The origins of genomic imprinting in mammals
Carol A. Edwards A B , Nozomi Takahashi A , Jennifer A. Corish A and Anne C. Ferguson-Smith A
A B , Nozomi Takahashi A , Jennifer A. Corish A and Anne C. Ferguson-Smith A
A Department of Genetics, University of Cambridge, Downing Street, Cambridge CB2 3EH, UK.
B Corresponding author. Email: cae28@cam.ac.uk
Reproduction, Fertility and Development 31(7) 1203-1218 https://doi.org/10.1071/RD18176
Submitted: 10 May 2018 Accepted: 1 October 2018 Published: 8 January 2019
Journal Compilation © CSIRO 2019 Open Access CC BY
Abstract
Genomic imprinting is a process that causes genes to be expressed according to their parental origin. Imprinting appears to have evolved gradually in two of the three mammalian subclasses, with no imprinted genes yet identified in prototheria and only six found to be imprinted in marsupials to date. By interrogating the genomes of eutherian suborders, we determine that imprinting evolved at the majority of eutherian specific genes before the eutherian radiation. Theories considering the evolution of imprinting often relate to resource allocation and recently consider maternal–offspring interactions more generally, which, in marsupials, places a greater emphasis on lactation. In eutherians, the imprint memory is retained at least in part by zinc finger protein 57 (ZFP57), a Kruppel associated box (KRAB) zinc finger protein that binds specifically to methylated imprinting control regions. Some imprints are less dependent on ZFP57 in vivo and it may be no coincidence that these are the imprints that are found in marsupials. Because marsupials lack ZFP57, this suggests another more ancestral protein evolved to regulate imprints in non-eutherian subclasses, and contributes to imprinting control in eutherians. Hence, understanding the mechanisms acting at imprinting control regions across mammals has the potential to provide valuable insights into our understanding of the origins and evolution of genomic imprinting.
Additional keywords : epigenetics, evolution, marsupials.
Introduction
Genomic imprinting is an epigenetic process whereby genes are expressed from one of the two chromosome homologues in a parent-of-origin-specific manner. Because imprinted genes are, in effect, functionally haploid, they lose the protection that diploidy provides against deleterious mutations. This poses many questions about the evolution of this epigenetic process. Herein we discuss the current knowledge as to when imprinting arose in vertebrates, why the imprinting process may have evolved and how the mechanism of imprinting may have emerged.
Imprinting first arose in the therian lineage
To date, the only vertebrates in which genomic imprinting has been reported are mammals. In the mouse, >130 imprinted genes have been identified, most of which are found in clusters located at particular chromosomal regions in the genome. To identify when imprinting evolved in the mammalian lineage, various known eutherian imprinted genes have been tested in marsupials and monotremes (Table 1). Six imprinted genes have been identified in marsupials, namely insulin-like growth factor 2 (IGF2), the non-coding RNA gene H19, insulin (INS), IGF2 receptor (IGF2R), paternally expressed 10 (PEG10) and mesoderm specific transcript/paternally expressed 1 (MEST/PEG1; Killian et al. 2000; O’Neill et al. 2000; Suzuki et al. 2005, 2007; Ager et al. 2007; Smits et al. 2008), whereas no imprinted gene has been found in monotremes (Killian et al. 2000, 2001; Edwards et al. 2008).
The most intact imprinted gene cluster in marsupials is the IGF2/H19 domain. In eutherians, this region contains two protein coding genes, IGF2 and INS, and the long non-coding RNA (lncRNA) gene H19. In mice, Igf2 is paternally expressed in the developing embryo except in the choroid plexus and leptomeninges (DeChiara et al. 1991; Ferguson-Smith et al. 1991). IGF2 was first shown to be expressed from the paternally inherited chromosome in pouch young of the grey short-tailed opossum (Monodelphis domestica; O’Neill et al. 2000). More extensive analysis in the tammar wallaby (Macropus eugenii) found it to be paternally expressed in the fetal body and adult mammary gland, but biased paternal expression was found in the fetal brain and placenta (Suzuki et al. 2005; Stringer et al. 2012b, 2012c). The insulin gene (INS or Ins2 in mouse) is only imprinted in the human and mouse yolk sac where it too is expressed from the paternally inherited chromosome (Deltour et al. 1995; Moore et al. 2001). In tammar wallaby, INS is paternally expressed in the yolk sac membrane (YSM), the principal placenta in most marsupials (Ager et al. 2007), and in the adult mammary gland (Stringer et al. 2012c). The lncRNA gene H19 is maternally expressed in mice (Bartolomei et al. 1991) and an orthologous region has been identified in tammar wallaby that is also expressed exclusively from the maternal allele in the yolk sac, fetal and pouch young liver and pouch young brain (Smits et al. 2008). IGF2 expression has also been studied in monotremes: both the short-beaked echidna (Tachyglossus aculeatus) and platypus (Ornithorhynchus anatinus) express IGF2 biallelically in adult tissues (Killian et al. 2001); hence, it is not imprinted, at least in adult prototherians.
The Igf2r gene encodes IGF2R and a mannose-6-phosphate (M6P) receptor. Igf2r is expressed from the maternally inherited chromosome in the mouse (Barlow et al. 1991), but imprinting in human is polymorphic, occurring in only a small proportion of the population (Xu et al. 1993). However, this scenario may not be the case for all primates because, in the Cynomolgus macaque, IGF2R was found to be imprinted in all individuals analysed (Cheong et al. 2015). In the mouse, the Igf2r gene lies within an imprinted cluster with solute carrier family 22 members 2 (Slc22a2) and 3 (Slc22a3), which are expressed from the maternally inherited alleles in the placenta. Their imprinting is regulated by the reciprocally imprinted, paternally expressed lncRNA, Airn, which is an antisense transcript to Igf2r (Zwart et al. 2001). In marsupials, IGF2R is maternally expressed in Virginia opossum (Didelphis virginiana) but lacks the differentially methylated region (DMR) in intron 2 that is the promoter for Airn in eutherians (Killian et al. 2000). M6P/IGF2R is biallelically expressed in the echidna and platypus. Interestingly, the monotreme orthologues lack the IGF2 binding domains and therefore produce proteins that only have M6P-binding properties, perhaps negating the need to be imprinted in these species. (Killian et al. 2000). Evidence for a marsupial Airn transcript has not been found (Weidman et al. 2006a), hence the mechanism regulating IGF2R imprinting in marsupials is not known.
Peg10 is a retrotransposon derived gene that arose in the genome after the divergence of the therians from the monotremes (Suzuki et al. 2007). In the mouse, Peg10 resides in a large imprinted cluster that also contains sarcoglycan, epsilon (Sgce), which is also paternally expressed, ankyrin repeat and SOCS box-containing 4 (Asb4), which is expressed from the maternally inherited chromosome and protein phosphatase 1, regulatory subunit 9A (Ppp1r9a), which exhibits maternally biased expression in the placenta (Ono et al. 2003). In the tammar wallaby, PEG10 is paternally expressed in the embryo and yolk sac, but the other genes are biallelically expressed (Suzuki et al. 2007). The final gene that has been suggested as imprinted in marsupials is MEST/PEG1. This gene is paternally expressed in eutherians (Kaneko-Ishino et al. 1995), but shows biallelic but paternally biased expression in multiple different tammar tissues (Suzuki et al. 2005).
Together, these data indicate that genomic imprinting first arose in the therian lineage. No imprinted genes have been identified in the monotremes, but only four genes have been experimentally assessed (IGF2, IGF2R, DLK1 (delta like non-canonical Notch ligand 1) and DIO3 (deiodinase, iodothyronine type III); Killian et al. 2000, 2001; Edwards et al. 2008) and, furthermore, due to scarcity of material, no genes have been tested in embryos. Hence, it is possible that there is imprinted expression in monotreme fetuses that becomes biallelic in adults, as is the case for IGF2 in humans (Issa et al. 1996).
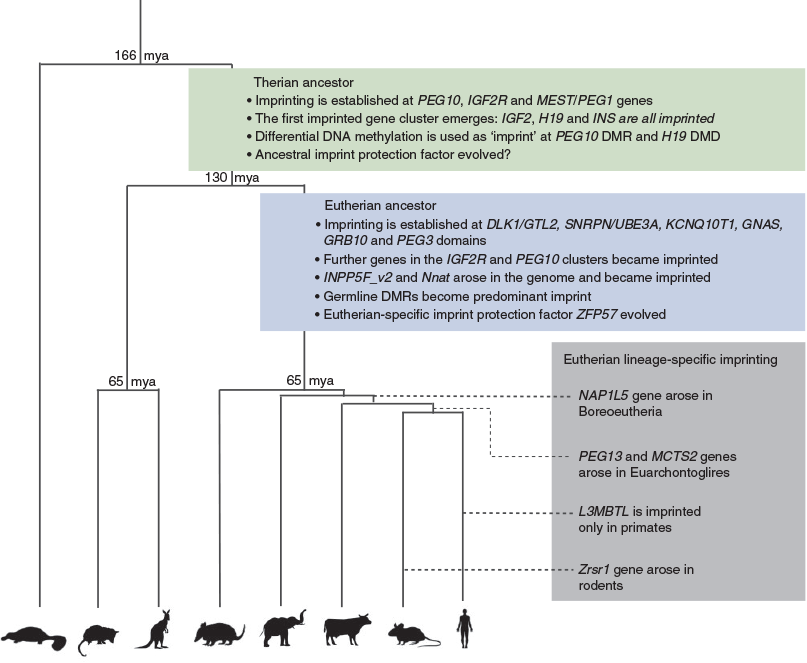
Most imprinted clusters were established in the eutherian ancestor before radiation
Genes that are imprinted in eutherians but not in marsupials fall into two categories: those that have a marsupial orthologue and those that do not. Thirteen genes fall into the first category and 11 fall into the second (Table 1).
The DLK1/GTL2 cluster contains genes from both categories (Fig. 1). The paternally expressed protein coding genes DLK1 and DIO3 are both present in the marsupials and monotremes but are biallelically expressed (Weidman et al. 2006b; Edwards et al. 2008). In addition, remnants of the retrotransposition event that formed the retrotransposon Gag like 1/paternally expressed 11 (RTL1/PEG11) gene in eutherians are present in marsupials but they lack the eutherian-specific open reading frames (Edwards et al. 2008). In contrast, the maternally expressed lncRNA, gene trap locus 2 /maternally expressed 3 (GTL2/MEG3) and the long arrays of imprinted microRNAs (miRNAs) and small nucleolar RNAs (snoRNAs) embedded within the DLK1/GTL2 cluster have no orthologues in marsupials or monotremes.

|
The Snrpn imprinted cluster is the largest eutherian imprinted domain, being over 3 Mb in the mouse. It is not present in marsupials (Rapkins et al. 2006). Misregulation of imprinting in this region in humans leads to two distinct neurological disorders, Prader–Willi syndrome and Angelman syndrome (Buiting 2010). In eutherians, the region contains two maternally expressed genes, ubiquitin protein ligase E3A (Ube3a) and ATPase, class V, type 10A (Atp10a), four paternally expressed protein-coding genes (makorin, ring finger protein, 3 (Mkrn3), melanoma antigen, family L, 2 (Magel2), necdin (Ndn) and small nuclear ribonucleoprotein N (Snrpn)) and paternally expressed non-coding (nc)RNAs including large arrays of snoRNAs. Genomic studies of this region in marsupials reveal that the region is eutherian specific. SNRPN is present in the tammar wallaby and opossum, but it is tandemly repeated next to its parent gene small nuclear ribonucleoprotein B (SNRPB) (Rapkins et al. 2006). In the tammar wallaby, SNRPN is on chromosome 1q, whereas UBE3A is on chromosome 5. MKRN3, MAGEL2, NDN and the snoRNAs are all absent in marsupials, indicating that region only came together in the eutherians perhaps along with the evolution of its imprinting (Rapkins et al. 2006).
In eutherians, the CDKN1C imprinted domain lies directly next to the IGF2/H19 cluster and this synteny is conserved in the tammar wallaby (Ager et al. 2008a). The mouse Cdkn1c region contains 10 imprinted maternally expressed protein coding genes including: achaete-scute family bHLH transcription factor 2 (Ascl2), cyclin dependent kinase inhibitor 1C (Cdkn1c), potassium voltage-gated channel, subfamily Q, member 1 (Kcnq1) and pleckstrin homology like domain family A member 2 (Phlda2). The paternally expressed ncRNA gene Kcnq1ot1, which is an antisense transcript to Kcnq1, is known to regulate imprinted gene expression across the cluster (Fitzpatrick et al. 2002). Interestingly, KCNQ1OT1 is expressed in the tammar wallaby, but is biallelically expressed and lacks the differentially methylated promoter that acts as the imprinting control region in eutherians (Ager et al. 2008a). Consistent with this, CDKN1C and PHLDA2 have both been shown to be biallelically expressed in the tammar wallaby (Ager et al. 2008a; Suzuki et al. 2011), hence imprinting is also not conserved at this region.
Data from these studies indicate that most genes acquired imprinted regulation in the eutherian lineage. However, eutherian studies have only been performed on mammals belonging to the suborders Euarchontoglires (e.g. primates and rodents) or Laurasiatheria (e.g. canines and ungulates), whereas the Xenarthra and Afrotheria have not been studied. By studying available genomes of Xenarthra and Afrotheria species we are able to ascertain when genes missing from marsupial genomes first arose in the eutherian lineage. In addition, by looking at the conservation of elements, such as known imprinting control regions (ICRs) in mouse and human through eutherian evolution, we can infer whether imprinting arose before or after the eutherian radiation.
For example, we know that the DLK1/DIO3 region in marsupials lacks the maternally expressed ncRNAs as well as an intact copy of RTL1 (Edwards et al. 2008). In contrast, by performing sequence analysis on the DLK1/DIO3 region in the elephant (Afrotheria) and armadillo (Xenarthra) we identified GTL2, snoRNAs and miRNA orthologues within the domain (UCSC genome browser; Karolchik et al. 2012). Both species also contain intact copies of RTL1. Furthermore, regions of homology to the ICR, the intergenic DMR (IG-DMR), were found in both elephant and armadillo (Fig. 1). Bisulfite pyrosequencing of the IG-DMR conserved region in elephant samples shows the region is approximately 50% methylated, indicating that this region is likely to be a DMR in the elephant (Fig. 1).
Using comparative sequence analysis, we can predict when the majority of imprinted domains were established (Fig. 2). For example, some sequence conservation between the ICRs of the SNRPN, GRB10 (growth factor receptor bound protein 10) and GNAS (guanine nucleotide binding protein, alpha stimulating) clusters was identified in at least one species from all eutherian clades (UCSC genome browser conservation track; Karolchik et al. 2012), suggesting that imprinting regulation was established in the common eutherian ancestor for each of these domains. However, to confirm imprinting in all these domains, expression and methylation analyses would need to be performed in relevant species. Four of the genes that have no marsupial orthologue are retrotransposed copies of X-linked genes that have arisen at various points in eutherian evolution (Wood et al. 2007). Taken together, the data indicate that most of the imprinting that has been characterised in eutherians was established after their divergence from marsupials but before the eutherian radiation (between 65 and 130 million years ago), but that imprinting at other loci has arisen subsequently, for example when a retrotransposition event has occurred, and perhaps governed by novel selective pressures.
|
Function of imprinted genes in eutherians
In order to understand why the imprinting process arose in mammals, it is essential to analyse the functions of known imprinted genes. Several human syndromes are caused by dysregulation of imprinted regions through either uniparental disomies (UPDs), microdeletions or defects in imprinting regulation. The phenotypes of these syndromes provide some clues to the functions of imprinted genes, such as in growth and neural function. Silver-Russell syndrome (dysregulation of multiple clusters), Temple syndrome (DLK1/GTL2 domain) and transient neonatal diabetes mellitus (pleiomorphic adenoma gene-like 1 - PLAGL1 domain) all lead to intrauterine growth retardation (Temple et al. 2000; Abu-Amero et al. 2008; Ioannides et al. 2014), whereas patients with Beckwith–Wiedemann syndrome (IGF2 and CDKN1C clusters) have fetal and postnatal overgrowth (Weksberg et al. 2010). Prader–Willi syndrome leads to hyperphagia and obsessive compulsive behaviour with mild mental retardation and is caused by maternal UPD15 or paternal deletions at 15q11–13, which contains the SNRPN cluster. Paternal UPD15 or maternal deletions at this domain lead to Angelman syndrome, another neurological disorder, but in this case exemplified by severe mental retardation hyperactivity and constant laughter (Nicholls and Knepper 2001). Mental retardation is also evident in patients with Kagami–Ogata syndrome (paternal UPD14), and mild intellectual disability is associated with Temple syndrome (maternal UPD14; Ioannides et al. 2014; Kagami et al. 2015).
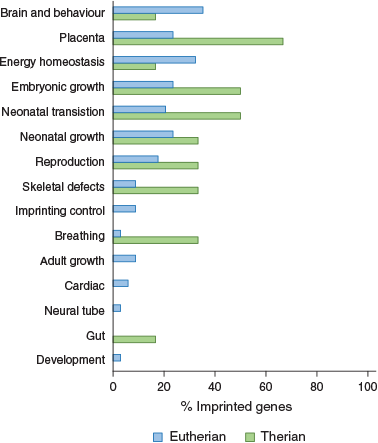
An extensive review of phenotypes of mouse models of imprinted genes found that most imprinted genes function in neonatal transitions and metabolism in addition to fetal and neonatal growth, placentation and behaviour (Cleaton et al. 2014). Brain and behaviour was the most common function of murine imprinted genes, followed by placentation and energy homeostasis (Fig. 3). Interestingly, when we look at marsupial imprinted genes we see that they do not fall into the category associated with brain and behaviour, but rather most of them have a role in placentation and all are known to be expressed in the marsupial YSM placenta (Fig. 3; Suzuki et al. 2005, 2007; Ager et al. 2007, 2008b; Smits et al. 2008; Stringer et al. 2012b). This suggests that the major role of imprinting in marsupials is the control of nutritional resources, whereas in eutherians imprinting is also important in controlling behaviour and postnatal adaptations during the life course.
|
Imprinted genes have major roles in placentation
An important role for imprinted genes in human and mouse placentation is well established (for reviews, see Coan et al. 2005; Monk 2015). In mice, early experiments using pronuclear transfer demonstrated that a full complement of both maternally and paternally derived genomes was necessary for mammalian development to term (Barton et al. 1984; McGrath and Solter 1984; Surani et al. 1984). Gynogenetic and parthenogenetic conceptuses generally develop only as far as the 25 somite stage and have poorly developed extra-embryonic tissues (Surani and Barton 1983; Surani et al. 1984). In contrast, androgenotes have well developed extra-embryonic tissues but very retarded embryos (Barton et al. 1984), indicating roles for imprinted genes in both placental and fetal development.
One of the first imprinted genes discovered, Igf2, has a critical role in nutrient transfer in the placenta. Mice with a global paternal deletion of the gene have small placentas with fewer glycogen cells and increased spongiotrophoblast cells in the junctional zone (Lopez et al. 1996). These placentas, in the absence of fetal IGF2, have reduced levels of the amino acid transporter solute carrier family 38 member 2 (Slc38a2) (Constância et al. 2005). In addition, deletion of a labyrinthine trophoblast-specific transcript which derives from the P0 promoter (Igf2-P0) in mice leads to reduced diffusion and permeability from embryonic day (E) 15.5. However, in the presence of fetal IGF2, these placentas upregulate the glucose transporter gene solute carrier family 2 member 3 (Slc2a3) and the imprinted amino acid transporter solute carrier family 38 member 4 (Slc38a4) (Constância et al. 2002, 2005; Sibley et al. 2004). The Igf2r gene is also expressed in the placenta. Maternal deletion of this gene leads to placentomegaly and larger fetuses that cannot survive to term (Lau et al. 1994; Wang et al. 1994). Fetuses lacking a maternal copy of Igf2r have higher levels of circulating IGF2 (Lau et al. 1994) and the overgrowth phenotype is corrected in mice also lacking a functional Igf2 gene (Filson et al. 1993). Thus, although the primary function of IGF2 is in the regulation of nutrient supply and demand in the placenta and fetus, the primary role for IGF2R is in modulating IGF2 levels (Filson et al. 1993). Other genes involved in nutrient transfer are also imprinted in the placenta, including Slc22a2 and Slc22a3 in the Igf2r cluster (Zwart et al. 2001) and solute carrier family 22 member 18 (Slc22a18) in the Kcnq1 domain (Dao et al. 1998). Genes in the Dlk1/Gtl2 cluster regulate placental development and function (Georgiades et al. 2001; Sekita et al. 2008; Ito et al. 2015). Furthermore, Grb10 has been shown to influence fetal resource acquisition because deletion of this maternally expressed gene results in placental overgrowth and increased placental efficiency (Charalambous et al. 2010).
The placenta also has a critical endocrine role inducing and maintaining physiological changes in the mother. Mothers need to undergo substantial changes in food intake and metabolism, the cardiovascular system, immune system and mammary glands during pregnancy, all of which are controlled, in part, by endocrine secretions by the conceptus. Recently, it was shown that fetus-derived DLK1 is found at high levels in maternal blood in mouse and human, where it plays a significant role in maternal metabolic adaptations (Cleaton et al. 2016). It has also been proposed that by regulating the size of different compartments of the placenta, imprinted genes are regulating not only resource allocation, but also the amount of hormones received by the mother, which, in turn, can influence the mother’s adaptation to pregnancy (John 2013). Seven imprinted genes were identified that affect placental endocrine lineages in the mouse: Ascl2, Phlda2, Cdkn1c and Igf2r, which are expressed from the maternally inherited copy, and paternally expressed 3 (Peg3), Peg10 and Igf2, which are paternally expressed (John 2013).
Deletion of Igf2 causes growth restriction in all endocrine lineages. Placental growth restriction has been reported for mice lacking Peg3 (Curley et al. 2004), and more recent expression data suggest that lack of Peg3 does have an effect on endocrine lineages (Broad and Keverne 2011; John 2013). Peg10 has a critical role in placentation; knockout mouse placentas completely lack spongiotrophoblast and embryos die at E10.5 due to placental failure (Ono et al. 2006). Igf2r affects the placenta by acting on IGF2, as discussed above (Filson et al. 1993). The three other maternally expressed genes that influence the endocrine compartments in the placenta are all located within the coordinately regulated Cdkn1c imprinted cluster. Phlda2 encodes a pleckstrin homology-like domain protein that acts to negatively regulate the spongiotrophoblast lineage (Frank et al. 2002) and Ascl2 encodes a transcription factor that is believed to repress the formation of parietal trophoblast giant cells, as well as acting upstream of Phlda2 to repress spongiotrophoblast formation (Tunster et al. 2016). Cdkn1c also represses spongiotrophoblast and labyrinthine trophoblast proliferation (Takahashi et al. 2000). Together, these data indicate that of the major site of imprinted gene expression is the placenta and their function in eutherians is to control the supply and demand of prenatal resources.
Placentation and imprinting evolution
Because imprinted loci repress one of their two gene copies, they exhibit functional haploidy and lose the protection that diploidy provides against deleterious mutations. This has led to much speculation about why the process arose. In this section we discuss three of the most popular theories of imprinting evolution, the conflict/kinship theory, the supply and demand theory and the maternal–offspring coadaptation model, and how they relate to what we currently know about marsupial and eutherian imprinting.
The reciprocal functions and imprinting status of Igf2 and Igf2r led to perhaps the most prevailing theory of imprinting evolution, the conflict/kinship theory (Moore and Haig 1991). This theory argues that imprinting arose as a consequence of a conflict of interest between maternally and paternally derived genomes driven by prenatal resource control. It suggests that paternally expressed genes, such as Igf2, would be growth enhancing, favouring greater resource allocation from the mother both in utero and perinatally, perhaps to the detriment of later offspring of the mother. Conversely, imprinted genes expressed from maternally inherited chromosomes, such as Igf2r, would be growth limiting, allowing her offspring to traverse the birth canal and to conserve her resources for future offspring. Most imprinted genes with a placental phenotype in mouse knockout models follow this prediction. Deletion of six paternally expressed genes, namely Igf2, Peg1/Mest, Peg3, Peg10, Plagl1 and Rtl1, causes placental growth restriction, and deletion of six imprinted genes expressed from maternally inherited chromosomes, namely Igf2r, Cdkn1c, Phlda2, Grb10, Rtl1as and H19, causes placentomegaly (Cleaton et al. 2014). Interestingly, deletion of one maternally expressed gene, namely Ascl2, leads to complete loss of spongiotrophoblast and is embryonic lethal at E10.5, similar to the Peg10-knockout mouse, which also lacks spongiotrophoblast and dies at E10.5, which would argue against the conflict hypothesis (Guillemot et al. 1994; Ono et al. 2006). However, Ascl2-null mice have an increased number of parietal trophoblast giant cells and mice expressing an Ascl2 transgene show a reduction in the spongiotrophoblast compartment, suggesting that ASCL2 does have a growth-limiting function in these cell lineages (Tunster et al. 2016). Furthermore, a larger placenta does not necessarily mean a more efficient placenta. For example, mice with paternal UPD of chromosome 12 (i.e. two paternally inherited Dlk1/Gtl2 domains including Rtl1) show placentomegaly, but have defects in all three layers of the placenta, including reduced fetal capillary volume (Georgiades et al. 2000, 2001). The conflict/kinship theory seems only to apply to the subset of imprinted genes regulating growth and placentation, including those that evolved imprinting in marsupials.
The supply and demand theory of imprinting proposes that imprinted genes in the placenta are controlling the supply of nutrients to the placenta, whereas imprinted genes in the fetus are controlling the demand for nutrients (Reik et al. 2003). For example, in the fetus paternally derived Igf2 is controlling demand by promoting growth, whereas in the placenta it is controlling supply through its effects on diffusion, permeability and transport. Conversely, maternally expressed genes Phlda2 and Cdkn1c in the adjacent imprinting cluster can counteract the effects of Igf2 by reducing nutrient supply. Maternally expressed Igf2r would act to suppress supply and demand in the fetus and placenta through its negative regulation of IGF2 levels. Reik et al. (2003) propose that the regulation of placental supply and fetal demand is a particular function of imprinted genes and suggests the coevolution of imprinting and placentation. Again, this theory is formulated around the more ancestral placental-specific prenatal resource control functions of imprinted genes. The evolutionary pressure behind the direction of imprinting is predicted to be dependent, in part, on parental conflict, and the supply and demand theory can be seen as an extension to the conflict/kinship theory.
The maternal–offspring coadaptation model for imprinting evolution was first proposed by Wolf and Hager (2006). Coadaptation occurs when offspring genes evolve to function with a particular parentally supplied environment and when the parental genotype for this environment becomes associated with the offspring genes that are adapted to it (Wolf and Brodie 1998). The maternal–offspring coadaptation model suggests that the expression of genes from maternally inherited chromosomes increases the adaptive integration of mother and offspring genomes where there are close maternal–offspring interactions, such as in the placenta. This theory can explain the observation that all genes that are exclusively imprinted in the placenta are maternally expressed (Wolf and Hager 2006). However, although it can include postnatal maternal–offspring relationships that do not include the placenta, it is hard to reconcile this model to imprinted genes with key roles to play in postweaning functions.
Both the supply and demand and the conflict/kinship theories predict that imprinting would exist in animals with placentas but not in egg-laying animals. Phylogenetic data are in agreement with this and suggest imprinting and placentation evolved in parallel: the number of imprinted genes has increased as mammalian placentas became more complex and able to sustain longer in utero development. To date, no imprinted genes have been identified in non-mammalian vertebrates or monotremes (Killian et al. 2000, 2001; Edwards et al. 2008). Placentation in monotremes is the most rudimental. Although these are egg-laying species, fetuses receive endometrial secretions via the shell, suggesting the level of nutrients received by the fetus is indeed dictated by the mother. Eggs are laid in the short-beaked echidna at the 18–20 somite stage (~18 days after conception) and are incubated for 10–11 days.
Imprinting is only seen in marsupials and eutherians, mammals that have a direct apposition of the endometrium and fetal membranes. Early in gestation, marsupials receive nutrients via a shell membrane, but this ruptures later in gestation at approximately the same stage that monotreme eggs are laid (18 days after removal of pouch young in the tammar wallaby, which reactivates the diapause-arrested blastocyst). After the marsupial shell membrane ruptures, a direct contact between the YSM and the endometrium is formed, possibly allowing for the fetus to have some control over the levels of nutrients it receives. Interestingly, IGF2 and growth hormone expression in the tammar wallaby placenta increase at this time, indicating that the conceptus is able to secrete hormones and signal to the mother (Menzies et al. 2011). Once the shell membrane ruptures, gestation continues in the tammar wallaby for a further 8 days.
The six identified imprinted genes in marsupials are all expressed in the marsupial placenta, and five have been shown to be imprinted (IGF2R imprinting has not been assessed in marsupial placenta yet; Suzuki et al. 2005, 2007; Ager et al. 2007, 2008b; Smits et al. 2008; Stringer et al. 2012b). Although gene manipulation technologies are evolving for non-model organisms, to date it has been difficult to perform embryonic manipulations on marsupials, so the roles of these genes in marsupial placentation have not been assessed. However, we do know from mouse models that five of these genes affect size and/or efficiency of the eutherian placenta (IGF2, H19, IGF2R, PEG1/MEST and PEG10) and fit with the supply and demand model of imprinting evolution. The imprinting of these genes also agrees with the conflict hypothesis: IGF2, PEG1/MEST and PEG10 are growth enhancing and paternally expressed, whereas IGF2R and H19 are maternally expressed and growth suppressing. The maternal–offspring coadaptation model is harder to reconcile with the marsupial data. This theory predicts genes involved in placentation would be maternally expressed; however, only two maternally expressed genes have been identified in the marsupial placenta, IGF2R and H19, and major functions of both these genes are in modulating IGF2 levels in the fetus (Filson et al. 1993; Ripoche et al. 1997; Wilkin et al. 2000; Gabory et al. 2009). Of course, it is possible that a completely different set of genes regulates marsupial placentation, and these may exhibit metatherian-specific imprinting.
Mammary gland: a site for genomic imprinting?
The theories described above propose specific roles for imprinted genes in resource acquisition from mother to child, particularly in utero. However, mammalian young continue to acquire nutrients from their mother after birth via lactation. Conflict is unlikely to influence the evolution of imprinting in the mammary gland because, unlike the placenta, the genome of the offspring’s father is not represented here and the grandparental genomes, which are present, are unlikely to be in conflict because both are equally likely to be present in the neonates. However, conflict could still lead to imprinting in neonates, and one would expect genes involved in suckling and appetite to be imprinted if this is the case. The coadaptation model can also be allied to imprinting in postnatal resource allocation (Renfree et al. 2013). For example, coadaptation may lead to the imprinting of genes involved in suckling, because this would enhance the genetic integration of intimate maternal–offspring interactions (Stringer et al. 2014). However, this theory also would not predict genes in the mammary gland being imprinted because it only requires genes to be expressed from maternally inherited chromosomes in the pup to exhibit increased relatedness to the mother. Therefore, none of the models proffered to date predicts the mammary gland to be a major site for imprinting, implying that if imprinting does occur here, it would be due to different evolutionary pressures or the absence of pressures in the mammary gland selecting against an imprinting status established during development.
To date, the mammary gland has not been a tissue that has been extensively studied in the imprinting field. However, Grb10 has recently been shown to be expressed from the maternally inherited allele in lactating mammary glands in mice expressing a Grb10-driven LacZ reporter (Cowley et al. 2014). It was demonstrated that Grb10 performs complimentary functions in mothers and pups: in pups, GRB10 supresses growth, whereas in mothers Grb10 expression increases milk production. Thus, postnatal Grb10 expression fits with the supply and demand theory of imprinting because it controls the supply of nutrients from the mother and the pups’ demand for resources. The complementary and pleiotropic effects of Grb10 expression in mother and offspring also fit with coadaptation (Cowley et al. 2014). Studies specifically assessing imprinting in mammary glands have not been performed for any other genes. A recent study using RNA sequencing (RNA-seq) demonstrated that 25 known imprinted genes were imprinted in virginal and/or lactating mammary glands (Andergassen et al. 2017). The authors of that study concluded that because there was little difference in imprinting between virginal and lactating mammary glands, there was no specific role for imprinted expression in lactation. Interestingly, in that study expression was reported for Grb10 at similar levels for virgin and lactating mammary glands (Andergassen et al. 2017), whereas Cowley et al. (2014) found no Grb10 reporter expression in virginal mammary glands. It is likely that gross RNA-seq analysis of whole mammary glands fails to take into account the cellular complexity of this organ, indicating that a more detailed systematic approach is needed to fully understand the role of imprinted genes in the mammary gland and postnatal provisioning.
In addition to Grb10, six mouse imprinted models have been shown to affect postnatal maternal provision: null models of Peg3, GnasXl, Magel2, Cdkn1c and Igf2r show impaired suckling (reviewed in Cleaton et al. 2014). Deletion of three paternally expressed genes, namely Magel2, Peg1/Mest and Peg3, has been reported to cause defects in maternal care (Lefebvre et al. 1998; Li et al. 1999; Schaller et al. 2010). Interestingly, loss of function in Peg3 causes failure in postnatal feeding when the deletion is present in the mother or her offspring, suggesting coadaptation. When mothers lack a functional copy of the gene, they fail to respond to signals from the wild-type placenta, they increase food intake in early pregnancy and they show impaired milk let down, leading to growth-retarded wild-type pups. When the pup is lacking a functional copy of the gene, it has impaired suckling efficiency compared with wild-type litter mates (Curley et al. 2004). These observations led to the proposal of a modified coadaptation model suggesting that genes that are simultaneously expressed in the maternal hypothalamus, placenta and fetal hypothalamus, such as Peg3, would tend to be switched off on the maternal allele (Keverne and Curley 2008). This would allow rapid fixation of positive traits in the population. When an advantageous mutation is inherited from the father, all offspring would benefit from efficient placental transfer in utero and good maternal care after birth, which is primed by expression in the placenta. If the advantageous mutation is inherited from the mother, it will be silenced in the offspring but they would still benefit from good maternal care and milk let down via Peg3 action in the maternal hypothalamus (Keverne and Curley 2008).
A recent study questions this theory because a different deletion of Peg3 exon 9 (removing 90% of the coding sequence including all of the zinc fingers) found no maternal behaviour, lactation or suckling deficiencies (Denizot et al. 2016). This suggests that the previously reported phenotypes are due either specifically to the loss of the 5′ portion of the gene or to technical differences between experiments. The original Peg3 deletion left a beta-galactosidase-Neo (β-geo) cassette in exon 5 that could affect the expression of other genes in the region, but a more recent knockout that conditionally removed exon 6 in the mammary gland leaving only flippase recognition targets and Lox sites in place also showed that on paternal transmission, mothers had problems releasing milk (Li et al. 1999; Frey and Kim 2015). When Peg3 is truncated at exon 5 there is evidence that its maternally expressed downstream neighbour zinc finger, imprinted 1 (Zim1) is upregulated, but this is believed to be a trans process because expression of Zim1 is still predominantly from the maternal allele (Ye et al. 2014). Interestingly, no changes were seen in Zim1 expression in the Peg3 exon 9 deletion (Denizot et al. 2016). Genetic background can also influence the phenotype of mutations: the exon 9 deletion is on a C57Bl/6J background, whereas the original Peg3 deletion was generated in 129Sv mice (Li et al. 1999; Denizot et al. 2016). However, the model of Li et al. (1999) was later back-crossed onto a C57Bl/6J background and similar behavioural phenotypes were observed (Champagne et al. 2009). Furthermore, the mammary-specific mutation was also on a C57Bl/6J background (Frey and Kim 2015). Together, these observations indicate that the phenotypic differences in the models are most likely due the positioning of the deletion or different methods used to assess the phenotypes. Further work is necessary to confirm whether PEG3 does indeed influence maternal behaviours and lactation. This is especially relevant because the Keverne and Curley (2008) extension of the coadaptation theory has been developed around these functions.
Marsupials are altricial and rely more heavily on lactation for maternal provision of nutrients than eutherians. For example, the tammar wallaby has a 26.5-day gestation period followed by up to 350 days lactation (13.2-fold longer than gestation), whereas mice have a 20-day gestation followed by up to 24 days lactation (1.2-fold longer than gestation). By weaning, the average litter mass in marsupials is 55%, compared with 59% in eutherians, indicating that maternal investment is similar in both reproductive strategies (Hayssen et al. 1985). The mammary gland not only provides nutrition to offspring, but it also provides a biochemical signalling route between the mother and her young, with milk containing many signalling molecules including insulin and IGF2 (Malven et al. 1987; Prosser 1996). This is the ancestral mechanism of signalling between mother and child, and functions that are performed by the placenta in eutherians are thought to be performed via milk in the marsupial (Power and Schulkin 2013). In agreement with this are recent data from RNA-seq that indicate that the marsupial mammary gland shares many transcripts with the eutherian placenta, including genes involved in nutrient transport and IGF-binding protein 1 (Igfbp1), which is important in IGF regulation (Guernsey et al. 2017). This suggests that placentation and lactation are performing similar functions in resource acquisition in eutherians and marsupials respectively. If this is the case, then the main site of imprinting in the marsupial would be the mammary gland (Stringer et al. 2014) and eutherians may have less dependence on imprinting in the mammary gland. Imprinting analysis in marsupial mammary glands has been performed for three genes. GRB10 was biallelically expressed in adult tammar wallaby mammary glands (Stringer et al. 2012a). This is not unexpected because GRB10 in marsupials lacks the paternal-specific promoter required for central nervous system expression in eutherians, indicating that this is a eutherian-specific imprinted gene (Garfield et al. 2011; Stringer et al. 2012a). Both IGF2 and INS are monoallelically expressed in tammar wallaby mammary gland (Stringer et al. 2012c). In eutherians, Ins2/INS imprinting has only been reported in the yolk sac (Deltour et al. 1995; Moore et al. 2001); its more sustained imprinting in tammar wallaby indicates that marsupials may have a different repertoire of imprinted genes to eutherians, and that by simply testing known imprinted genes in these species we may be missing key examples.
The intimate relationship between mother and fetus or neonate in mammals provides the young with all their nutritional needs. Failure to establish and maintain this relationship during pregnancy or postnatally can have implications to offspring that last for the rest of their life. Imprinted gene expression is enriched at all stages of this relationship, and the role of imprinted genes in placentation is well established. Neither the coadaptation nor conflict hypotheses predict that the mammary gland would be a site for imprinted expression; however, there is evidence of imprinted expression in this tissue, although few genes have been studied to date. This suggests that either a different evolutionary pressure is present here or that imprinting in the mammary gland simply reflects the gene’s status from earlier in development. Because monotremes also have a long lactation period, it may be that these most distantly related mammals may also exhibit imprinting in mammary glands. Clearly, a systematic analysis of this organ in monotremes, marsupials and eutherians is necessary to fully understand the roles of imprinted genes in postnatal resource provisioning in mammals and the evolution of this remarkable process.
Evolution of the imprinting mechanisms: lessons from marsupial imprinting
In order for the transcriptional machinery of a cell to be able to distinguish the maternally and paternally inherited copies of imprinted genes, it is necessary for the chromosomes to be marked in some way. DNA methylation is the primary ‘imprint’ in eutherians. All known imprints are germline (g) DMRs that occur over an ICR. ICRs are genomic elements that control the imprinted expression of a singleton imprinted gene or of all imprinted genes within a coordinately regulated cluster. The majority of ICRs (23 are confirmed) are maternally methylated gene promoters that gain methylation during oogenesis but remain unmethylated in spermatozoa. Only three paternally methylated ICRs have been identified; these are intergenic elements that control the Igf2/H19, Dlk1/Gtl2 and Rasgrf1 (RAS protein-specific guanine nucleotide-releasing factor) domains and become methylated during spermatogenesis but remain unmethylated in ova.
There are only limited data indicating that the imprinting mechanism is conserved between eutherians and marsupials. The H19 differentially methylated domain (H19 DMD) in the tammar wallaby is differentially methylated in pouch young samples and hypermethylated in adult testis (Smits et al. 2008). The tammar H19 DMD also contains CCCTC-binding factor (CTCF) binding sites and has insulator activity similar to those seen at the mouse ICR, indicating the region is likely to be functionally conserved between marsupials and eutherians (Smits et al. 2008). The PEG10 promoter is a maternally methylated DMR in the tammar wallaby, as in the mouse (Suzuki et al. 2007). In the tammar wallaby, PEG10 is a singleton imprinted gene, whereas in eutherians the DMR controls the imprinted expression of neighbouring genes as well (Ono et al. 2003; Suzuki et al. 2007). In the mouse, the Igf2r imprinting domain is controlled by a maternally methylated ICR that is the promoter for the antisense transcript Airn located in intron 2 of Igf2r (Wutz et al. 1997). No DMR has been identified in the corresponding intron in marsupials although a maternally methylated DMR has been reported in intron 11 (Killian et al. 2000; Weidman et al. 2006a; Das et al. 2012). Interestingly, no orthologue for Airn has been reported in marsupials, suggesting a different mechanism of imprinting control in this region. With regard to other loci, incomplete imprinting of one MEST transcript has been reported in the tammar wallaby, but no differential methylation has been observed at the promoter for this transcript, which is where the ICR is found in eutherians (Suzuki et al. 2005).
Together, these data indicate that a different imprinting control repertoire may be used by marsupials to mark the maternal and paternal copies of IGF2R and MEST. Recently, researchers have found that maternal histone 3 lysine 27 trimethylation (H3K27me3) is associated with repression of some maternal alleles of paternally expressed genes in the preimplantation embryo (Inoue et al. 2017). Imprinting at these genes was lost in the embryo by the epiblast stage, but four genes appeared to retain paternal-specific expression in the placenta. It is therefore possible that histone modifications rather the DNA methylation may represent a more ancestral form of imprinting control. Although the establishment of imprinting by H3K27me3 had never been reported before, a role for H3K27me3 in maintaining imprinted expression in response to the germline-derived DNA methylation imprint has been reported previously (Lewis et al. 2004; Umlauf et al. 2004; Yamasaki-Ishizaki et al. 2007). Therefore, further analyses of the epigenetic profiles at marsupial IGF2R and MEST are necessary to explore possible mechanisms that may imprint these genes.
It was initially thought that gDMRs were established specifically at ICRs, but subsequent whole-methylome sequencing studies have indicated that there are many more gDMRs than there are imprints (Kobayashi et al. 2012). Therefore, what sets ICRs apart from these other gDMRs is their resistance to reprogramming after fertilisation. There are two waves of epigenetic reprogramming in eutherian development. The first takes place in the primordial germ cells (PGCs) as they migrate towards the genital ridge early in embryogenesis. In mice, this wave of demethylation is completed by E13.5 and all imprints are erased (Hajkova et al. 2002). De novo methylation then occurs in both germlines. In the tammar wallaby, PGCs complete migration to the genital ridge just before birth and continue to proliferate until 25 days after birth (Alcorn and Robinson 1983; Renfree et al. 1996; Ullmann et al. 1997). Analysis of the only two DMR imprints identified in marsupials found that the relative timing of reprogramming was conserved between eutherians and marsupials. The PEG10 DMR and H19 DMD became fully demethylated by Days 7 and 14 postpartum respectively, and de novo methylation at H19 DMD in the male germline started at Day 34, demonstrating that the basic mechanisms of the first wave of reprogramming are conserved within therians (Suzuki et al. 2013).
The second round of global demethylation occurs after fertilisation in the preimplantation embryo. Of considerable importance, the only gDMRs that are maintained during this wave of reprogramming are imprints that are protected by proteins that target the methylated copy. Postfertilisation reprogramming has not been studied in non-eutherian mammals, so it is not known whether the PEG10 DMR and H19 DMD are also protected from this genome-wide wave of demethylation.
The retention of imprints during this second wave of global demethylation is the key step in imprinting control because it preserves the epigenetic memory of parental origin. Li et al. (2008) demonstrated that zinc finger protein 57 (ZFP57) was necessary for the maintenance of several imprints during postfertilisation reprogramming. ZFP57 is a Kruppel associated box (KRAB) zinc finger protein (KZFP) that is highly expressed in the oocyte and, unlike most of the other approximately 280 KZFPs studied to date in the eutherian genome (Imbeault et al. 2017), it binds to methylated DNA. Chromatin immunoprecipitation (ChIP) analysis of C57BL/6J × Castaneus mouse hybrid embryonic stem cells demonstrated that ZFP57 binds to the methylated copy of all imprinted DMRs (Strogantsev et al. 2015). Zfp57 is a maternal–zygotic effect gene because mutant mice lacking both the oocyte and zygotically expressed copies of the gene (MZ−/−) die prenatally (Li et al. 2008; Takahashi et al. 2015). Mice lacking a zygotic copy of the gene have a much milder phenotype, indicating that maternal oocyte-derived ZFP57 may have an important role in protecting imprints in the early preimplantation embryo. This was confirmed by a comprehensive analysis of ICR methylation in E12.5 tissues from MZ−/− mice. Of the nine ICRs studied, seven showed a significant reduction in methylation in MZ−/− mutants (Table 2). However, the degree of methylation loss varied; for example the Snrpn DMR showed complete loss of methylation, whereas the Igf2r DMR showed only a 10–20% reduction (Takahashi et al. 2015). Two ICRs showed no loss of methylation in MZ−/− mice: (1) KvDMR, which controls the Kcnq1ot1 imprinted domain; and (2) the H19 DMR (Table 2). These analyses have since been extended, identifying two additional ICRs that remain considerably protected from demethylation in MZ−/− mouse mutants (Peg10 and Mest; N. Takahashi and A. C. Ferguson-Smith, unpubl. data). Therefore, the data indicate that murine ICRs can be divided into three classes based on their ability to retain imprints in the absence of ZFP57: those dependent on ZFP57, those partially dependent on ZFP57 and those not dependent on ZFP57 (Table 2). These data suggest that there is a least one other protein that is required to protect imprints in the preimplantation embryo. Importantly, none of the four domains that are imprinted in marsupials is completely protected by Zfp57 in eutherians, suggesting that such additional factors may be critical for the maintenance of marsupial imprinting and are likely to be more ancient than Zfp57, which is only found in eutherians (Imbeault et al. 2017).

|
It is possible that the more ancestral imprint protection factor belongs to the same gene family as Zfp57. The KRAB zinc finger family is one of the largest protein families in the human genome, containing over 350 members (Huntley et al. 2006; Imbeault et al. 2017). This is a rapidly evolving gene family that first arose in the Sarcopterygii lineage (tetrapods, coelacanths and lungfish; Imbeault et al. 2017). Most KZFPs have been shown to suppress transposable elements by recruiting KRAB-associated protein-1/tripartite motif containing 28 (KAP1/TRIM28) and establishing repressive chromatin marks such as H3K9me3 and DNA methylation (Wolf and Goff 2009; Quenneville et al. 2012; Jacobs et al. 2014; Imbeault et al. 2017). It has been proposed that KZFPs play a major role in host defence and that the rapid evolution of KZFPs is the result of an ‘arms race’ between transposable elements and the host genome (Jacobs et al. 2014). In agreement with this idea is the correlation between the number of long terminal repeat (LTR) retrotransposons and the number of KRAB zinc finger genes in mammalian genomes. For example, in the platypus genome, only 43 KZFPs have been identified and its genome only consists of 0.2% LTR retrotransposons. In the therian genomes the number of LTR retrotransposons and KZFP genes is much higher; 851 KZFPs have been identified in the marsupial opossum and 10% of its genome consists of LTR retrotransposons (Imbeault et al. 2017).
It is also possible that the rapid expansion of KZFPs at the same time as the evolution of imprinting facilitated the co-option of KZFPs for imprint protection in addition to a role in host defence. Indeed, it has long been proposed that the imprint mechanism evolved from a host defence strategy. Soon after DNA methylation was first identified as the imprint, it was suggested that the process may be an extension of its role in silencing foreign DNA (Barlow 1993). Multiple transgenes have been shown to become imprinted in a manner independent of their preintegration site (Reik et al. 1987; Sapienza et al. 1987; Swain et al. 1987; Chaillet et al. 1991; Sasaki et al. 1991). Furthermore, it was subsequently shown that several imprinted genes have themselves arisen from transposition events, including Peg10 and Rtl1/Peg11, which are both neogenes derived from Sushi-ichi retrotransposons (Ono et al. 2001; Seitz et al. 2003; Youngson et al. 2005) and Nap1L5 (nucleosome assembly protein 1-like 5), Zrsr1 (zinc finger, RNA binding motif and serine/arginine rich 1), Mcts2 (malignant T cell amplified sequence 2) and Inpp5f_v2 (inositol polyphosphate-5-phosphatase F variant 2), which are all retrocopies of X-linked genes (Wood et al. 2007). These theories of imprinting arising from a host defence mechanism are based on the fact that foreign DNA such as retrotransposons and retrocopies of genes are targeted for methylation to prevent erroneous expression and mobilisation.
Most KZFPs that have been characterised to date are involved in the establishment of repressive chromatin states and, in particular, recruiting H3K9me3. Although DNA methylation is the germline imprint, the relationship between DNA methylation and H3K9me3 is not fully defined temporally at imprints and, indeed, after fertilisation all ICRs are both DNA methylated and bound by H3K9 me3. A more in-depth study is required to identify conserved KZFPs that could bind and protect gDMRs at eutherian and marsupial imprinted domains.
Concluding remarks
By comparing and contrasting when, why and how imprinting arose in marsupials and eutherians, we can get an idea of the evolutionary processes that drove its acquisition in the therian ancestor. Both imprinting and viviparity first arose after the divergence of therians from the monotremes around 160 million years ago (O’Leary et al. 2013). In marsupials, six imprinted genes have been identified that represent the most ancestral imprinted genes. All these genes are expressed in the YSM, indicating that placentation and imprinting are closely linked. Moreover, PEG10 is a neogene derived from a retrotransposon that is only found in marsupials and eutherians. Its critical role in eutherian placentation and expression in the marsupial placenta suggest the emergence of this gene may be an important event in the evolution of placentation, perhaps driving the move towards viviparity (Ono et al. 2001; Suzuki et al. 2007).
Marsupial paternally expressed genes are growth enhancing, whereas maternally expressed genes are growth limiting, suggesting conflict/kinship and supply and demand were the major drivers of imprinting evolution at this stage in mammalian evolution. However, marsupials rely more heavily on lactation than eutherians to support the development of their young. It is of note that the INS gene is imprinted in the marsupial mammary gland, whereas in eutherians it is only imprinted in the yolk sac, supporting the idea that the mammary gland may be a particularly important site for imprinting in marsupials.
In eutherians, there is a switch of the major site of maternal resource allocation from the mammary gland to the placenta. That the majority of imprinted domains are evident in all four eutherian superorders along with long-lived, invasive chorioallantoic placentas once again points to the parallel evolution of imprinting with placentation. In this major wave of imprinting acquisition, more genes involved in resource allocation in the placenta became imprinted, including Cdkn1c, Phlda2, Ascl2, Grb10, as did transport genes such as Slc38a4 and Slc22a18. However, in addition to placentation, imprinted genes in eutherians have vital roles in maternal adaptation to pregnancy (Dlk1, Peg3), maternal care (Peg3, Magel2), metabolism (Gnas cluster) and behaviour (Snrpn cluster). This wide range of functions cannot easily be explained by a single evolutionary theory and suggests that more than one selective pressure may be acting at imprinted loci.
The mechanisms by which imprinted genes are marked and maintained appear to be different between marsupials and eutherians. In eutherians, all verified imprinted genes are marked by differential DNA methylation, which is established in the germline, but only two DMRs have been identified in marsupials, suggesting that a different more ancestral imprint mark may exist. The mechanisms by which methylation is maintained at ICRs appear to differ too; the majority of eutherian-specific ICRs are protected in the preimplantation embryo by Zfp57, whereas the more ancestral gDMRs are not. This also suggests that another more ancestral imprinting protection factor exists in marsupials.
We therefore hypothesise that mammalian clade-specific imprinted genes may exist that have evolved alongside the evolution of clade-specific mechanisms that target and maintain parental origin-specific epigenetic states at such loci and that the evolution of epigenetic pathways designed to control repressive states has contributed to their emergence. Further genome-wide systematic analysis of parental origin-specific gene expression in multiple species alongside detailed characterisation of epigenetic targeting mechanisms has the potential to test this hypothesis.
Materials and methods
Elephant samples
Elephant samples were collected at the Elephant Research Unit in the Lower Save Conservancy in Zimbabwe. Ele5 and Ele9 are placental tissues from two individuals, whereas Ele13 and Ele14 are amnion and umbilical cord from the same individual. DNA was extracted using standard phenol–chloroform protocols.
Bisulfite pyrosequencing
A 1-µg sample of genomic DNA was bisulfite converted using the Imprint DNA Modification Kit (Sigma Aldrich) according to the manufacturer’s instructions. Purified samples were amplified by polymerase chain reaction (PCR) performed in a final reaction volume of 10 mL containing 250 nM forward and reverse primers (forward, 5′-GGAAGTAGAGGGATGTTGGATGAA-3′; reverse, 5′-[Btn]CCCAAACTAACTCCATATCCTAAACC-3′), 0.25 U Taq (HotStarTaq DNA Polymerase; Qiagen) and 0.2 mM dNTPs. The PCR conditions were as follows: 95°C for 15 min, followed by 40 cycles of 94°C for 30 s, 48°C for 30 s and 72°C for 30 s, with a final step at 72°C for 5 min. Single-strand PCR products were purified (PyroMark Q96 Vacuum Prep Workstation; Qiagen) and pyrosequencing was performed on a PyroMark Q96MD (Qiagen) using PyroMark Gold Q96 Reagents (Qiagen) and the pyrosequencing primer 5′-GGGATGTTGGATGAAT-3′ in accordance with the manufacturer’s instructions.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The authors thank Twink Allen and Fiona Stansfield for supplying the elephant tissues. The work presented in the paper and the writing of the paper was supported by the Wellcome Trust (Grant no. WT095606).
References
Abu-Amero, S., Monk, D., Frost, J., Preece, M., Stanier, P., and Moore, G. E. (2008). The genetic aetiology of Silver–Russell syndrome. J. Med. Genet. 45, 193–199.| The genetic aetiology of Silver–Russell syndrome.Crossref | GoogleScholarGoogle Scholar | 18156438PubMed |
Ager, E., Suzuki, S., Pask, A., Shaw, G., Ishino, F., and Renfree, M. B. (2007). Insulin is imprinted in the placenta of the marsupial, Macropus eugenii. Dev. Biol. 309, 317–328.
| Insulin is imprinted in the placenta of the marsupial, Macropus eugenii.Crossref | GoogleScholarGoogle Scholar | 17706631PubMed |
Ager, E. I., Pask, A. J., Gehring, H. M., Shaw, G., and Renfree, M. B. (2008a). Evolution of the CDKN1C-KCNQ1 imprinted domain. BMC Evol. Biol. 8, 163.
| Evolution of the CDKN1C-KCNQ1 imprinted domain.Crossref | GoogleScholarGoogle Scholar | 18510768PubMed |
Ager, E. I., Pask, A. J., Shaw, G., and Renfree, M. B. (2008b). Expression and protein localisation of IGF2 in the marsupial placenta. BMC Dev. Biol. 8, 17.
| Expression and protein localisation of IGF2 in the marsupial placenta.Crossref | GoogleScholarGoogle Scholar | 18284703PubMed |
Alcorn, G. T., and Robinson, E. S. (1983). Germ cell development in female pouch young of the tammar wallaby (Macropus eugenii). J. Reprod. Fertil. 67, 319–325.
| Germ cell development in female pouch young of the tammar wallaby (Macropus eugenii).Crossref | GoogleScholarGoogle Scholar | 6834329PubMed |
Andergassen, D., Dotter, C. P., Wenzel, D., Sigl, V., Bammer, P. C., Muckenhuber, M., Mayer, D., Kulinski, T. M., Theussl, H. C., Penninger, J. M., Bock, C., Barlow, D. P., Pauler, F. M., and Hudson, Q. J. (2017). Mapping the mouse allelome reveals tissue-specific regulation of allelic expression. eLife 6, e25125.
| Mapping the mouse allelome reveals tissue-specific regulation of allelic expression.Crossref | GoogleScholarGoogle Scholar | 28806168PubMed |
Aziz, A., Baxter, E. J., Edwards, C., Cheong, C. Y., Ito, M., Bench, A., Kelley, R., Silber, Y., Beer, P. A., Chng, K., et al. (2013). Cooperativity of imprinted genes inactivated by acquired chromosome 20q deletions. J. Clin. Invest. 123, 2169–2182.
| Cooperativity of imprinted genes inactivated by acquired chromosome 20q deletions.Crossref | GoogleScholarGoogle Scholar | 23543057PubMed |
Barlow, D. P. (1993). Methylation and imprinting: from host defense to gene regulation?. Science 260, 309–310.
| Methylation and imprinting: from host defense to gene regulation?.Crossref | GoogleScholarGoogle Scholar | 8469984PubMed |
Barlow, D. P., Stoger, R., Herrmann, B. G., Saito, K., and Schweifer, N. (1991). The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349, 84–87.
| The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus.Crossref | GoogleScholarGoogle Scholar | 1845916PubMed |
Bartolomei, M. S., Zemel, S., and Tilghman, S. M. (1991). Parental imprinting of the mouse H19 gene. Nature 351, 153–155.
| Parental imprinting of the mouse H19 gene.Crossref | GoogleScholarGoogle Scholar | 1709450PubMed |
Barton, S. C., Surani, M. A., and Norris, M. L. (1984). Role of paternal and maternal genomes in mouse development. Nature 311, 374–376.
| Role of paternal and maternal genomes in mouse development.Crossref | GoogleScholarGoogle Scholar | 6482961PubMed |
Broad, K. D., and Keverne, E. B. (2011). Placental protection of the fetal brain during short-term food deprivation. Proc. Natl Acad. Sci. USA 108, 15237–15241.
| Placental protection of the fetal brain during short-term food deprivation.Crossref | GoogleScholarGoogle Scholar | 21810990PubMed |
Buiting, K. (2010). Prader–Willi syndrome and Angelman syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 154C, 365–376.
| Prader–Willi syndrome and Angelman syndrome.Crossref | GoogleScholarGoogle Scholar | 20803659PubMed |
Chaillet, J. R., Vogt, T. F., Beier, D. R., and Leder, P. (1991). Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell 66, 77–83.
| Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis.Crossref | GoogleScholarGoogle Scholar | 1649008PubMed |
Champagne, F. A., Curley, J. P., Swaney, W. T., Hasen, N. S., and Keverne, E. B. (2009). Paternal influence on female behavior: the role of Peg3 in exploration, olfaction, and neuroendocrine regulation of maternal behavior of female mice. Behav. Neurosci. 123, 469–480.
| Paternal influence on female behavior: the role of Peg3 in exploration, olfaction, and neuroendocrine regulation of maternal behavior of female mice.Crossref | GoogleScholarGoogle Scholar | 19485553PubMed |
Charalambous, M., Cowley, M., Geoghegan, F., Smith, F. M., Radford, E. J., Marlow, B. P., Graham, C. F., Hurst, L. D., and Ward, A. (2010). Maternally-inherited Grb10 reduces placental size and efficiency. Dev. Biol. 337, 1–8.
| Maternally-inherited Grb10 reduces placental size and efficiency.Crossref | GoogleScholarGoogle Scholar | 19833122PubMed |
Cheong, C. Y., Chng, K., Ng, S., Chew, S. B., Chan, L., and Ferguson-Smith, A. C. (2015). Germline and somatic imprinting in the nonhuman primate highlights species differences in oocyte methylation. Genome Res. 25, 611–623.
| Germline and somatic imprinting in the nonhuman primate highlights species differences in oocyte methylation.Crossref | GoogleScholarGoogle Scholar | 25862382PubMed |
Cleaton, M. A. M., Edwards, C. A., and Ferguson-Smith, A. C. (2014). Phenotypic outcomes of imprinted gene models in mice: elucidation of pre- and postnatal functions of imprinted genes. Annu. Rev. Genomics Hum. Genet. 15, 93–126.
| Phenotypic outcomes of imprinted gene models in mice: elucidation of pre- and postnatal functions of imprinted genes.Crossref | GoogleScholarGoogle Scholar |
Cleaton, M. A. M., Dent, C. L., Howard, M., Corish, J. A., Gutteridge, I., Sovio, U., Gaccioli, F., Takahashi, N., Bauer, S. R., Charnock-Jones, D. S., Powell, T. L., Smith, G. C. S., Ferguson-Smith, A. C., and Charalambous, M. (2016). Fetus-derived DLK1 is required for maternal metabolic adaptations to pregnancy and is associated with fetal growth restriction. Nat. Genet. 48, 1473–1480.
| Fetus-derived DLK1 is required for maternal metabolic adaptations to pregnancy and is associated with fetal growth restriction.Crossref | GoogleScholarGoogle Scholar |
Coan, P. M., Burton, G. J., and Ferguson-Smith, A. C. (2005). Imprinted genes in the placenta – a review. Placenta 26, S10–S20.
| Imprinted genes in the placenta – a review.Crossref | GoogleScholarGoogle Scholar | 15837057PubMed |
Constância, M., Hemberger, M., Hughes, J., Dean, W., Ferguson-Smith, A., Fundele, R., Stewart, F., Kelsey, G., Fowden, A., Sibley, C., and Reik, W. (2002). Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature 417, 945–948.
| Placental-specific IGF-II is a major modulator of placental and fetal growth.Crossref | GoogleScholarGoogle Scholar | 12087403PubMed |
Constância, M., Angiolini, E., Sandovici, I., Smith, P., Smith, R., Kelsey, G., Dean, W., Ferguson-Smith, A., Sibley, C. P., Reik, W., and Fowden, A. (2005). Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems. Proc. Natl Acad. Sci. USA 102, 19219–19224.
| Adaptation of nutrient supply to fetal demand in the mouse involves interaction between the Igf2 gene and placental transporter systems.Crossref | GoogleScholarGoogle Scholar | 16365304PubMed |
Cowley, M., Garfield, A. S., Madon-Simon, M., Charalambous, M., Clarkson, R. W., Smalley, M. J., Kendrick, H., Isles, A. R., Parry, A. J., Carney, S., Oakey, R. J., Heisler, L. K., Moorwood, K., Wolf, J. B., and Ward, A. (2014). Developmental programming mediated by complementary roles of imprinted Grb10 in mother and pup. PLoS Biol. 12, e1001799.
| Developmental programming mediated by complementary roles of imprinted Grb10 in mother and pup.Crossref | GoogleScholarGoogle Scholar | 24586114PubMed |
Curley, J. P., Barton, S., Surani, A., and Keverne, E. B. (2004). Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proc. Biol. Sci. 271, 1303–1309.
| Coadaptation in mother and infant regulated by a paternally expressed imprinted gene.Crossref | GoogleScholarGoogle Scholar | 15306355PubMed |
Dao, D., Frank, D., Qian, N., O’Keefe, D., Vosatka, R. J., Walsh, C. P., and Tycko, B. (1998). IMPT1, an imprinted gene similar to polyspecific transporter and multi-drug resistance genes. Hum. Mol. Genet. 7, 597–608.
| IMPT1, an imprinted gene similar to polyspecific transporter and multi-drug resistance genes.Crossref | GoogleScholarGoogle Scholar | 9499412PubMed |
Das, R., Anderson, N., Koran, M. I., Weidman, J. R., Mikkelsen, T. S., Kamal, M., Murphy, S. K., Linblad-Toh, K., Greally, J. M., and Jirtle, R. L. (2012). Convergent and divergent evolution of genomic imprinting in the marsupial Monodelphis domestica. BMC Genomics 13, 394.
| Convergent and divergent evolution of genomic imprinting in the marsupial Monodelphis domestica.Crossref | GoogleScholarGoogle Scholar | 22899817PubMed |
DeChiara, T. M., Robertson, E. J., and Efstratiadis, A. (1991). Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64, 849–859.
| Parental imprinting of the mouse insulin-like growth factor II gene.Crossref | GoogleScholarGoogle Scholar | 1997210PubMed |
Deltour, L., Montagutelli, X., Guenet, J. L., Jami, J., and Páldi, A. (1995). Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2. Dev. Biol. 168, 686–688.
| Tissue- and developmental stage-specific imprinting of the mouse proinsulin gene, Ins2.Crossref | GoogleScholarGoogle Scholar | 7729600PubMed |
Denizot, A. L., Besson, V., Correra, R. M., Mazzola, A., Lopes, I., Courbard, J. R., Marazzi, G., and Sassoon, D. A. (2016). A novel mutant allele of Pw1/Peg3 does not affect maternal behavior or nursing behavior. PLoS Genet. 12, e1006053.
| A novel mutant allele of Pw1/Peg3 does not affect maternal behavior or nursing behavior.Crossref | GoogleScholarGoogle Scholar | 27187722PubMed |
Dindot, S. V., Kent, K. C., Evers, B., Loskutoff, N., Womack, J., and Piedrahita, J. A. (2004). Conservation of genomic imprinting at the XIST, IGF2, and GTL2 loci in the bovine. Mamm. Genome 15, 966–974.
| Conservation of genomic imprinting at the XIST, IGF2, and GTL2 loci in the bovine.Crossref | GoogleScholarGoogle Scholar | 15599555PubMed |
Edwards, C. A., Mungall, A. J., Matthews, L., Ryder, E., Gray, D. J., Pask, A. J., Shaw, G., Graves, J. A., Rogers, J., Dunham, I., Renfree, M. B., and Ferguson-Smith, A. C. (2008). The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 6, e135.
| The evolution of the DLK1-DIO3 imprinted domain in mammals.Crossref | GoogleScholarGoogle Scholar | 18532878PubMed |
Evans, H. K., Weidman, J. R., Cowley, D. O., and Jirtle, R. L. (2005). Comparative phylogenetic analysis of blcap/nnat reveals eutherian-specific imprinted gene. Mol. Biol. Evol. 22, 1740–1748.
| Comparative phylogenetic analysis of blcap/nnat reveals eutherian-specific imprinted gene.Crossref | GoogleScholarGoogle Scholar | 15901842PubMed |
Ferguson-Smith, A. C., Cattanach, B. M., Barton, S. C., Beechey, C. V., and Surani, M. A. (1991). Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature 351, 667–670.
| Embryological and molecular investigations of parental imprinting on mouse chromosome 7.Crossref | GoogleScholarGoogle Scholar | 2052093PubMed |
Filson, A. J., Louvi, A., Efstratiadis, A., and Robertson, E. J. (1993). Rescue of the T-associated maternal effect in mice carrying null mutations in Igf-2 and Igf2r, two reciprocally imprinted genes. Development 118, 731–736.
| 8076514PubMed |
Fitzpatrick, G. V., Soloway, P. D., and Higgins, M. J. (2002). Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32, 426–431.
| Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1.Crossref | GoogleScholarGoogle Scholar | 12410230PubMed |
Frank, D., Fortino, W., Clark, L., Musalo, R., Wang, W., Saxena, A., Li, C. M., Reik, W., Ludwig, T., and Tycko, B. (2002). Placental overgrowth in mice lacking the imprinted gene Ipl. Proc. Natl Acad. Sci. USA 99, 7490–7495.
| Placental overgrowth in mice lacking the imprinted gene Ipl.Crossref | GoogleScholarGoogle Scholar | 12032310PubMed |
Frey, W. D., and Kim, J. (2015). Tissue-specific contributions of paternally expressed gene 3 in lactation and maternal care of Mus musculus. PLoS One 10, e0144459.
| Tissue-specific contributions of paternally expressed gene 3 in lactation and maternal care of Mus musculus.Crossref | GoogleScholarGoogle Scholar | 26640945PubMed |
Gabory, A., Ripoche, M. A., Le Digarcher, A., Watrin, F., Ziyyat, A., Forne, T., Jammes, H., Ainscough, J. F., Surani, M. A., Journot, L., and Dandolo, L. (2009). H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 136, 3413–3421.
| H19 acts as a trans regulator of the imprinted gene network controlling growth in mice.Crossref | GoogleScholarGoogle Scholar | 19762426PubMed |
Garfield, A. S., Cowley, M., Smith, F. M., Moorwood, K., Stewart-Cox, J. E., Gilroy, K., Baker, S., Xia, J., Dalley, J. W., Hurst, L. D., Wilkinson, L. S., Isles, A. R., and Ward, A. (2011). Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature 469, 534–538.
| Distinct physiological and behavioural functions for parental alleles of imprinted Grb10.Crossref | GoogleScholarGoogle Scholar | 21270893PubMed |
Georgiades, P., Watkins, M., Surani, M. A., and Ferguson-Smith, A. C. (2000). Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 127, 4719–4728.
| 11023874PubMed |
Georgiades, P., Watkins, M., Burton, G. J., and Ferguson-Smith, A. C. (2001). Roles for genomic imprinting and the zygotic genome in placental development. Proc. Natl Acad. Sci. USA 98, 4522–4527.
| Roles for genomic imprinting and the zygotic genome in placental development.Crossref | GoogleScholarGoogle Scholar | 11274372PubMed |
Guernsey, M. W., Chuong, E. B., Cornelis, G., Renfree, M. B., and Baker, J. C. (2017). Molecular conservation of marsupial and eutherian placentation and lactation. eLife 6, e27450.
| Molecular conservation of marsupial and eutherian placentation and lactation.Crossref | GoogleScholarGoogle Scholar | 28895534PubMed |
Guillemot, F., Nagy, A., Auerbach, A., Rossant, J., and Joyner, A. L. (1994). Essential role of Mash-2 in extraembryonic development. Nature 371, 333–336.
| Essential role of Mash-2 in extraembryonic development.Crossref | GoogleScholarGoogle Scholar | 8090202PubMed |
Hajkova, P., Erhardt, S., Lane, N., Haaf, T., El-Maarri, O., Reik, W., Walter, J., and Surani, M. A. (2002). Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 117, 15–23.
| Epigenetic reprogramming in mouse primordial germ cells.Crossref | GoogleScholarGoogle Scholar | 12204247PubMed |
Hayssen, V., Lacy, R. C., and Parker, P. J. (1985). Metatherian reproduction: transitional or transcending?. Am. Nat. 126, 617–632.
| Metatherian reproduction: transitional or transcending?.Crossref | GoogleScholarGoogle Scholar |
Huntley, S., Baggott, D. M., Hamilton, A. T., Tran-Gyamfi, M., Yang, S., Kim, J., Gordon, L., Branscomb, E., and Stubbs, L. (2006). A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 16, 669–677.
| A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors.Crossref | GoogleScholarGoogle Scholar | 16606702PubMed |
Imbeault, M., Helleboid, P.-Y., and Trono, D. (2017). KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature 543, 550–554.
| KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks.Crossref | GoogleScholarGoogle Scholar | 28273063PubMed |
Inoue, A., Jiang, L., Lu, F., Suzuki, T., and Zhang, Y. (2017). Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547, 419–424.
| Maternal H3K27me3 controls DNA methylation-independent imprinting.Crossref | GoogleScholarGoogle Scholar | 28723896PubMed |
Ioannides, Y., Lokulo-Sodipe, K., Mackay, D. J. G., Davies, J. H., and Temple, I. K. (2014). Temple syndrome: improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: an analysis of 51 published cases. J. Med. Genet. 51, 495–501.
| Temple syndrome: improving the recognition of an underdiagnosed chromosome 14 imprinting disorder: an analysis of 51 published cases.Crossref | GoogleScholarGoogle Scholar | 24891339PubMed |
Issa, J. P., Vertino, P. M., Boehm, C. D., Newsham, I. F., and Baylin, S. B. (1996). Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proc. Natl Acad. Sci. USA 93, 11757–11762.
| Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis.Crossref | GoogleScholarGoogle Scholar | 8876210PubMed |
Ito, M., Sferruzzi-Perri, A. N., Edwards, C. A., Adalsteinsson, B. T., Allen, S. E., Loo, T.-H., Kitazawa, M., Kaneko-Ishino, T., Ishino, F., Stewart, C. L., and Ferguson-Smith, A. C. (2015). A trans-homologue interaction between reciprocally imprinted miR-127 and Rtl1 regulates placenta development. Development 142, 2425–2430.
| A trans-homologue interaction between reciprocally imprinted miR-127 and Rtl1 regulates placenta development.Crossref | GoogleScholarGoogle Scholar | 26138477PubMed |
Jacobs, F. M. J., Greenberg, D., Nguyen, N., Haeussler, M., Ewing, A. D., Katzman, S., Paten, B., Salama, S. R., and Haussler, D. (2014). An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature 516, 242–245.
| An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons.Crossref | GoogleScholarGoogle Scholar |
John, R. M. (2013). Epigenetic regulation of placental endocrine lineages and complications of pregnancy. Biochem. Soc. Trans. 41, 701–709.
| Epigenetic regulation of placental endocrine lineages and complications of pregnancy.Crossref | GoogleScholarGoogle Scholar | 23697929PubMed |
Kagami, M., Kurosawa, K., Miyazaki, O., Ishino, F., Matsuoka, K., and Ogata, T. (2015). Comprehensive clinical studies in 34 patients with molecularly defined UPD(14)pat and related conditions (Kagami–Ogata syndrome). Eur. J. Hum. Genet. 23, 1488–1498.
| Comprehensive clinical studies in 34 patients with molecularly defined UPD(14)pat and related conditions (Kagami–Ogata syndrome).Crossref | GoogleScholarGoogle Scholar | 25689926PubMed |
Kaneko-Ishino, T., Kuroiwa, Y., Miyoshi, N., Kohda, T., Suzuki, R., Yokoyama, M., Viville, S., Barton, S. C., Ishino, F., and Surani, M. A. (1995). Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat. Genet. 11, 52–59.
| Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization.Crossref | GoogleScholarGoogle Scholar | 7550314PubMed |
Karolchik, D., Hinrichs, A. S., and Kent, W. J. (2012). The UCSC genome browser. Current Protocols in Bioinformatics 40(1) 1.4.1–1.4.33.
Keverne, E. B., and Curley, J. P. (2008). Epigenetics, brain evolution and behaviour. Front. Neuroendocrinol. 29, 398–412.
| Epigenetics, brain evolution and behaviour.Crossref | GoogleScholarGoogle Scholar | 18439660PubMed |
Killian, J. K., Byrd, J. C., Jirtle, J. V., Munday, B. L., Stoskopf, M. K., MacDonald, R. G., and Jirtle, R. L. (2000). M6P/IGF2R imprinting evolution in mammals. Mol. Cell 5, 707–716.
| M6P/IGF2R imprinting evolution in mammals.Crossref | GoogleScholarGoogle Scholar | 10882106PubMed |
Killian, J. K., Nolan, C. M., Stewart, N., Munday, B. L., Andersen, N. A., Nicol, S., and Jirtle, R. L. (2001). Monotreme IGF2 expression and ancestral origin of genomic imprinting. J. Exp. Zool. 291, 205–212.
| Monotreme IGF2 expression and ancestral origin of genomic imprinting.Crossref | GoogleScholarGoogle Scholar | 11479919PubMed |
Kobayashi, H., Sakurai, T., Imai, M., Takahashi, N., Fukuda, A., Yayoi, O., Sato, S., Nakabayashi, K., Hata, K., Sotomaru, Y., Suzuki, Y., and Kono, T. (2012). Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 8, e1002440.
| Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks.Crossref | GoogleScholarGoogle Scholar | 22242016PubMed |
Lau, M. M., Stewart, C. E., Liu, Z., Bhatt, H., Rotwein, P., and Stewart, C. L. (1994). Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev. 8, 2953–2963.
| Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality.Crossref | GoogleScholarGoogle Scholar | 8001817PubMed |
Lefebvre, L., Viville, S., Barton, S. C., Ishino, F., Keverne, E. B., and Surani, M. A. (1998). Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat. Genet. 20, 163–169.
| Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest.Crossref | GoogleScholarGoogle Scholar | 9771709PubMed |
Lewis, A., Mitsuya, K., Umlauf, D., Smith, P., Dean, W., Walter, J., Higgins, M., Feil, R., and Reik, W. (2004). Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 36, 1291–1295.
| Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation.Crossref | GoogleScholarGoogle Scholar | 15516931PubMed |
Li, L.-L., Keverne, E. B., Aparicio, S. A., Ishino, F., Barton, S. C., and Surani, M. A. (1999). Regulation of maternal behavior and offspring growth by paternally expressed Peg3. Science 284, 330–334.
| Regulation of maternal behavior and offspring growth by paternally expressed Peg3.Crossref | GoogleScholarGoogle Scholar |
Li, X., Ito, M., Zhou, F., Youngson, N., Zuo, X., Leder, P., and Ferguson-Smith, A. C. (2008). A maternal–zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547–557.
| A maternal–zygotic effect gene, Zfp57, maintains both maternal and paternal imprints.Crossref | GoogleScholarGoogle Scholar | 18854139PubMed |
Lopez, M. F., Dikkes, P., Zurakowski, D., and Villa-Komaroff, L. (1996). Insulin-like growth factor II affects the appearance and glycogen content of glycogen cells in the murine placenta. Endocrinology 137, 2100–2108.
| Insulin-like growth factor II affects the appearance and glycogen content of glycogen cells in the murine placenta.Crossref | GoogleScholarGoogle Scholar | 8612553PubMed |
Malven, P. V., Head, H. H., Collier, R. J., and Buonomo, F. C. (1987). Periparturient changes in secretion and mammary uptake of insulin and in concentrations of insulin and insulin-like growth factors in milk of dairy cows. J. Dairy Sci. 70, 2254–2265.
| Periparturient changes in secretion and mammary uptake of insulin and in concentrations of insulin and insulin-like growth factors in milk of dairy cows.Crossref | GoogleScholarGoogle Scholar | 3320114PubMed |
McGrath, J., and Solter, D. (1984). Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183.
| Completion of mouse embryogenesis requires both the maternal and paternal genomes.Crossref | GoogleScholarGoogle Scholar | 6722870PubMed |
Menzies, B. R., Pask, A. J., and Renfree, M. B. (2011). Placental expression of pituitary hormones is an ancestral feature of therian mammals. Evodevo 2, 16.
| Placental expression of pituitary hormones is an ancestral feature of therian mammals.Crossref | GoogleScholarGoogle Scholar | 21854600PubMed |
Monk, D. (2015). Genomic imprinting in the human placenta. Am. J. Obstet. Gynecol. 213, S152–S162.
| Genomic imprinting in the human placenta.Crossref | GoogleScholarGoogle Scholar | 26428495PubMed |
Moore, T., and Haig, D. (1991). Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 7, 45–49.
| Genomic imprinting in mammalian development: a parental tug-of-war.Crossref | GoogleScholarGoogle Scholar | 2035190PubMed |
Moore, G. E., Abu-Amero, S. N., Bell, G., Wakeling, E. L., Kingsnorth, A., Stanier, P., Jauniaux, E., and Bennett, S. T. (2001). Evidence that insulin is imprinted in the human yolk sac. Diabetes 50, 199–203.
| Evidence that insulin is imprinted in the human yolk sac.Crossref | GoogleScholarGoogle Scholar | 11147788PubMed |
Nicholls, R. D., and Knepper, J. L. (2001). Genome organization, function, and imprinting in Prader–Willi and Angelman syndromes. Annu. Rev. Genomics Hum. Genet. 2, 153–175.
| Genome organization, function, and imprinting in Prader–Willi and Angelman syndromes.Crossref | GoogleScholarGoogle Scholar | 11701647PubMed |
O’Leary, M. A., Bloch, J. I., Flynn, J. J., Gaudin, T. J., Giallombardo, A., Giannini, N. P., Goldberg, S. L., Kraatz, B. P., Luo, Z. X., Meng, J., et al. (2013). The placental mammal ancestor and the Post-K-Pg radiation of placentals. Science 339, 662–667.
| The placental mammal ancestor and the Post-K-Pg radiation of placentals.Crossref | GoogleScholarGoogle Scholar | 23393258PubMed |
O’Neill, M. J., Ingram, R. S., Vrana, P. B., and Tilghman, S. M. (2000). Allelic expression of IGF2 in marsupials and birds. Dev. Genes Evol. 210, 18–20.
| Allelic expression of IGF2 in marsupials and birds.Crossref | GoogleScholarGoogle Scholar | 10603082PubMed |
Ono, R., Kobayashi, S., Wagatsuma, H., Aisaka, K., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2001). A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237.
| A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21.Crossref | GoogleScholarGoogle Scholar | 11318613PubMed |
Ono, R., Shiura, H., Aburatani, H., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2003). Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6. Genome Res. 13, 1696–1705.
| Identification of a large novel imprinted gene cluster on mouse proximal chromosome 6.Crossref | GoogleScholarGoogle Scholar | 12840045PubMed |
Ono, R., Nakamura, K., Inoue, K., Naruse, M., Usami, T., Wakisaka-Saito, N., Hino, T., Suzuki-Migishima, R., Ogonuki, N., Miki, H., Kohda, T., Ogura, A., Yokoyama, M., Kaneko-Ishino, T., and Ishino, F. (2006). Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106.
| Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality.Crossref | GoogleScholarGoogle Scholar | 16341224PubMed |
Power, M. L., and Schulkin, J. (2013). Maternal regulation of offspring development in mammals is an ancient adaptation tied to lactation. Appl. Transl. Genom. 2, 55–63.
| Maternal regulation of offspring development in mammals is an ancient adaptation tied to lactation.Crossref | GoogleScholarGoogle Scholar | 27896056PubMed |
Prosser, C. G. (1996). Insulin-like growth factors in milk and mammary gland. J. Mammary Gland Biol. Neoplasia 1, 297–306.
| Insulin-like growth factors in milk and mammary gland.Crossref | GoogleScholarGoogle Scholar | 10887503PubMed |
Quenneville, S., Turelli, P., Bojkowska, K., Raclot, C., Offner, S., Kapopoulou, A., and Trono, D. (2012). The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development. Cell Reports 2, 766–773.
| The KRAB-ZFP/KAP1 system contributes to the early embryonic establishment of site-specific DNA methylation patterns maintained during development.Crossref | GoogleScholarGoogle Scholar | 23041315PubMed |
Rapkins, R. W., Hore, T., Smithwick, M., Ager, E., Pask, A. J., Renfree, M. B., Kohn, M., Hameister, H., Nicholls, R. D., Deakin, J. E., and Graves, J. A. (2006). Recent assembly of an imprinted domain from non-imprinted components. PLoS Genet. 2, e182.
| Recent assembly of an imprinted domain from non-imprinted components.Crossref | GoogleScholarGoogle Scholar | 17069464PubMed |
Reik, W., Collick, A., Norris, M. L., Barton, S. C., and Surani, M. A. (1987). Genomic imprinting determines methylation of parental alleles in transgenic mice. Nature 328, 248–251.
| Genomic imprinting determines methylation of parental alleles in transgenic mice.Crossref | GoogleScholarGoogle Scholar | 3600805PubMed |
Reik, W., Constancia, M., Fowden, A., Anderson, N., Dean, W., Ferguson-Smith, A., Tycko, B., and Sibley, C. (2003). Regulation of supply and demand for maternal nutrients in mammals by imprinted genes. J. Physiol. 547, 35–44.
| Regulation of supply and demand for maternal nutrients in mammals by imprinted genes.Crossref | GoogleScholarGoogle Scholar | 12562908PubMed |
Renfree, M. B., O, W. S., Short, R. V., and Shaw, G. (1996). Sexual differentiation of the urogenital system of the fetal and neonatal tammar wallaby, Macropus eugenii. Anat. Embryol. (Berl.) 194, 111–134.
| Sexual differentiation of the urogenital system of the fetal and neonatal tammar wallaby, Macropus eugenii.Crossref | GoogleScholarGoogle Scholar | 8827321PubMed |
Renfree, M. B., Suzuki, S., and Kaneko-Ishino, T. (2013). The origin and evolution of genomic imprinting and viviparity in mammals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120151.
| The origin and evolution of genomic imprinting and viviparity in mammals.Crossref | GoogleScholarGoogle Scholar | 23166401PubMed |
Ripoche, M. A., Kress, C., Poirier, F., and Dandolo, L. (1997). Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 11, 1596–1604.
| Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element.Crossref | GoogleScholarGoogle Scholar | 9203585PubMed |
Sapienza, C., Peterson, A. C., Rossant, J., and Balling, R. (1987). Degree of methylation of transgenes is dependent on gamete of origin. Nature 328, 251–254.
| Degree of methylation of transgenes is dependent on gamete of origin.Crossref | GoogleScholarGoogle Scholar | 3600806PubMed |
Sasaki, H., Hamada, T., Ueda, T., Seki, R., Higashinakagawa, T., and Sakaki, Y. (1991). Inherited type of allelic methylation variations in a mouse chromosome region where an integrated transgene shows methylation imprinting. Development 111, 573–581.
| 1680050PubMed |
Schaller, F., Watrin, F., Sturny, R., Massacrier, A., Szepetowski, P., and Muscatelli, F. (2010). A single postnatal injection of oxytocin rescues the lethal feeding behaviour in mouse newborns deficient for the imprinted Magel2 gene. Hum. Mol. Genet. 19, 4895–4905.
| A single postnatal injection of oxytocin rescues the lethal feeding behaviour in mouse newborns deficient for the imprinted Magel2 gene.Crossref | GoogleScholarGoogle Scholar | 20876615PubMed |
Seitz, H., Youngson, N., Lin, S. P., Dalbert, S., Paulsen, M., Bachellerie, J. P., Ferguson-Smith, A. C., and Cavaille, J. (2003). Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34, 261–262.
| Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene.Crossref | GoogleScholarGoogle Scholar | 12796779PubMed |
Sekita, Y., Wagatsuma, H., Nakamura, K., Ono, R., Kagami, M., Wakisaka, N., Hino, T., Suzuki-Migishima, R., Kohda, T., Ogura, A., Ogata, T., Yokoyama, M., Kaneko-Ishino, T., and Ishino, F. (2008). Role of retrotransposon-derived imprinted gene, Rtl1, in the feto–maternal interface of mouse placenta. Nat. Genet. 40, 243–248.
| Role of retrotransposon-derived imprinted gene, Rtl1, in the feto–maternal interface of mouse placenta.Crossref | GoogleScholarGoogle Scholar | 18176565PubMed |
Sibley, C. P., Coan, P. M., Ferguson-Smith, A. C., Dean, W., Hughes, J., Smith, P., Reik, W., Burton, G. J., Fowden, A. L., and Constancia, M. (2004). Placental-specific insulin-like growth factor 2 (Igf2) regulates the diffusional exchange characteristics of the mouse placenta. Proc. Natl Acad. Sci. USA 101, 8204–8208.
| Placental-specific insulin-like growth factor 2 (Igf2) regulates the diffusional exchange characteristics of the mouse placenta.Crossref | GoogleScholarGoogle Scholar | 15150410PubMed |
Smits, G., Mungall, A. J., Griffiths-Jones, S., Smith, P., Beury, D., Matthews, L., Rogers, J., Pask, A. J., Shaw, G., VandeBerg, J. L., McCarrey, J. R., Renfree, M. B., Reik, W., and Dunham, I. (2008). Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat. Genet. 40, 971–976.
| Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians.Crossref | GoogleScholarGoogle Scholar | 18587395PubMed |
Stringer, J. M., Suzuki, S., Pask, A. J., Shaw, G., and Renfree, M. B. (2012a). GRB10 imprinting is eutherian mammal specific. Mol. Biol. Evol. 29, 3711–3719.
| GRB10 imprinting is eutherian mammal specific.Crossref | GoogleScholarGoogle Scholar | 22787282PubMed |
Stringer, J. M., Suzuki, S., Pask, A. J., Shaw, G., and Renfree, M. B. (2012b). Promoter-specific expression and imprint status of marsupial IGF2. PLoS One 7, e41690.
| Promoter-specific expression and imprint status of marsupial IGF2.Crossref | GoogleScholarGoogle Scholar | 22848567PubMed |
Stringer, J. M., Suzuki, S., Pask, A. J., Shaw, G., and Renfree, M. B. (2012c). Selected imprinting of INS in the marsupial. Epigenetics Chromatin 5, 14.
| Selected imprinting of INS in the marsupial.Crossref | GoogleScholarGoogle Scholar | 22929229PubMed |
Stringer, J. M., Pask, A. J., Shaw, G., and Renfree, M. B. (2014). Post-natal imprinting: evidence from marsupials. Heredity 113, 145–155.
| Post-natal imprinting: evidence from marsupials.Crossref | GoogleScholarGoogle Scholar | 24595366PubMed |
Strogantsev, R., Krueger, F., Yamazawa, K., Shi, H., Gould, P., Goldman-Roberts, M., McEwen, K., Sun, B., Pedersen, R., and Ferguson-Smith, A. C. (2015). Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol. 16, 112.
| Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression.Crossref | GoogleScholarGoogle Scholar | 26025256PubMed |
Surani, M. A. H., and Barton, S. C. (1983). Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos. Science 222, 1034–1036.
| Development of gynogenetic eggs in the mouse: implications for parthenogenetic embryos.Crossref | GoogleScholarGoogle Scholar |
Surani, M. A., Barton, S. C., and Norris, M. L. (1984). Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550.
| Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis.Crossref | GoogleScholarGoogle Scholar | 6709062PubMed |
Suzuki, S., Renfree, M. B., Pask, A. J., Shaw, G., Kobayashi, S., Kohda, T., Kaneko-Ishino, T., and Ishino, F. (2005). Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mech. Dev. 122, 213–222.
| Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby.Crossref | GoogleScholarGoogle Scholar | 15652708PubMed |
Suzuki, S., Ono, R., Narita, T., Pask, A. J., Shaw, G., Wang, C., Kohda, T., Alsop, A. E., Marshall Graves, J. A., Kohara, Y., Ishino, F., Renfree, M. B., and Kaneko-Ishino, T. (2007). Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3, e55.
| Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting.Crossref | GoogleScholarGoogle Scholar | 17432937PubMed |
Suzuki, S., Shaw, G., Kaneko-Ishino, T., Ishino, F., and Renfree, M. B. (2011). Characterisation of marsupial PHLDA2 reveals eutherian specific acquisition of imprinting. BMC Evol. Biol. 11, 244.
| Characterisation of marsupial PHLDA2 reveals eutherian specific acquisition of imprinting.Crossref | GoogleScholarGoogle Scholar | 21854573PubMed |
Suzuki, S., Shaw, G., and Renfree, M. B. (2013). Postnatal epigenetic reprogramming in the germline of a marsupial, the tammar wallaby. Epigenetics Chromatin 6, 14.
| Postnatal epigenetic reprogramming in the germline of a marsupial, the tammar wallaby.Crossref | GoogleScholarGoogle Scholar | 23732002PubMed |
Swain, J. L., Stewart, T. A., and Leder, P. (1987). Parental legacy determines methylation and expression of an autosomal transgene: a molecular mechanism for parental imprinting. Cell 50, 719–727.
| Parental legacy determines methylation and expression of an autosomal transgene: a molecular mechanism for parental imprinting.Crossref | GoogleScholarGoogle Scholar | 3040259PubMed |
Takahashi, K., Kobayashi, T., and Kanayama, N. (2000). p57(Kip2) regulates the proper development of labyrinthine and spongiotrophoblasts. Mol. Hum. Reprod. 6, 1019–1025.
| p57(Kip2) regulates the proper development of labyrinthine and spongiotrophoblasts.Crossref | GoogleScholarGoogle Scholar | 11044465PubMed |
Takahashi, N., Gray, D., Strogantsev, R., Noon, A., Delahaye, C., Skarnes, W. C., Tate, P. H., and Ferguson-Smith, A. C. (2015). ZFP57 and the targeted maintenance of postfertilization genomic imprints. Cold Spring Harb. Symp. Quant. Biol. 80, 177–187.
| ZFP57 and the targeted maintenance of postfertilization genomic imprints.Crossref | GoogleScholarGoogle Scholar | 27325708PubMed |
Temple, I. K., Gardner, R. J., Mackay, D. J. G., Barber, J. C. K., Robinson, D. O., and Shield, J. P. H. (2000). Transient neonatal diabetes: widening the understanding of the etiopathogenesis of diabetes. Diabetes 49, 1359–1366.
| Transient neonatal diabetes: widening the understanding of the etiopathogenesis of diabetes.Crossref | GoogleScholarGoogle Scholar | 10923638PubMed |
Tunster, S. J., Creeth, H. D. J., and John, R. M. (2016). The imprinted Phlda2 gene modulates a major endocrine compartment of the placenta to regulate placental demands for maternal resources. Dev. Biol. 409, 251–260.
| The imprinted Phlda2 gene modulates a major endocrine compartment of the placenta to regulate placental demands for maternal resources.Crossref | GoogleScholarGoogle Scholar | 26476147PubMed |
Ullmann, S. L., Shaw, G., Alcorn, G. T., and Renfree, M. B. (1997). Migration of primordial germ cells to the developing gonadal ridges in the tammar wallaby Macropus eugenii. J. Reprod. Fertil. 110, 135–143.
| Migration of primordial germ cells to the developing gonadal ridges in the tammar wallaby Macropus eugenii.Crossref | GoogleScholarGoogle Scholar | 9227367PubMed |
Umlauf, D., Goto, Y., Cao, R., Cerqueira, F., Wagschal, A., Zhang, Y., and Feil, R. (2004). Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 36, 1296–1300.
| Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes.Crossref | GoogleScholarGoogle Scholar | 15516932PubMed |
Wang, Z. Q., Fung, M. R., Barlow, D. P., and Wagner, E. F. (1994). Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature 372, 464–467.
| Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene.Crossref | GoogleScholarGoogle Scholar | 7984240PubMed |
Weidman, J. R., Dolinoy, D. C., Maloney, K. A., Cheng, J. F., and Jirtle, R. L. (2006a). Imprinting of opossum Igf2r in the absence of differential methylation and air. Epigenetics 1, 50–55.
| Imprinting of opossum Igf2r in the absence of differential methylation and air.Crossref | GoogleScholarGoogle Scholar |
Weidman, J. R., Maloney, K. A., and Jirtle, R. L. (2006b). Comparative phylogenetic analysis reveals multiple non-imprinted isoforms of opossum Dlk1. Mamm. Genome 17, 157–167.
| Comparative phylogenetic analysis reveals multiple non-imprinted isoforms of opossum Dlk1.Crossref | GoogleScholarGoogle Scholar | 16465595PubMed |
Weksberg, R., Shuman, C., and Beckwith, J. B. (2010). Beckwith–Wiedemann syndrome. Eur. J. Hum. Genet. 18, 8–14.
| Beckwith–Wiedemann syndrome.Crossref | GoogleScholarGoogle Scholar | 19550435PubMed |
Wilkin, F., Paquette, J., Ledru, E., Mamelin, C., Pollak, M., and Deal, C. L. (2000). H19 sense and antisense transgenes modify insulin-like growth factor-II mRNA levels. Eur. J. Biochem. 267, 4020–4027.
| H19 sense and antisense transgenes modify insulin-like growth factor-II mRNA levels.Crossref | GoogleScholarGoogle Scholar | 10866801PubMed |
Wolf, J. B., and Brodie, E. D. (1998). The coadaptation of parental and offspring characters. Evolution 52, 299–308.
| The coadaptation of parental and offspring characters.Crossref | GoogleScholarGoogle Scholar | 28568322PubMed |
Wolf, D., and Goff, S. P. (2009). Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458, 1201–1204.
| Embryonic stem cells use ZFP809 to silence retroviral DNAs.Crossref | GoogleScholarGoogle Scholar | 19270682PubMed |
Wolf, J. B., and Hager, R. (2006). A maternal–offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 4, e380.
| A maternal–offspring coadaptation theory for the evolution of genomic imprinting.Crossref | GoogleScholarGoogle Scholar | 17105351PubMed |
Wood, A. J., Roberts, R. G., Monk, D., Moore, G. E., Schulz, R., and Oakey, R. J. (2007). A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 3, e20.
| A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation.Crossref | GoogleScholarGoogle Scholar | 17291163PubMed |
Wutz, A., Smrzka, O. W., Schweifer, N., Schellander, K., Wagner, E. F., and Barlow, D. P. (1997). Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389, 745–749.
| Imprinted expression of the Igf2r gene depends on an intronic CpG island.Crossref | GoogleScholarGoogle Scholar | 9338788PubMed |
Xu, Y. Q., Goodyer, C. G., Deal, C., and Polychronakos, C. (1993). Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochem. Biophys. Res. Commun. 197, 747–754.
| Functional polymorphism in the parental imprinting of the human IGF2R gene.Crossref | GoogleScholarGoogle Scholar |
Yamasaki-Ishizaki, Y., Kayashima, T., Mapendano, C. K., Soejima, H., Ohta, T., Masuzaki, H., Kinoshita, A., Urano, T., Yoshiura, K.-i., Matsumoto, N., Ishimaru, T., Mukai, T., Niikawa, N., and Kishino, T. (2007). Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol. Cell. Biol. 27, 732–742.
| Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10.Crossref | GoogleScholarGoogle Scholar | 17101788PubMed |
Ye, A., He, H., and Kim, J. (2014). Paternally expressed Peg3 controls maternally expressed Zim1 as a trans factor. PLoS One 9, e108596.
| Paternally expressed Peg3 controls maternally expressed Zim1 as a trans factor.Crossref | GoogleScholarGoogle Scholar | 25396734PubMed |
Youngson, N. A., Kocialkowski, S., Peel, N., and Ferguson-Smith, A. C. (2005). A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 61, 481–490.
| A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting.Crossref | GoogleScholarGoogle Scholar | 16155747PubMed |
Zwart, R., Sleutels, F., Wutz, A., Schinkel, A. H., and Barlow, D. P. (2001). Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev. 15, 2361–2366.
| Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes.Crossref | GoogleScholarGoogle Scholar | 11562346PubMed |



