357 EARLY FETAL DEVELOPMENT OF NUCLEAR TRANSFER BOVINE EMBRYOS GENERATED FROM FIBROBLASTS GENETICALLY MODIFIED BY piggyBac TRANSPOSITION
A. Alessio A , A. Fili A , D. Forcato A , M. F. Olmos-Nicotra A , F. Alustiza A , N. Rodriguez A , R. V. Sampaio B , J. Sangalli B , F. Bressan B , P. Fantinato-Neto B , F. Meirelles B , J. Owens C , S. Moisyadi C , W. A. Kues D and P. Bosch AA Department of Molecular Biology, FCEFQyN, National University of Río Cuarto, Río Cuarto, Córdoba, Argentina;
B Department of Veterinary Medicine, Faculty of Animal Sciences and Food Engineering, University of São Paulo, São Paulo, Brazil;
C Department of Anatomy, Biochemistry and Physiology; John A. Burns School of Medicine, Honolulu, HI, USA;
D Friedrich-Loeffler-Institut, Institut für Nutztiergenetik, Mariensee, Germany
Reproduction, Fertility and Development 27(1) 266-267 https://doi.org/10.1071/RDv27n1Ab357
Published: 4 December 2014
Abstract
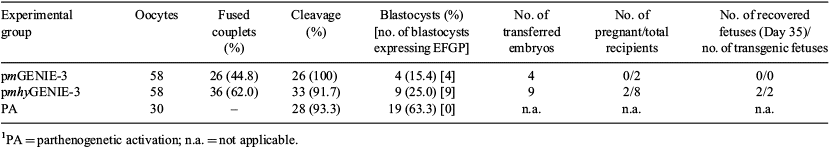
Transposon-mediated transgenesis is a well-established tool for genome manipulation in small animal models. However, translation of this active transgenesis method to the large animal setting requires further investigation. We have previously demonstrated that a helper-independent piggyBac (PB) transposon system can efficiently transpose transgenes into the bovine genome [Alessio et al. 2014 Reprod. Domest. Anim. 49 (Suppl. 1), 8]. The aims of the current study were a) to investigate the effectiveness of a hyperactive version of the PB transposase, and b) to determine the ability of the genetically modified cells to support early embryo and fetal development upon somatic cell nuclear transfer (SCNT). Bovine fetal fibroblasts (BFF) were chemically transfected with either pmGENIE-3 (a helper-independent PB transposon conferring genes for hygromycin resistance and enhanced green fluorescent protein (EGFP); Urschitz et al. 2010 PNAS USA 107, 8117–8122), pmhyGENIE-3 (carrying an hyperactive version of the PB transposase; Marh et al. 2012 PNAS USA 109, 19 184–19 189), or pmGENIE-3/Δ PB (a control plasmid lacking a functional PB transposase). Upon transfection, cell cultures were subjected to 14 days of hygromycin selection. Antibiotic-resistant and EGFP+ colonies were counted and data analysed by ANOVA and Tukey's test. For SCNT, pmhyGENIE-3 and pmGENIE-3 polyclonal cell lines were selected by FACS and individual cells used as nuclear donors. Day 7 blastocysts were nonsurgically transferred to synchronized recipients. Conceptuses were recovered by Day 35 of gestation, observed under fluorescence excitation, and genotyped. The mean number of colonies in pmhyGENIE-3 group was significantly higher than those in pmGENIE-3 and the control group (324.0 ± 17.8 v. 100.0 ± 16.1 and 2.8 ± 0.8 respectively, n = 4–7; P < 0.05). The hyperactive transposase increased transgene integration efficiency 3.24 times compared with the conventional PB transposase. The SCNT and early fetal development data are summarised in Table 1. Phenotypic analysis revealed that both transgenic fetuses and the extraembryonic membranes expressed EGFP with no macroscopic evidence of variegated transgene expression. Molecular analysis by PCR confirmed that both fetuses carried the transposon DNA. Here, we demonstrate that a hyperactive version of the PB transposase is more active in bovine cells than the conventional PB transposase. In addition, SCNT embryos generated from genetically modified cells by the pGENIE transposon system can progress to early stages of fetal development.
|
The financial support of UNRC, CONICET and ANPCyT from Argentina is gratefully acknowledged.


