just behind Elim Beach, east of Hopevale, 18 June 2021, M.T.Mathieson MTM3346 & R.Booth (BRI AQ 1023568).
Line diagram of Gompholobium nitidum. (a) Flowering branchlet. (b) Leaf. (c) Standard petal. (d) Wing petal. (e) Keel petal. (f) Ovary and style. All from M.T.Mathieson MTM3325 & R.Booth (BRI AQ 1023559) spirit material, except b. from M.T.Mathieson MTM3316 & R.Booth (BRI AQ 1023538) spirit material.
2. Gompholobium papuanum Merr. & L.M.Perry, J. Arnold. Arbor. 23: 400 (1942)
Distribution and ecology
Type: British [Papua] New Guinea: [upper] Wassi [Mai] Kussa River, Tumbuke [S of Mata, E of Morehead], [13] December 1936, L.J.Brass 8432 (holo: A 00066210*; iso: BM 000550500*, BRI AQ0022864!, L 0019006*).
Gompholobium sp. (Tozers Gap C.H.Gittins 1030): A.E.Holland in R.J.F.Henderson (ed.), Queensland Pl.: Names & Dist. 81 (1997).
Erect shrubs 0.75–1.2 m tall, not viscid. Branchlets straight, terete, with faint ribs, not tuberculate, glabrous or with sparse, spreading straight hairs up to 0.3 mm long. Stipules subulate, erect, 1.0–1.7 × 0.3–0.5 mm. Leaves imparipinnate; petiole 0.7–2.1 mm long; petiolules 0.5–0.8 mm long. Leaflets glabrous, 7–11(–13), lanceolate to oblanceolate, 6–14 × 1.5–4.5 mm, not tuberculate, discolourous, glossy green adaxially, paler and papillate abaxially; bases cuneate, rarely attenuate, margin entire, thickened abaxially, rarely slightly revolute; apices acute, attenuate to apiculate, apiculum 0.1–0.2 mm. Flowers solitary or extremely rarely two, at apex of branchlets, subtending leaves usually longer than inflorescence. Peduncles 0.5–1.0 mm long, glabrous. Pedicels 1.5–4 mm long, glabrous. Bracts subulate, 1.5–1.7 × 0.5–0.6 mm, glabrous. Bracteoles persistent on centre of the pedicel, subulate, 1.2–1.4 × 0.3–0.4 mm, glabrous, margin entire. Buds ellipsoid, ecostate, green, 6–8 × 3–5 mm, glabrous, apiculum straight, 0.2–0.4 mm long, tips of calyx fused. Hypanthium ~1 mm long. Calyx with adaxial lobes fused at base for 2.5–3 mm, symmetrical, 8–9.5 × 3.5–4 mm in total length, abaxial lobes fused at base for 2.4–3 mm, symmetrical, 8–9 × 3–3.5 mm in total length. Petals: standard claw ~1.2 × 0.9 mm, lamina broadly obovate, 10.1–11.0 × 11.0–14.2 mm (width × height), without auricles, pale yellow on the front and grey–green on the back, eye marking absent, deeply emarginate with notch 2–3 mm deep; wings straight, narrowly obovate, 8.5–9 × 4–4.6 mm including claw, auricle present, pale yellow with grey–brown on back, apex rounded to obtuse; keel 8.9–9.4 × 4.4–4.7 mm including claw, dark green–brown from centre to the apex, pale green–yellow proximally, apex obtuse with margins minutely serrulate. Stamens with filaments orange–red, 7.2–9 × 0.3–0.4 mm; anthers sub-basifixed, ovoid, uniform in size, 1.2–1.5 × 0.3–0.4 mm, pale yellow with a narrow red connective. Gynoecium with a stipe 0.5–0.6 mm long, ovary 3.7–4.5 × 1.5–1.7 mm, glabrous, style 2.9–3.3 × 0.2–0.3 mm, glabrous; ovules 2–6. Fruit ovoid, 7.1–7.5 × 5.2–5.7 mm, glabrous, funicle 2–2.6 mm long. Seeds brown, glabrous, 2.9–3.2 × 1.8–2.2 mm, cuticular wrinkles present, rim aril absent. Fig. 9.
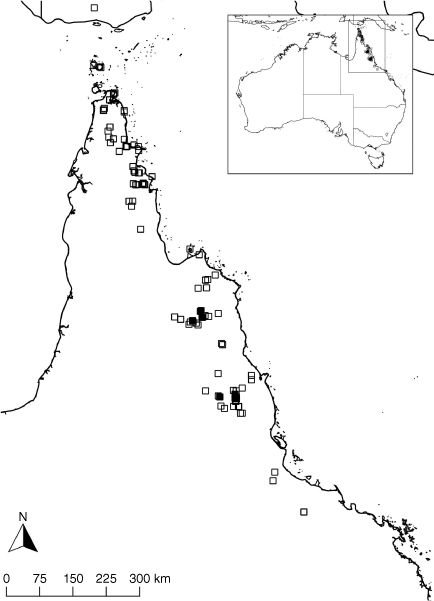
This widespread species can be found in a variety of sclerophyll woodlands and open forests, closed Acacia woodlands and heathlands occurring on a broad range of landforms including consolidated dunes, alluvial flats, deeply weathered plains, sedimentary and metamorphic formations, and granites and rhyolites. The distribution covers the drier western slopes of the Wet Tropics bioregion west of Townsville, continues northwards across the undulating ranges and plains of eastern Cape York Peninsula, and extends into some of the Torres Strait Islands and Papua New Guinea (Fig. 10). Little is known of the geological characteristics of the type locality in Papua New Guinea. Flowering occurs from late in the wet season to the drier months of March–August, apparently never profusely.
Conservation status
The extensive distribution across north-eastern Queensland and lack of documented threats and decline qualify this reasonably abundant species as being of Least Concern under IUCN red list criteria (IUCN Standards and Petitions Committee 2022).
Selected specimens examined
Queensland [Cook District]: Stannary Hills, west of Herberton, 21 Apr. 2021, M.T.Mathieson MTM3305 & S.L.Thompson (BRI AQ 1022134); Split Rock Gallery, south of Laura, 12 Apr. 2021, M.T.Mathieson MTM3299, N. Owens, S.L.Thompson & C. Musgrave (BRI AQ 1022128); Roadside NE of Laura, towards ‘Welcome’ Station, 11 Apr. 2021, M.T.Mathieson MTM3292 & S.L.Thompson (BRI AQ 1021927); off Old Coach Rd, Jowalbinna area, 12 Apr. 2021, M.T.Mathieson MTM3298, C.Musgrave, S.L.Thompson & N.Owens (BRI AQ 1022127); West of Bakerville, 14 Apr. 2021, M.T.Mathieson MTM3304 & S.L.Thompson (BRI AQ 1022132); Mungkan Kandju NP, N of Coen, 10 Aug. 2006, K.R.McDonald KRM5547 (BRI AQ 619705); Stanley Island, 29 May 1995, J. LeCussan 474 (BRI AQ 584747, MEL); Iron Range Rd, before Garraway Creek crossing, 16 Apr 1988, P.I.Forster 4245 (BRI AQ 429615, DNA, MEL, MEXU); Cape York Peninsula, NE of Bamaga airstrip, 4 Aug. 1978, A.Kanis 2063 (BRI AQ 383513, CANB, L, MO, US); Davies Ck – E of falls, 1 Mar. 1998, B.S.Wannan 628 & N.Weston (BRI AQ 536444); Olive River Environmental Reserve, Cape York Peninsula, 15 June 2007, P.I.Forster PIF32653 & K.R.McDonald (BRI AQ 728083); Jardine River, Cape York Peninsula, 19 May 1948, L.J.Brass s.n. (BRI AQ 324988); WSW of Badu Airport, Badu Island, Torres Strait, 24 July 2008, D.G.Fell DGF9709 & D.J.Stanton (BRI AQ 951364); Dulhunty River, Gulf side of Cape York Peninsula, F.W.Whitehouse s.n. (BRI AQ 231537); Dismal Ck, Kutchera, NE of Croydon, 13 Aug. 2007, R.Cumming 24797 & C.Appelman (BRI AQ 860797); NE of Innot Hot Springs, 27 Jan. 2001, B.S.Wannan 2036 & E.J.Younghusband (BRI AQ 731551); SE of Lakefield ranger base, 1 Aug. 2014, S.L.Thompson SLT14336, K.Henderson, C.Henderson, J.Anderson & D.G.Fell (BRI AQ 915966); 16 km from Mt Garnet on Lappa Road, 7 Mar. 2004, K.R.McDonald KRM1829 (BRI AQ 765960); Mt Mulligan, NW of Dimbulah, 15 Apr. 1985, J.R.Clarkson 5774 (BRI AQ 399156).
3. Gompholobium cinctum M.T.Mathieson & C.L.Simmons, sp. nov.
Type: Queensland: Cook District: Yuku Baja Muliku (Annan River) Resources Reserve, SSE of Cooktown, 9 April 2021, M.T.Mathieson MTM3266, S.L.Thomson & L.Naylor (holo: BRI AQ 1021909; iso: CNS distribuendi).
Gompholobium sp. (Point Archer J.Wrigley + NQ1301): A.E.Holland in R.J.F.Henderson (ed.), Queensland Pl.: Names & Dist. 81 (1997).
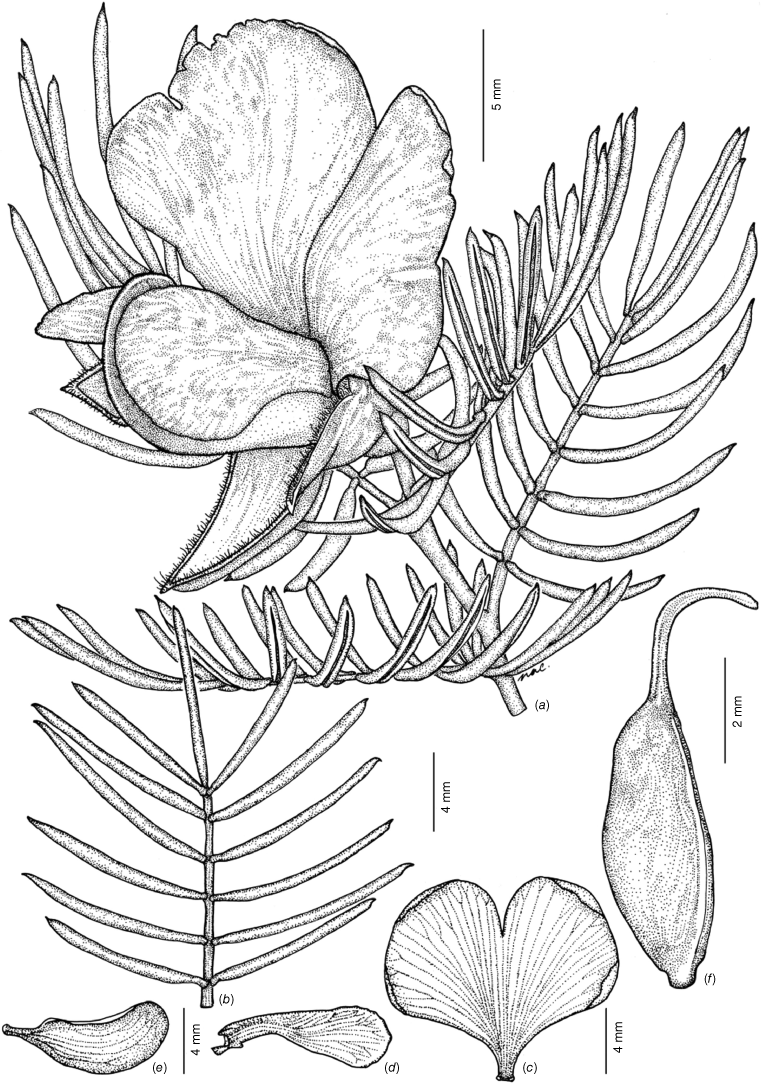
Low, erect shrubs 0.2–0.5 m tall, not viscid. Branchlets straight, terete, with faint ribs, not tuberculate, glabrous or with sparse, spreading straight hairs ~0.4 mm long. Stipules subulate, erect, 1–2.2 × 0.3–0.4 mm. Leaves imparipinnate; petiole 0.8–2.0 mm long; petiolules 0.4–0.6 mm long. Leaflets glabrous, 9–13(–15), linear, (8–)9.5–13(–15) × (0.4–)0.6–0.7(–0.9) mm, not tuberculate, discolourous, dark glossy green adaxially, grey–green and papillate abaxially, bases cuneate, margin entire and tightly revolute, mostly hiding the abaxial surface of the leaflet; apices apiculate, apiculum 0.1–0.3 mm long. Flowers solitary at apex of branchlets, subtending leaves usually longer than inflorescence. Peduncle 0.3–0.5 mm long, glabrous. Pedicels 1.2–1.6 mm long, glabrous. Bracts subulate, 1.5–1.7 × 0.2–0.4 mm, glabrous. Bracteoles persistent on centre of the pedicel, subulate, 1.6–1.8 × 0.2–0.3 mm, glabrous, margin entire. Buds ellipsoid, ecostate, green, 5–7 × 7.5–9 mm, glabrous, apiculum straight, 0.15–0.2 mm long, tips of calyx fused. Hypanthium ~1 mm long. Calyx with adaxial lobes fused at base for 3.2–3.4 mm, symmetrical, 8.5–10 × 4–5 mm in total length, abaxial lobes fused at base for 2.5–3 mm, symmetrical, 8–10.4 × 3.2–4 mm in total length. Petals: standard claw 1.7 × 1.5 mm, lamina broadly obovate, 9–12.5 × 10–16 mm (width × height), without auricles, pale yellow on the front and grey–green on the back, eye marking absent, deeply emarginate with notch ~3 mm deep; wings straight, narrowly obovate, 9–10 × 3–3.5 mm including claw, auricle small but present, pale yellow with grey–brown on back, apex rounded to obtuse; keel 9.5–10 × 3.5 mm including claw, dark green–brown from centre to the apex, pale green–yellow proximally, apex obtuse with margins minutely serrulate. Stamens with filaments yellow–orange, 7–7.5 × 0.3–0.4 mm; anthers sub-basifixed, ovoid, uniform in size, 0.8–1.0 × 0.25–0.35 mm, pale yellow, with a distinct red connective. Gynoecium with stipe 0.4–0.5 mm long, ovary 5.5–5.9 × 3–3.4 mm, glabrous, style 4.2–4.8 × 0.2–0.3 mm, glabrous; ovules 2–6. Fruit ovoid, 7–7.6 × 5–6.3 mm, glabrous, funicle 1.5–1.9 mm long. Seeds brown, glabrous, 2.8–3.1 × 1.5–1.9 mm, cuticular wrinkles present, rim aril absent. Fig. 11.
This short-statured shrub species is currently known predominantly from the confined area of dissected, coarse-grained sandstone ranges and plateaux west of Cooktown and Hopevale from the Isabella Falls area to south of Laura. The species grows in low open woodland with a sparse to well-developed shrub layer on sandy loams. A coastal population is found near Archer Point south of Cooktown, occurring in the sparse shrub layer of low open woodlands on sand-topped metamorphic substrate (Fig. 12). An apparent outlier was collected in similar habitat on Snapper Island some 70 km to the south, scattered throughout the range, yet locally abundant. Sparse, sporadic flowering has been noted throughout the year with marked increase in frequency during the winter and spring months of May–October.
Despite this species having a reasonably small distribution range, the abundance of suitable habitat, presence of the species in numerous protected areas including Biniirr NP, Yuku Baja Muliku (Annan River) Resources Reserve and Quinkan Reserve 2 (East Quinkan), and the apparent lack of immediate threatening processes, this locally abundant species qualifies as being of Least Concern under IUCN red list criteria (IUCN Standards and Petitions Committee 2022).
The specific epithet is derived from the Latin cinctus, encircled, referring to the distribution of this species that is surrounded by the distributions of Gompholobium nitidum and G. papuanum.
Queensland [Cook District]: Battlecamp Rd, Biniirr NP, 10 Apr. 2021, M.T.Mathieson MTM3282, E.Harrigan, J.Harrigan, S.L.Thompson & N.G.Harrigan (BRI AQ 1021922); Battlecamp Rd, Biniirr NP, west of Isabella Falls, 8 Apr. 2021, M.T.Mathieson MTM3260, J.Harrigan, S.L.Thompson & E.Harrigan (BRI AQ 1021877); Northern end of Crocodile Station, SE of Laura, off Peninsula Development Rd, 11 Apr. 2021, M.T.Mathieson MTM3290 & S.L.Thompson (BRI AQ 1021926); Battlecamp Rd, Biniirr NP (CYPAL), 10 Apr. 2021, M.T.Mathieson MTM3283, N.G.Harrigan, S.L.Thompson, J.Harrigan & E.Harrigan (BRI AQ 1021923); Battlecamp Rd, Biniirr NP (CYPAL), 10 Apr. 2021, M.T.Mathieson MTM3284, N.G.Harrigan, J.Harrigan, E.Harrigan & S.L.Thompson (BRI AQ 1021924); Battlecamp Rd, Biniirr NP (CYPAL), 10 Apr. 2021, M.T.Mathieson MTM3285, N.G.Harrigan, S.L.Thompson, E.Harrigan & J.Harrigan (BRI AQ 1021925); Battlecamp Rd, west of Hopevale, 19 June 2021, M.T.Mathieson MTM3357 & R.Booth (BRI AQ 1023571); 5 km from Point Archer along road towards Cooktown, 17 June 1972, J.Wrigley 1301 & I.Telford (BRI AQ 511507); Yuku Baja Muliku (Annan River) Resources Reserve, 9 Apr. 2021, M.T.Mathieson MTM3267, L.Naylor & S.L.Thompson (BRI AQ 1023914); Yuku Baja Muliku (Annan River) Resources Reserve, 9 Apr. 2021, M.T.Mathieson MTM3268, L.Naylor & S.L.Thompson (BRI AQ 1023917); Kings Plains Holding, 21 km NNW of Kings Plains Homestead, 4 July 2015, D.G.Fell DGFKK90/4 & K.Smith (BRI AQ 980658); Kings Plains Station, just E of the Quinkan Reserve 2 (East Quinkan) boundary, WSW of Cooktown, 14 Mar. 2017, P.I.Forster PIF45006 & K.R.McDonald (BRI AQ 941247); Quinkan Reserve 2 (East Quinkan), SE of Laura, Lot/Plan: 3/BS169, 7 Mar. 2017, P.I.Forster PIF44557 & K.R.McDonald (BRI AQ 941339); Snapper Island, 19 Mar. 2002, A.Freeman s.n. (BRI 556224).
Line diagram of Gompholobium cinctum. (a) Flowering branchlet. (b) Leaf. (c) Standard petal. (d) Wing petal. (e) Keel petal. (f) Ovary and style. All from M.T.Mathieson MTM3260 et al. (BRI AQ 1021877) spirit material except (a) from photograph of M.T.Mathieson MTM3292 & S.L.Thompson (BRI AQ 1021927).
Data availability
Sequencing data produced from the ddRAD library are available on the Bioplatforms Australia, Genomics for Australian Plants website (https://data.bioplatforms.com/organization/bpa-plants) under Dataset 83668.
Declaration of funding
We acknowledge the contribution of the Genomics for Australian Plants Framework Initiative consortium (https://www.genomicsforaustralianplants.com/consortium/) in the generation of data used in this publication. The Initiative is supported by funding from Bioplatforms Australia (enabled by NCRIS), the Ian Potter Foundation, Royal Botanic Gardens Foundation (Victoria), Royal Botanic Gardens Victoria, the Royal Botanic Gardens and Domain Trust, the Council of Heads of Australasian Herbaria, CSIRO, Centre for Australian National Biodiversity Research and the Department of Biodiversity, Conservation and Attractions, Western Australia. The funders did not participate in the analysis of data or preparation of the paper. Fieldwork and illustrations were partially funded by a Bush Blitz Taxonomy Research Grant (DNP-BSS-1920-002) awarded to M. T. Mathieson. DNA extractions were funded by the Queensland Herbarium, Department of Environment and Science, Queensland and the Royal Botanic Gardens Victoria.
Acknowledgements
We are grateful to Mabel Lum, Lalita Simpson and the Genomics for Australian Plants Conservation Genomics team for project support. The bioinformatics processing on the Royal Botanic Gardens Victoria high computing cluster resource (Volvox) was supported by Chris Jackson. David Halford and Peter Bostock provided helpful comments, useful discussion, editing and typification advice for the taxonomy sections of the manuscript that is greatly appreciated, as are the botanical illustrations drawn by Nicole Crosswell (http://www.nicolecrosswell.com/). We are grateful to the Elders, managers and rangers of the Yuku Baja Muliku Landowner and Reserves Ltd, Ang-Gnarra Aboriginal Corporation, Hopevale Congress Aboriginal Corporation, Waranthurin Aboriginal Corporation, Mbarbarum Aboriginal Corporation and Western Yalanji Aboriginal Corporation for support of and participation in this project, and permission to work on their lands. We thank our valued and experienced colleagues Ron Booth (BRI), Simon Thompson (Cape York Tenure Resolution, Department of Environment, Science and Innovation), Mark Newton (Queensland Herbarium, Cairns) and Frank Zich (Australian Tropical Herbarium) for support, knowledge and companionship in the field.
References
Anderson BM, Binks R, Byrne M, Crawford AD, Shepherd KA (2023) Using RADseq to resolve species boundaries in a morphologically complex group of yellow-flowered shrubs (Geleznowia, Rutaceae). Australian Systematic Botany 36, 277-311.
| Crossref | Google Scholar |
Aronne G, Giovanetti M, De Micco V (2012) Morphofunctional traits and pollination mechanisms of Coronilla emerus L. flowers (Fabaceae). The Scientific World Journal 2012, 381575.
| Crossref | Google Scholar | PubMed |
Barrett RL, Clugston JAR, Cook LG, Crisp MD, Jobson PC, Lepschi BJ, Renner MAM, Weston PH (2021) Understanding diversity and systematics in Australian Fabaceae Tribe Mirbelieae. Diversity 13, 391.
| Crossref | Google Scholar |
Barrett RL, Clugston JAR, Albrecht DE, Elkan L, Hosking JR, Jobson PC, McCune SF, Orme AE, Palsson RL, Renner MAM, Wardrop C, Weston PH (2024) Revision of the Pultenaea setulosa species complex (Fabaceae: Mirbelieae) including 14 new species. Australian Systematic Botany 37, SB23014.
| Crossref | Google Scholar |
Bello A, Stirton CH, Chimphango SBM, Muasya AM (2018) Morphological evidence for introgressive hybridization in the genus Psoralea L. (Psoraleeae, Fabaceae). South African Journal of Botany 118, 321-328.
| Crossref | Google Scholar |
Binks RM, Wilkins CF, Markey AS, Lyons MN, Byrne M (2020) Genomic data and morphological re-assessment reveals synonymy and hybridisation among Seringia taxa (Lasiopetaleae, Malvaceae) in remote north-western Australia. Taxon 69, 307-320.
| Crossref | Google Scholar |
Brown GK, Halford JJ (2024) Leguminosae. In ‘Census of the Queensland Flora and Fungi 2023 (Print)’. (Ed. AR Bean) pp. 54–67. (Queensland Department of Environment, Science and Innovation, Queensland Government) Available at http://www.data.qld.gov.au/dataset/census-of-the-queensland-flora-and-fungi-2023 [Verified 20 December 2024]
Cardoso D, De Queiroz LP, Pennington RT, De Lima HC, Fonty E, Wojciechowski MF, Lavin M (2012) Revisiting the phylogeny of papilionoid legumes: New insights from comprehensively sampled early-branching lineages. American Journal of Botany 99, 1991-2013.
| Crossref | Google Scholar | PubMed |
Chappill JA, Wilkins CF, Crisp MD (2008) Taxonomic revision of Gompholobium (Leguminosae: Mirbelieae). Australian Systematic Botany 21, 67-151.
| Crossref | Google Scholar |
Chifman J, Kubatko L (2014) Quartet inference from SNP data under the coalescent model. Bioinformatics 30, 3317-3324.
| Crossref | Google Scholar | PubMed |
Council of Heads of Australasian Herbaria (2011) Gompholobium Sm. In ‘Australian Plant Name Index: Vascular Plants’. Available at https://biodiversity.org.au/nsl/services/rest/name/apni/64809/api/apni-format [Verified 5 February 2024]
Crisp MD, Cook LG (2003a) Phylogeny and evolution of anomalous roots in Daviesia (Fabaceae: Mirbelieae). International Journal of Plant Sciences 164, 603-612.
| Crossref | Google Scholar |
de Queiroz K (2005) A unified concept of species and its consequences for the future of taxonomy. Proceedings of the California Academy of Sciences 56, 196-221.
| Google Scholar |
de Queiroz K (2020) An updated concept of subspecies resolves a dispute about the taxonomy of incompletely separated lineages. Herpetological Review 51, 459-461.
| Google Scholar |
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19, 11-15.
| Google Scholar |
Eaton DAR, Overcast I (2020) Ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 36, 2592-2594.
| Crossref | Google Scholar | PubMed |
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14, 2611-2620.
| Crossref | Google Scholar | PubMed |
Fahey PS, Udovicic F, Cantrill DJ, Nicolle D, McLay TGB, Bayly MJ (2022) A phylogenetic investigation of the taxonomically problematic Eucalyptus odorata complex (E. section Adnataria series Subbuxeales): evidence for extensive interspecific gene flow and reticulate evolution. Australian Systematic Botany 35, 403-435.
| Crossref | Google Scholar |
Gruber B, Unmack PJ, Berry OF, Georges A (2018) dartR: an R package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Molecular Ecology Resources 18, 691-699.
| Crossref | Google Scholar | PubMed |
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307-321.
| Crossref | Google Scholar | PubMed |
Harrison RG, Larson EL (2014) Hybridization, introgression, and the nature of species boundaries. Journal of Heredity 105, 795-809.
| Crossref | Google Scholar | PubMed |
Heenan P, Mitchell C, Houliston G (2018) Genetic variation and hybridisation among eight species of kōwhai (Sophora: Fabaceae) from New Zealand revealed by microsatellite markers. Genes 9, 111.
| Crossref | Google Scholar | PubMed |
Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35, 518-522.
| Crossref | Google Scholar | PubMed |
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23, 254-267.
| Crossref | Google Scholar | PubMed |
IUCN Standards and Petitions Committee (2022) ‘Guidelines for Using the IUCN Red List Categories and Criteria. Version 15.1.’ (International Union for Conservation of Nature and Natural Resources: Gland, Switzerland) Available at https://www.iucnredlist.org/documents/RedListGuidelines.pdf [Verified 26 September 2023]
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403-1405.
| Crossref | Google Scholar | PubMed |
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587-589.
| Crossref | Google Scholar | PubMed |
Leppik EE (1966) Floral evolution and pollination in the Leguminosae. Annales Botanici Fennici 3, 299-308.
| Google Scholar |
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30(5), 1188-1195.
| Crossref | Google Scholar |
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32, 268-274.
| Crossref | Google Scholar | PubMed |
Orel HK, McLay TGB, Guja LK, Duretto MF, Bayly MJ (2023) Genomic data inform taxonomy and conservation of Critically Endangered shrubs: a case study of Zieria (Rutaceae) species from eastern Australia. Botanical Journal of the Linnean Society 205, 292-312.
| Crossref | Google Scholar |
Orthia LA, Cook LG, Crisp MD (2005) Generic delimitation and phylogenetic uncertainty: an example from a group that has undergone an explosive radiation. Australian Systematic Botany 18, 41-47.
| Crossref | Google Scholar |
Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE (2012) Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7, e37135.
| Crossref | Google Scholar | PubMed |
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155, 945-959.
| Crossref | Google Scholar | PubMed |
Renner MAM, Barrett RL, Clarke S, Clugston JAR, Wilson TC, Weston PH (2022) Morphological and molecular evidence refute a broad circumscription for Pultenaea glabra (Fabaceae: Mirbelieae), with implications for taxonomy, biogeography, and conservation. Australian Systematic Botany 35, 127-179.
| Crossref | Google Scholar |
Scaccabarozzi D, Dixon KW, Tomlinson S, Milne L, Bohman B, Phillips RD, Cozzolino S (2020) Pronounced differences in visitation by potential pollinators to co-occurring species of Fabaceae in the southwest Australian biodiversity hotspot. Botanical Journal of the Linnean Society 194, 308-325.
| Crossref | Google Scholar |
Threatened Species Scientific Committee (2021) Guidelines for assessing the conservation status of native species according to the Environment Protection and Biodiversity Conservation Act 1999 and Environment Protection and Biodiversity Conservation Regulations 2000. Available at https://www.dcceew.gov.au/sites/default/files/env/pages/d72dfd1a-f0d8-4699-8d43-5d95bbb02428/files/tssc-guidelines-assessing-species-2021.pdf [Verified 26 September 2023]
Uluer DA, Forest F, Armbruster S, Hawkins JA (2022) Reconstructing an historical pollination syndrome: keel flowers. BMC Ecology and Evolution 22, 45.
| Crossref | Google Scholar | PubMed |
Westerkamp C, Weber A (1999) Keel flowers of the Polygalaceae and Fabaceae: a functional comparison. Botanical Journal of the Linnean Society 129, 207-221.
| Crossref | Google Scholar |
Wiens JJ (2007) Species delimitation: New approaches for discovering diversity. Systematic Biology 56, 875-878.
| Crossref | Google Scholar | PubMed |
Wilkins CF, Sandiford EM (2020) Gompholobium glabristylum (Fabaceae), a new native pea from montane habitats in Stirling Range National Park. Nuytsia 31, 223-227.
| Crossref | Google Scholar |
Wilkins CF, Trudgen ME (2012) A new species of Gompholobium (Fabaceae: Mirbelieae) from the Pilbara bioregion of Western Australia. Nuytsia 22, 31-40.
| Crossref | Google Scholar |