Increased DNA strand breaks in spermatozoa of Pxt1 knockout mice
Bernadetta Pawlicka A , Michał Duliban B , Mateusz Zięba A , Michał Bochenek C , Kamila Zięba A , Ibrahim Adham D , Maja Studencka-Turski D , Andreas Meinhardt E and Paweł Grzmil
B , Mateusz Zięba A , Michał Bochenek C , Kamila Zięba A , Ibrahim Adham D , Maja Studencka-Turski D , Andreas Meinhardt E and Paweł Grzmil  A *
A *
A Laboratory of Genetics and Evolutionism, Institute of Zoology and Biomedical Research, Jagiellonian University in Kraków, Kraków, Poland.
B Department of Endocrinology, Institute of Zoology and Biomedical Research, Jagiellonian University in Kraków, Kraków, Poland.
C Malopolska Centre of Biotechnology, Jagiellonian University in Kraków, Kraków, Poland.
D Institute of Human Genetics, University Medical Center Göttingen, Göttingen, Germany.
E Institute of Anatomy and Cell Biology, Justus-Liebig-University, Giessen, Germany.
Reproduction, Fertility and Development 35(11) 589-600 https://doi.org/10.1071/RD23061
Published online: 3 July 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: The Pxt1 gene encodes a male germ cell–specific protein and its overexpression results in male germ cell degeneration and male infertility in transgenic mice.
Aims: The analysis of the function of Pxt1 during mouse spermatogenesis.
Methods: The phenotype of Pxt1 knockout mice was characterised by testicular histology, assessment of semen parameters including sperm motility, and DNA fragmentation by flow cytometry. Gene expression was analysed using RT-PCR. Fertility of mutants was checked by standard breeding and competition breeding tests.
Key results: In Pxt1−/− mice, a strong increase in the sperm DNA fragmentation index (DFI) was observed, while other sperm parameters were comparable to those of control animals. Despite enhanced DFI, mutants were fertile and able to mate in competition with wild type males.
Conclusions: Pxt1 induces cell death; thus, the higher sperm DFI of mice with targeted deletion of Pxt1 suggests some function for this gene in the elimination of male germ cells with chromatin damage.
Implications: Ablation of mouse Pxt1 results in enhanced DFI. In humans, the homologous PXT1 gene shares 74% similarity with the mouse gene; thus, it can be considered a candidate for mutation screening in patients with increased DFI.
Keywords: competition breeding test, gene expression, gene knockout, male fertility, Pxt1, spermatogenesis, sperm chromatin damage, sperm DFI.
Introduction
The study of spermatogenesis is hampered by the lack of suitable cell lines mimicking this process in vitro. Therefore, a widely used strategy to explain molecular mechanisms regulating male gamete production is the generation and use of knockout mouse models. This approach has led to the discovery of many spermatogenesis-related genes (reviewed in Matzuk and Lamb 2002, 2008; Yatsenko et al. 2010).
The peroxisomal testis-specific 1 gene (Pxt1, Gene ID: 69307) was isolated and characterised as a gene with testis-specific expression (Grzmil et al. 2007). Past studies showed that the expression of Pxt1 is developmentally regulated during spermatogenesis and restricted to male germ cells in the mouse testis. The open reading frame of this gene encodes a small protein consisting of 51 amino acids. At the N-terminus, PXT1 contains a domain similar to the BH3 domain and it has been demonstrated that in transiently transfected cells PXT1 induces cell death (Kaczmarek et al. 2011). Moreover, in a transgenic mouse line overexpressing Pxt1 under the control of the Pgk-2 promoter, massive degeneration of male germ cells was observed, leading to complete infertility of adult males (Kaczmarek et al. 2011). While excessive or induced apoptosis can lead to male infertility, controlled apoptotic loss of germ cells is a feature of normal spermatogenesis in humans and rodents (Sinha Hikim et al. 1998; De Felici and Klinger 2015) and is regarded as critical for the maintenance of the species-specific ratio of Sertoli cells to germ cells (Tripathi et al. 2009; Shukla et al. 2012).
Apoptosis is an active, highly regulated biological process that enables maintenance of tissue homeostasis by elimination of aged, overproduced or dysfunctional cells. Despite an increasing number of reports about pro-apoptotic factors acting during spermatogenesis, the mechanisms underlying this important event in the male gonad remain poorly understood. Elimination of defective germ cells is of pivotal importance for proper spermatogenesis as production of aberrant spermatozoa negatively impacts male fertility. Because Pxt1 induces cell death (Kaczmarek et al. 2011) and its expression is restricted to male germ cells in mice (Grzmil et al. 2007), we hypothesised that this gene may be involved in the elimination of defective gametes and thus it may help to preserve sperm quality. Interaction between PXT1 and the protein encoded by BCL2-associated athanogene 6 (Bag6) protected transfected cells from PXT1-induced cell death (Kaczmarek et al. 2011). Bag6 is ubiquitously expressed with the highest levels seen in the testis, indicating a role in male germ cell differentiation (Wang and Liew 1994; Ozaki et al. 1999). Considering the expression profile and interaction of PXT1 and BAG6 proteins, we hypothesised that in normally developing male germ cells BAG6 can bind to PXT1, protecting the cell from PXT1 cytotoxic activity, wheras PXT1 is released from the BAG6–PXT1 complex in defective cells to elicit cell death (Kaczmarek et al. 2011). However, the exact role of Pxt1 in spermatogenesis was unknown.
In this study, we generated a Pxt1 knockout mouse model to elucidate the function of Pxt1. The data obtained from this mouse model indicate that the main function of Pxt1 is related to the elimination of male germ cells with DNA strand breaks.
Materials and methods
Animals
Mice were maintained at the Laboratory of Genetics and Evolutionism, Institute of Zoology and Biomedical Research, Jagiellonian University, Kraków, Poland. Mice were kept under a 12 h light–dark cycle with free access to water and standard laboratory diet. According to local regulations, post-mortem tissue collection and all procedures used in this work do not require the approval of the local ethics committee. The generation and use of genetically modified organisms, namely the Pxt1 knockout mice line, were approved by the Polish Ministry of Climate and Environment under the licence number 21/2018.
Generation of Pxt1 knockout mice
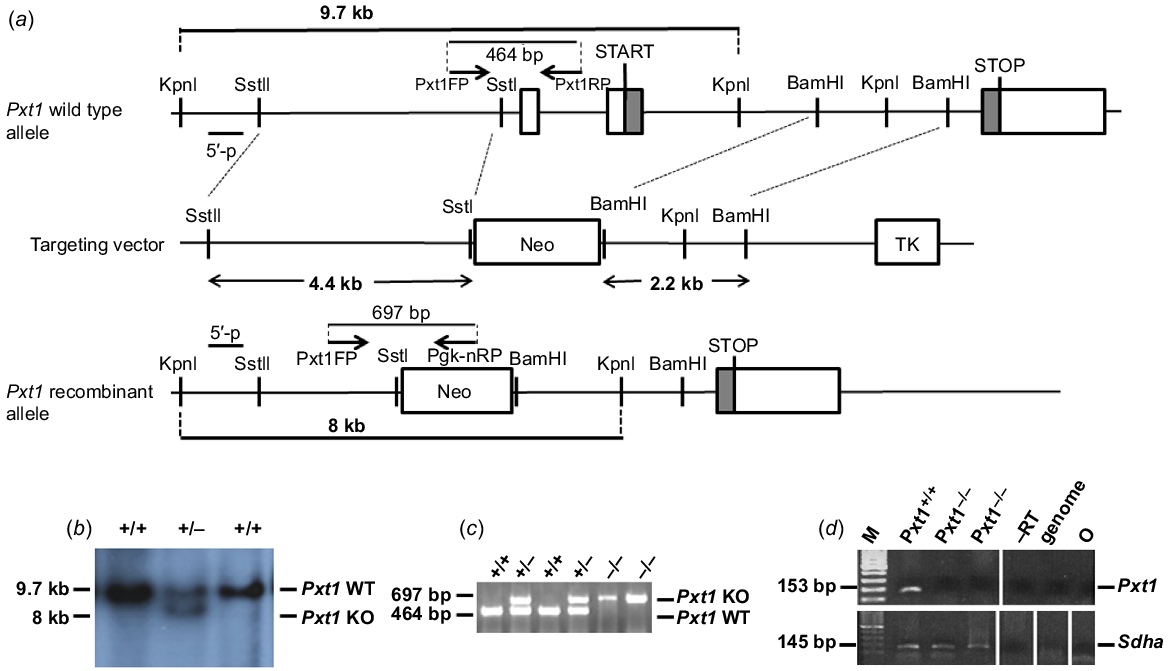
To generate the Pxt1-knockout construct, two fragments of mouse chromosome 17 were isolated from cosmid clone no. MGc121A20214Q2 of the 129ola library (RZPD, Germany), which was screened with a molecular probe spanning exon 3 of the mouse Pxt1 gene. The strategy to generate the mouse knockout line is given in Fig. 1. The 4.4-kb genomic fragment containing the 5′ flanking region of the Pxt1 gene was isolated with SstI and SstII restriction enzymes. The isolated 4.4-kb SstI/SstII fragment was cloned into the pTKNeo vector (Rosahl et al. 1995). The 2.2-kb genomic fragment containing the 3′ flanking region of the Pxt1 gene was isolated with the BamHI restriction enzyme and cloned into the pTKNeo vector. The linearised DNA construct was electroporated into 129/Sv-derived R1 embryonic stem (ES) cells (Nayernia et al. 2002). Genomic DNA was isolated from colonies resistant to G418 (400 μg/mL) and gancyclovir (2 μM), digested with KpnI, and hybridised with the 5′ external probe. The external probe was generated in PCR reaction with 5′Pxt1FP and 5′Pxt1RP primers. Homologous recombined ES cells were injected into C57Bl/6J blastocysts to produce chimeric animals (Joyner 1993). Chimeric males were mated to 129/Sv females and F1 offspring were genotyped by PCR with primers Pxt1FP, Pgk-nRP and Pxt1RP. The product size was 464 bp for the wild type allele and 697 bp for the mutant allele. The sequences of primers are given in Table 1. DNA for PCR genotyping was isolated from mouse tail biopsies using the standard phenol/chloroform isolation technique. Heterozygous mice from the 129/Sv background were then backcrossed on the ICR strain. Heterozygous males and females from generation B7 were intercrossed to produce homozygous mutants. All analyses presented in this manuscript were obtained on the ICR genetic background.
Targeted disruption of the Pxt1 gene. (a) Structure of the wild type and recombinant alleles, relevant restriction sites and applied primers are shown. The knockout strategy was designed to replace exons 1 and 2 of the Pxt1 gene with the pgk-neo cassette (Neo). TK – thymidine kinase cassette. (b) Southern blot analysis with KpnI-digested genomic DNA isolated from recombinant ES cells. The external probe detected 9.7-kb wild type and 8-kb recombinant KpnI fragments. (c) PCR genotyping of mice obtained from heterozygous mating. Using Pxt1FP, Pxt1RP and Pgk-nRP primers, a 464 bp wild type fragment and a 697 bp recombinant fragment were amplified. (d) RT-PCR analysis of total RNA isolated from the testes of Pxt1+/+ and Pxt1−/− males using the Pxt1-specific primers. No Pxt1 expression could be detected in Pxt1–/– testes, whereas an expected 153 bp product was observed in the Pxt1+/+ sample. Synthesised cDNA quality was verified using Sdha-specific primers. –RT – primer specificity control; in this sample, cDNA was synthesised without the use of reverse transcriptase. Genome – genomic DNA was used as a template. O – negative control, where water was added instead of cDNA template.
| Primer | Sequence (5′ to 3′) | Tm (°C) |
|---|---|---|
| 5′Pxt1FP | 5′-TGAACCCAGTGAGGTTGTCGCAGAG-3′ | 65 |
| 5′Pxt1RP | 5′-CAGAACCCAGCACCAGCCATTG-3′ | |
| Pxt1FP | 5′-GAAGAACGGGAGGAACAGAA-3′ | 58 |
| Pxt1RP | 5′-CAGACAGCGGTTTACAACCAT-3′ | |
| Pgk-nRP | 5′-AGGAGCAAGGTGAGATGACAG-3′ | |
| Pxt1_ex2_FP | 5′-CAGCTTAGACACATTGGGGACA-3′ | 60 |
| Pxt1_ex3_RP | 5′-ACCTGGCCTCTCACGAACAC-3′ | |
| Uchl1FP | 5′-GCCAGTGTCGGGTAGATGACAAAGT-3′ | 60 |
| Uchl1RP | 5′-GGCTGGTTCTCTCTCCCCAGACTTA-3′ | |
| MeiocFP | 5′-TCCAAGACTGACTTCCAACCCATC-3′ | 62 |
| MeiocRP | 5′-GGCACCTAGGCACTCCTTGTCTTT-3′ | |
| Pttg1FP | 5′-GAAAGGCTTTGGGGACAGTCAACAG-3′ | 60 |
| Pttg1RP | 5′-GTAGGCATCATCAGGAGCAGGAACA-3′ | |
| Acrv1FP | 5′-CACCACTCAAAACTCCCAGCAGTGCA-3′ | 60 |
| Acrv1RP | 5′-AGTCAGAGCAAGAAGTAGGGCACGGG-3′ | |
| Tnp2Ex1FP | 5′-CACCAAGATGCAGAGCCTTCCCAC-3′ | 60 |
| Tnp2Ex2RP | 5′-GTGACATCATCCCAACAGTCCCCTAGT-3′ | |
| SdhaFP | 5′-GCTTGCGAGCTGCATTTGG-3′ | 60 |
| SdhaRP | 5′-CATCTCCAGTTGTCCTCTTCCA-3′ | |
| mTBP_F | 5′-CCCACAACTCTTCCATTCTCAAAC-3′ | 56 |
| mTBP_R | 5′-TCAAGTTTACAGCCAAGATTCACG-3′ |
RNA isolation and RT-PCR analysis
Testes and epididymes of adult animals were collected post-mortem, frozen in liquid nitrogen and stored at −80°C until further analysis. Caput, corpus and cauda epididymis were collected separately. RNA was isolated with NucleoSpin RNA mini kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instructions. One microgram of RNA was oligo-T primed and cDNA synthesis was performed using the High Capacity Reverse Transcriptase Kit (Applied Biosystems, USA) according to the manufacturer’s instructions. Expression of the Pxt1 gene was analysed using gene-specific primers Pxt1_ex2_FP and Pxt1_ex3_RP and the Kapa Taq PCR Kit (Kapa Biosystems, Switzerland). The expected product was 153 bp in size. To analyse the expression of Uchl1, Meioc, Pttg1, Acrv1 and Tnp2, gene-specific primers (Table 1) were used, amplifying 246 bp, 239 bp, 152 bp, 201 bp and 428 bp products, respectively. cDNA quality was checked with Sdha-specific primers (SdhaFP and SdhaRP), amplifying a 145 bp product, and with Tbp-specific primers (mTBP_F and mTBP_R), amplifying a 183 bp PCR product. The sequences of all primers used in this work are given in Table 1. To test primer specificity, cDNA generated without the use of reverse transcription, and genomic DNA were used as templates in PCR amplification.
Fertility testing
To analyse the consequences of Pxt1 ablation for male fertility, first heterozygous animals were intercrossed. Next, five Pxt1−/− males and five Pxt1+/+ males were mated with wild type or heterozygous females. The offspring were genotyped using DNA isolated from mouse tails and PCR reaction using Pxt1FP, Pxt1RP and Pgk-nRP primers.
Testis histology
For the histological analysis, male testes from Pxt1−/− and Pxt1+/+ mice were fixed in Bouin and embedded in paraffin using routine methods. Sections of 7-μm thickness were cut using a microtome (Reichert-Jung, Germany), deparaffinised, rehydrated and stained with haematoxylin and eosin (H-E). The stained sections were analysed using a light microscope (Opta-tech, Warsaw, Poland).
Determination of sperm number
The day before the experiment, IVF medium (Origio) was coated with mineral oil (Sigma Aldrich) and placed in an incubator for equilibration (37°C, 5% CO2). The cauda epididymis dissected from Pxt1−/− and Pxt1+/+ mice was suspended in 0.5 mL of IVF medium, punctured 15 times and gently pressed with forceps without harming blood vessels. Then, spermatozoa were left for 10 min in an incubator (37°C and 5% CO2) to allow them to disperse. To estimate the sperm count, 10 μL of IVF sperm suspension was placed in a Petri dish. The samples were then diluted by adding 10 μL of M2 medium. Sperm samples were then counted in a Neubauer chamber using a light microscope (Opta-tech).
Hypo-osmotic swelling test
To assess the integrity of the sperm tail membrane, 20 μL of sperm suspension in IVF medium was placed in a Petri dish. Then spermatozoa were suspended in 100 μL of sterile water. Following 5 min of incubation at 37°C, samples of 10 μL were collected and analysed with a light microscope (Opta-tech).
Assessment of sperm viability
The proportion of live/dead spermatozoa was analysed with 20 μL of sperm suspension in IVF medium and 20 μL of 0.2% eosin Y solution. After 10 min of incubation at 37°C, sperm viability was analysed with a light microscope.
Assessment of sperm morphology
To assess sperm morphology, 20 μL of sperm suspension in IVF medium, obtained immediately from the cauda epididymis was spread onto a glass slide and air-dried. Afterwards the cells were fixed in a mixture of acetic acid and ethyl alcohol at a ratio of 1:3 and left for 24 h in 0.2% eosine solution. Slides were photographed using a light microscope (Opta-tech), and the images were analysed using the ‘cell counter’ plugin of ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Chromomycin A3 staining
Chromomycin A3 (CMA3) staining allows the detection of protamine deficiency in spermatozoa as described previously (Lolis et al. 1996). For CMA3 staining, 50 μL of sperm suspension was centrifuged at 300g then spermatozoa were fixed for 5 min in a mixture of acetic acid and ethyl alcohol at a ratio of 1:3 at 4°C. Then 20 μL of sperm suspension was spread onto a glass slide and air-dried. Next, 100 μL of CMA3 (0.25 mg/mL in McIlvain buffer, pH 7 with 10 mM MgCl2) was added and slides were incubated for 20 min in the dark at room temperature with a cover glass, followed by three times washing with McIlvain buffer, pH 7. Spermatozoa were analysed with a fluorescent microscope. Light yellow spermatozoa were assigned as positive for protamine deficiency, whereas dark yellow spermatozoa were assigned as negative for protamine deficiency. At least 400 spermatozoa were counted for each animal.
Aniline blue staining
Aniline blue (AB) is an acidic dye with affinity for proteins in loose chromatin and it is used to detect residual histones in spermatozoa (Auger et al. 1990). For AB staining, 20 μL of sperm suspension was spread onto a glass slide and fixed in 3% glutaraldehyde solution in PBS for 30 min at room temperature. Then slides were air dried and stained with AB solution (Riedel-de Haën, Seelze, Germany) for 7 min at room temperature, followed by three times washing in PBS. Slides were analysed under the microscope and dark blue and blue stained spermatozoa were assigned as positive for residual histones, whereas light blue spermatozoa were assigned as negative for residual histones. At least 400 spermatozoa were counted for each animal.
Computer assisted semen analysis
Sperm motility was analysed using the computer assisted semen analysis (CASA) system with spermatozoa collected from the cauda epididymis. Epididymides were punctured with a needle, gently pressed with forceps without harming blood vessels and spermatozoa were left for 10 min in IVF medium at 37°C (SPL Life Sciences, South Korea). Epididymides were discarded and 13 μL of the sperm suspension was transferred to the incubation chamber at 37°C. Sperm movement was quantified using the CEROS system (ver. 10; Hamilton Thorne Research, Beverly, MA, USA). Spermatozoa from five mutant mice and five wild type control males were analysed using the following parameters: negative phase-contrast optics recording 60 frames per second, minimum contrast 60, minimum cell size 6 pixels and minimum static contrast 15 pixels. Slow motile cells and low-velocity nonprogressive spermatozoa were not recorded. For statistical analysis, frequencies of the six sperm motility parameters – amplitude of lateral head displacement (ALH, μm), beat cross frequency (BCF, Hz), average path velocity (VAP, μm/s), straight line velocity (VSL, μm/s), curvilinear velocity (VCL, μm/s) and straightness (STR, calculated as a ratio of VSL to VAP, expressed in %) – were examined by probability plots categorised by mouse type (wild type/mutant). In total, 5218 spermatozoa of Pxt1+/+ and 5675 spermatozoa of Pxt1−/− mice were analysed.
Sperm chromatin structure assay
Examination of sperm DNA fragmentation and chromatin condensation was performed on the same semen samples used for CASA examination. The sperm chromatin structure assay (SCSA) method (Evenson et al. 2002) was used. Briefly, after 30 s of sperm DNA denaturation at pH 1.5, the samples were stained with acridine orange. The green (525 nm) and red (620 nm) fluorescence from normal and fragmented DNA, respectively, were measured using a Navios (Beckman Coulter) flow cytometer. For DNA fragmentation the following calculation was used: ‘sperm DNA fragmentation’ = [red/(red + green)] fluorescence ratio. The percentage of spermatozoa with fragmented DNA was expressed as DNA fragmentation index (DFI), i.e. population with strong [red/(red + green)] values. Moreover, the DFI population was split into DFI moderate and DFI high subpopulations to reflect the level of DNA fragmentation. Sperm chromatin condensation was determined from a dot plot of green fluorescence vs sperm DNA fragmentation parameter. Spermatozoa with poorly condensed chromatin showed high green fluorescence and normal DNA fragmentation ratio. The percentage of spermatozoa with low chromatin condensation was expressed as high DNA stainability (HDS). Flow cytometric data analysis was performed with Kaluza (Beckman Coulter) software.
Competition breeding test
In order to determine whether the targeted deletion of the Pxt1 gene results in any disadvantages in mutants as compared with wild type males, a competition breeding test was performed as described previously (Grzmil et al. 2008). Briefly, mutant and wild type male mice were put in one cage at the age of 25 days and allowed to grow up together to avoid aggressive behaviour against each other in adulthood. At the age of 3 months, one Pxt1−/− and one Pxt1+/+ male were put together into a large test cage (200 cm × 100 cm, 150 cm high) with four wild type females. Animals were maintained under a 12 h light–dark cycle with free access to standard laboratory diet and water. The cage was equipped with multiple hiding places. After 17 days, females were individually transferred to normal breeding cages and allowed to deliver. Genotypes of all offspring were determined by PCR with Pxt1FP, Pgk-nRP and Pxt1RP primers. To avoid misleading results, each male was transferred to a normal breeding cage after the competition breeding test and mated with two wild type females. Only males that produced offspring within 2 months in this post-competition fertility test were considered for data analysis of the competition test.
Statistical analyses
All data were initially tested for normality using the Shapiro–Wilk test. All data that were not normally distributed and could not be normalised by transformation, such as live/dead sperm ratio, all CASA parameters and DFI high, were analysed with the nonparametric alternative for the t-test, the U-Mann–Whitney test. Data that were not significantly different from normal distribution, such as litter size, sperm number, hypo-osmotic test, sperm morphology, DFI total, DFI moderate, HDS, CMA3 and AB were analysed using the t-test. Proportions of wild type, heterozygous and homozygous animals produced in heterozygous breeding and competition test results were analysed using the chi-square test. A P-value below 0.05 was considered significant. All statistical analyses were performed using the Statistica v.13 software package (TIBICO Software Inc., USA).
Results
Targeted disruption of the Pxt1 gene in mice
Overexpression of the Pxt1 gene was previously shown to result in the complete degeneration of germ cells (Kaczmarek et al. 2011). To elucidate the function of this gene, a gene targeting approach was applied to generate homozygous Pxt1−/− mice. The mouse Pxt1 gene was disrupted in 129/Sv-derived R1 ES cells by homologous recombination using a replacement-targeting vector containing neomycin resistant (Neo) and thymidine kinase expression cassettes. The Neo cassette replaced exons 1 and 2, the latter containing the start codon ATG (Fig. 1a). Targeted R1 ES cells were identified using Southern blot analysis and an external probe located upstream of the 5′ flanking region of the targeting construct. The external probe detected the wild type allele as a 9.7-kb fragment and the recombinant allele as an 8-kb fragment in genomic DNA digested with the KpnI restriction enzyme (Fig. 1b). The ES cells heterozygous at the targeted loci were injected into C57BL/6J blastocysts to generate chimeric mice. Male chimeric mice transmitting the targeted mutation into the germ line were bred with female mice from the 129/Sv strain to obtain the Pxt1 knockout model. Heterozygous animals were intercrossed to generate homozygous mice. Animals were genotyped by PCR analysis of DNA obtained from mouse tails and with Pxt1FP, Pxt1RP and Pgk-nRP primers (Fig. 1c). Identified mutants from 129/Sv strain were then backcrossed on the ICR genetic background. Heterozygous animals from the B7 generation were then intercrossed to establish a mutant line on the ICR background (for the purpose of further analyses not included in this manuscript) and to generate homozygous mutant males. All results presented in this work were obtained on the ICR background. RNA was isolated from testes of adult Pxt1−/− mice and RT-PCR was performed with Pxt1-specific primers (Pxt1_ex2_FP and Pxt1_ex3_RP) to demonstrate the lack of Pxt1 gene expression (Fig. 1d, upper panel). No expression was observed in testes of Pxt1−/− mice, whereas the expected 153 bp band was present in samples from the testes of Pxt1+/+ control animals. The quality and integrity of the RNA samples was verified by RT-PCR reaction and Sdha-specific primers (Fig. 1d, bottom panel).
Pxt1 mutant males are fertile
Breeding of Pxt1−/− males and females with wild type animals revealed that both mutant males and females were fertile. Pxt1 is exclusively expressed in mouse testes (Grzmil et al. 2007). Therefore, all further analyses were performed to investigate the consequences of Pxt1 disruption for spermatogenesis. From five pairs of heterozygous males and females, 140 offspring were obtained. Among them, 31 animals were +/+, 81 were +/− and 28 were −/−. These genotype ratios did not differ significantly from the expected Mendelian ratio (P = 0.17). All offspring were viable and did not demonstrate any obvious phenotype. Next, five Pxt1−/− and five Pxt+/+ males were intercrossed with wild type or heterozygous females to analyse the fertility of Pxt1-deficient males. All tested males were fertile and no significant differences in litter size between mutants and controls were detected (Table 2). Moreover, no significant differences were observed in litter frequencies or offspring survival from mutants and control males. Histological analysis of the testis of Pxt1−/− mutants did not reveal any obvious changes as compared to the wild type control. (Fig. 2a). In the testis of Pxt1−/− mice, histology was normal, with all germ cell types and stages of spermatogenesis present and comparable to control tissue.
| Male genotype | No. of litters | Total no. of offspring | Mean no. of offspring per litter ± s.d. | P |
|---|---|---|---|---|
| Pxt1+/+ | 23 | 104 | 4.5 ± 1.5 | |
| Pxt1−/− | 17 | 82 | 4.8 ± 1.5 | 0.56 |
Testicular histology and the analysis of expression of markers of different germ cell types in Pxt1−/− mice. Spermatogenesis proceeds normally in testes of Pxt1 knockout mice. (a) Representative micrographs of H-E stained testes of adult Pxt1−/− and Pxt1+/+ mice. Spermatogenesis in Pxt1−/− testes is not affected as indicated by the presence of high numbers of late elongated spermatids similar to the histology seen in testes of wild type mice. (b) The expression of all analysed markers characteristic of subsequent spermatogenesis stages was detected in testes of Pxt1−/− mice by RT-PCR. –RT – control without reverse transcriptase in the reaction. gDNA – genomic DNA. O – negative control, where water was added instead of cDNA template.
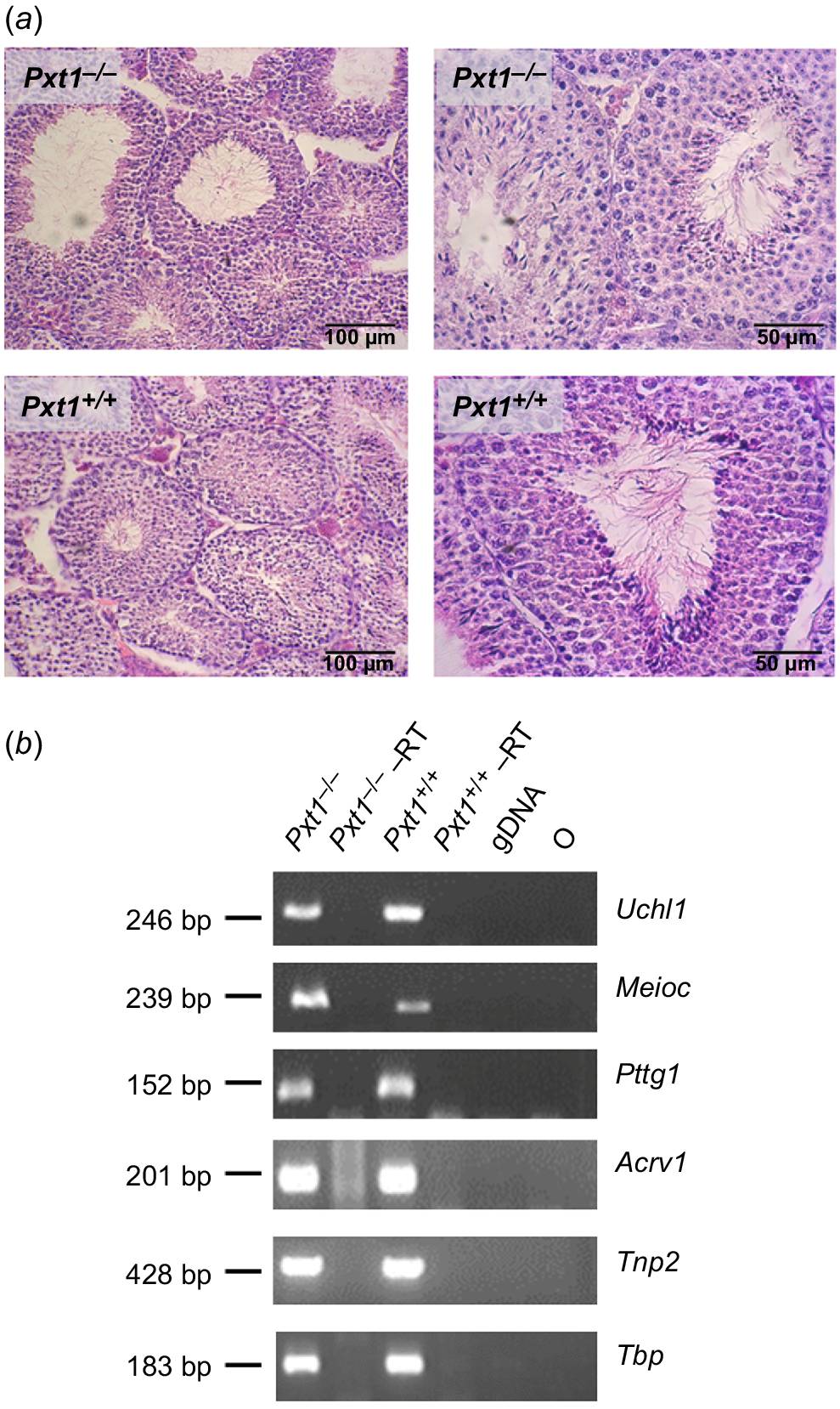
To extend the histological observations, RNA was isolated from testes of adult Pxt1−/− and Pxt1+/+ mice and RT-PCR analysis with primers specific for known markers of different germ cell types was performed. As identified previously in single-cell RNA sequencing, Uchl1 expression starts when differentiated spermatogonia are present in mouse testes, Meioc transcripts are present in early spermatocytes, Pttg1 in late spermatocytes, Acrv1 in round spermatids and Tnp2 in elongated spermatids (reviewed in Du et al. 2021). The expression of all analysed markers was observed in the testes of Pxt1−/− mice (Fig. 2b). The quality and integrity of the RNA samples was verified by RT-PCR reaction and Tbp-specific primers (Fig. 2b, bottom panel).
Next, epididymides of three Pxt1−/− mutant males and three Pxt1+/+ control animals were collected and dissected in IVF medium. Sperm numbers in cauda epididymidis were determined using the Neubauer cell chamber. A hypo-osmotic swelling test was performed to analyse the integrity of the spermatozoa tail membrane. Morphology was analysed using light microscopy on spermatozoa stained with eosine. In addition, live/dead spermatozoa were also counted by light microscopy. For each analysis of sperm parameters, at least 400 spermatozoa were counted per animal. Statistical analysis revealed that sperm numbers were slightly but significantly reduced in Pxt1−/− mice as compared to the Pxt1+/+ control. No significant differences were observed in the hypo-osmotic swelling test, sperm morphology or live/dead sperm ratio (Table 3).
| Parameter | Genotype | P | |||
|---|---|---|---|---|---|
| Pxt1+/+ (N = 3) | Pxt1−/− (N = 3) | ||||
| Mean | s.d. | Mean | s.d. | ||
| Sperm number (×107) | 1.7 | 0.07 | 1.5 | 0.06 | 0.02* |
| Hypo-osmotic swelling test (%) | 75 | 2.1 | 74 | 2.5 | 0.66 |
| Sperm head morphology (%) | 78 | 1.3 | 75 | 2.0 | 0.19 |
| Live sperm rate (%) | 77 | 0.6 | 75 | 1.4 | 0.08 |
*P < 0.05.
Sperm motility was analysed using the CASA system. Six Pxt1−/− and five Pxt1+/+ males were utilised in this analysis. The following parameters were analysed: ALH and BCF describe sperm head activity; VAP, VSL, and VCL measure average, straight line and curvilineal sperm velocity parameters, respectively; and STR provides information about path shape (Table 4). Statistical analysis of the results revealed that mutant and control animals did not differ significantly in any of the analysed motility parameters.
| Parameter | Pxt1+/+ (N = 5) | Pxt1−/− (N = 6) | P | ||
|---|---|---|---|---|---|
| Mean | s.d. | Mean | s.d. | ||
| VAP (μm/s) | 111.62 | 21.86 | 121.81 | 21.42 | 0.65 |
| VSL (μm/s) | 76.46 | 16.43 | 83.92 | 12.92 | 0.65 |
| VCL (μm/s) | 190.47 | 38.10 | 207.22 | 46.07 | 0.65 |
| ALH (μm) | 8.56 | 1.20 | 8.89 | 1.69 | 0.93 |
| BCF (Hz) | 19.90 | 1.88 | 19.28 | 1.14 | 0.65 |
| STR (%) | 62.78 | 1.46 | 63.06 | 2.11 | 0.93 |
Targeted disruption of Pxt1 results in an enhanced sperm DNA fragmentation index
Pxt1-deficient males were fertile and demonstrated near normal sperm parameters and testis histology. Our previous work demonstrated that overexpression of Pxt1 induces male germ cell death in transgenic mice (Kaczmarek et al. 2011). Therefore, we asked whether the lack of Pxt1 activity could result in decreased elimination of damaged germ cells. One sperm quality parameter regarded as relevant for male fecundity (Westerman 2020) is the sperm DFI. Spermatozoa were isolated from cauda epididymidis of five Pxt1−/− males and five control males and SCSA was performed with flow cytometry. From a single animal, 10 000 to 15 000 spermatozoa were counted. Results are given as total DFI according to the fluorescence signal ratio (red/red + green). In addition, spermatozoa with increased DFI were divided into two groups: DFI moderate and DFI high. Rules of spermatozoa assignment to a particular category are presented in Supplementary Fig. S1A and B. Spermatozoa of Pxt1−/− males demonstrated a significantly higher total DFI as compared to control males (P < 0.001, Fig. 3a). When spermatozoa with increased DFI were divided into DFI moderate and DFI high groups, mutant spermatozoa with DFI high appeared as the dominant population, whereas the proportion of DFI moderate spermatozoa was significantly lower as compared to control mice (P < 0.001, Fig. 3a).
SCSA and Pxt1 expression analyses. Spermatozoa of Pxt1+/+ and Pxt1−/− mice were analysed using SCSA and flow cytometry. The DFI parameter was calculated as ratio of green and red fluorescence according to the formula: [red/(green + red)]. (a) Spermatozoa from Pxt1−/− mice demonstrated a significant increase in DFI as compared to spermatozoa from Pxt1+/+ mice. DFI total – sum of DFI high and DFI moderate. (b) HDS classification of immature spermatozoa. Pxt1−/− mice demonstrate a significant reduction of spermatozoa classified as HDS as compared to control mice. (c) The expression of Pxt1 was analysed in epididymis and testis of adult Pxt1+/+ mice by RT-PCR. No expression was detected in caput, corpus or in cauda epididymidis, whereas the expected 153-bp PCR product was observed in testis. –RT – control without reverse transcriptase in the reaction. gDNA – genomic DNA. O – negative control, where water was added instead of cDNA template. ***P < 0.001.
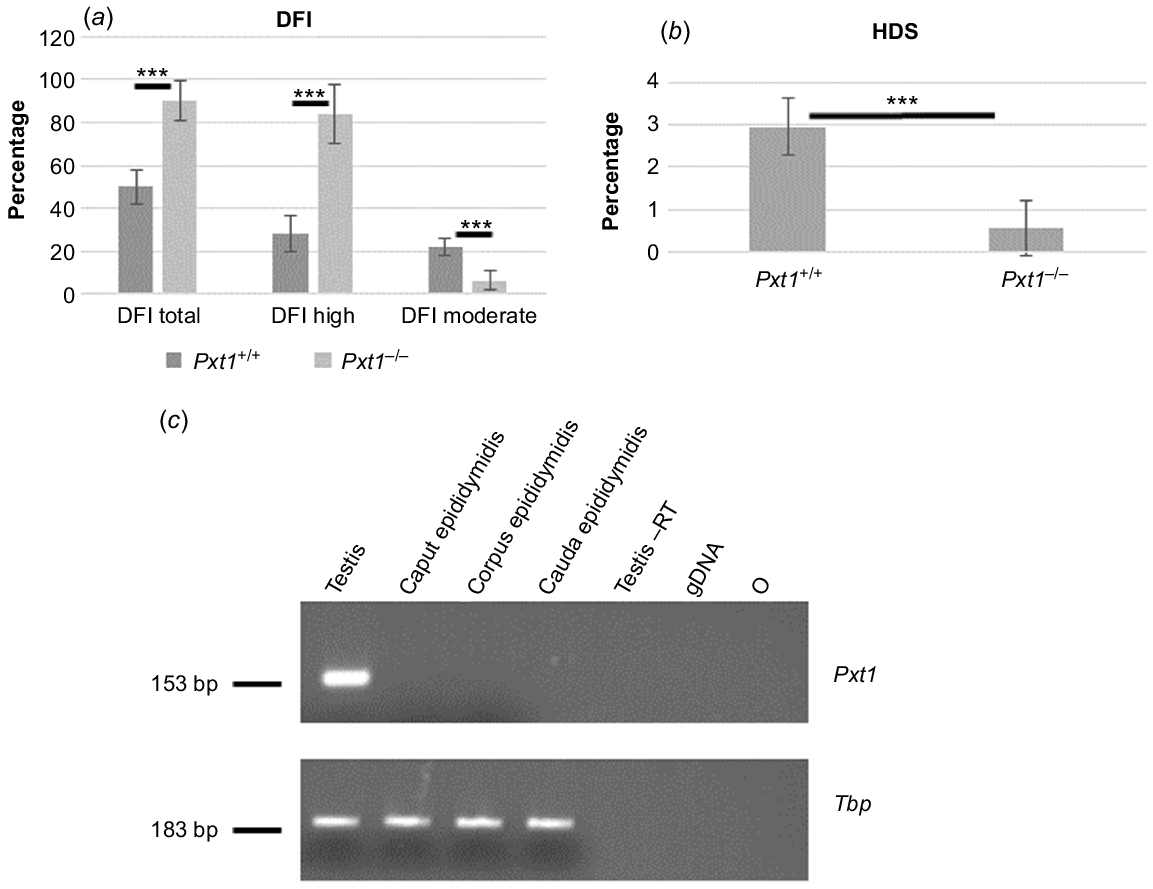
DNA condensation was also analysed from the plot (Supplementary Fig. S1C and D) as green vs red fluorescence. Spermatozoa with poor DNA condensation showed higher green fluorescence expressed as HDS. Classification rules of immature sperm (showing HDS) are shown in Fig. S1C and D. Pxt1−/− derived spermatozoa demonstrated significantly lower HDS as compared to Pxt1+/+ derived spermatozoa (P < 0.001, Fig. 3b).
The expression of the Pxt1 gene was previously demonstrated in mouse testis (Grzmil et al. 2007). To analyse whether this gene is expressed in mouse epididymis, RNA was isolated from caput, corpus and cauda epididymidis of adult Pxt1+/+ mice and RT-PCR analysis was performed with Pxt1-specific primers (Pxt1_ex2_FP and Pxt1_ex3_RP). No expression of Pxt1 was detected in any analysed epididymal samples, whereas the expected 153-bp fragment was amplified in cDNA synthesised from testicular RNA that served as a positive control (Fig. 3c). The quality and integrity of the RNA samples were verified by RT-PCR reaction and Tbp-specific primers (Fig. 3c, bottom panel).
During spermatogenesis, histons are partially replaced by protamines and altered protamination may lead to increased spermatozoa susceptibility to DNA damage (reviewed in Agarwal et al. 2020). To analyse the protamination process in spermatozoa of Pxt1 knockout mice, CMA3 and AB staining were performed. The CMA3 staining detects protamine deficiency (Lolis et al. 1996) and AB staining allows the detection of residual histones (Auger et al. 1990). No significant differences were detected between Pxt1−/− and Pxt1+/+ spermatozoa stained with CMA3 and AB (P = 0.33 and P = 0.58, respectively, Table 5).
Pxt1-deficient males are able to mate in the presence of wild type competitors
Enhanced sperm DFI in Pxt1−/− mice suggests that their spermatozoa may be of lower quality as compared to control animals. However, homozygous mutants were fertile. A typical test of fertility is performed in standard breeding cages with unlimited access of tested males to females and without the presence of other males. We asked whether Pxt1−/− males would be able to mate when in competition with wild type males. In a competitive-breeding experiment, Pxt1−/− and Pxt1+/+ males were bred with four wild type females in a large cage (2 m2) for 17 days. Thereafter, the females were transferred individually to normal breeding cages and allowed to deliver, and the genotypes of the resulting newborns were determined by PCR. This experiment was performed five times, using different males and females in each experiment. From 20 females used in this experiment, six females did not become pregnant and 14 females delivered 53 offspring. Eight females produced 30 wild type offspring (litter size: 2, 7, 2, 7, 1, 3, 3, 5), all descended from Pxt1+/+ males, and six females produced 23 heterozygous offspring (litter size: 4, 4, 7, 5, 2, 1), all descended from Pxt1−/− males. No litters contained mixtures of Pxt1+/+ and Pxt1+/− offspring (Table 6). Statistical analysis revealed that there is no significant difference between the numbers of litters or offspring descended from Pxt1−/− and Pxt1+/+ males.
| Genotype of males | Litters | No. of offspring |
|---|---|---|
| Pxt1+/+ | 8 | 30 |
| Pxt1−/− | 6 | 23 |
| P = 0.59 | P = 0.34 |
Pxt1−/− and Pxt1+/+ males were mated with four wild type females in a large breeding cage. This competition breeding test was repeated five times. The number of litters and the number of total offspring from Pxt1+/+ (genotyped as +/+) and from Pxt1−/− (genotyped as +/−) males are given.
Discussion
Of all analysed tissues in the mouse, the expression of Pxt1 was restricted to the testis. Pxt1 expression was higher in germ cells co-cultured with Sertoli cells as compared with cultured germ cells only and this gene was not expressed in testes of W/Wv mutant mice (Grzmil et al. 2007), which lack germ cells altogether (Lyon and Searle 1989). These findings suggest that Pxt1 is a male germ cell–specific gene and that its expression may be regulated during spermatogenesis. Indeed, the analysis of the expression profile of Pxt1 in mouse testes at different postnatal ages revealed that expression commences between 15 and 17 days postpartum. Previous in situ expression analyses demonstrated that Pxt1 transcripts were mainly detected in spermatocytes (Grzmil et al. 2007).
To understand the function of the Pxt1 gene in mouse spermatogenesis, we generated and analysed Pxt1−/− mice. Mating of heterozygous males and females produced the expected Mendelian ratio of all three genotypes (Pxt1+/+, Pxt1+/− and Pxt1−/−). Pxt1−/− males were viable and did not exhibit an obvious phenotype. Homozygous mutant males were fertile and produced a normal number of offspring. Testicular histology did not reveal any abnormalities. The analysis of sperm quality parameters demonstrated that besides a slight reduction in sperm number, the most prominent phenotype was an increased proportion of spermatozoa with DNA strand breaks (DFI). However, this phenotype did not affect the fecundity of mutants as they were able to mate in competition with wild type males.
The number of spermatozoa isolated from mutant cauda epididymidis was significantly lower as compared to wild type animals, yet the observed mean number of spermatozoa (1.5 × 107) was not drastically reduced. We and others have described that the usual number of spermatozoa isolated from cauda epididymidis in 0.5 mL of medium (as in the present work) varies between 0.8 and 2.4 × 107 without consequences for male fertility (Mannan et al. 2003; Nayernia et al. 2003; Adham et al. 2005; Grzmil et al. 2008). In bulls it has been demonstrated that a slight but significant reduction of sperm numbers has no effect on insemination success (Januskauskas et al. 1996). Therefore, we conclude that although the number of spermatozoa isolated from mutants was significantly reduced as compared to wild type control, this phenotype has limited or even no biological consequence.
The most prominent phenotype in Pxt1−/− males is reflected by more than double increase of total DFI and almost three times increase in proportion of spermatozoa with high DFI as compared to control animals. Defective chromatin condensation, abortive apoptosis and oxidative stress are common causes of sperm DNA strand breaks (Agarwal et al. 2020). We have demonstrated that in Pxt1 knockout mice the protamination process is not affected; therefore, the observed enhanced DFI is not a consequence of defective chromatin condensation but may result from abortive cell elimination. In humans, the DFI has been shown to be a critical factor in predicting the clinical outcome in assisted reproduction (Oleszczuk et al. 2016; Evenson 2017, 2018). Based on our previous work demonstrating that Pxt1 overexpression induces cell death in transfected cells as well as in germ cells of transgenic mice (Kaczmarek et al. 2011), Pxt1 expression profiles (Grzmil et al. 2007, this work) and the observation that ablation of Pxt1 results in an increased proportion of spermatozoa with DNA strand breaks, we suggest that the function of this gene in spermatogenesis is related to the elimination of germ cells with aberrant chromatin structure. The N-terminal domain of PXT1 is similar to the BH3 motif. Proteins containing only the BH3 domain serve as apoptosis inducers (reviewed in Lomonosova and Chinnadurai 2008). Even though the BH3-similar motif of PXT1 does not exactly match the classical BH3 consensus (Aouacheria et al. 2015), the overexpression of PXT1 triggers typical signs of apoptosis, including nuclear fragmentation, cell rounding and shrinkage, plasma membrane blebbing and externalisation of phosphatidylserine residues to the outer plasma membrane (Casciola-Rosen et al. 1996; Mund et al. 2003; Nozawa et al. 2009). Deletion of this BH3-similar motif resulted in a significant reduction of PXT1 cytotoxic activity (Kaczmarek et al. 2011). Moreover, human (but not mouse) PXT1 binds BCL-XL, an antiapoptotic protein (Lim et al. 2022). Recently, human PXT1 was reported to activate the apoptotic effector BAK (Aguilar et al. 2023). Previously, we have demonstrated that the mouse BCL2-associated athanogene 6 (BAG6), a known anti-apoptotic protein (Wu et al. 2004; Kikukawa et al. 2005), binds mouse PXT1, with this interaction protecting cells from PXT1-induced degeneration (Kaczmarek et al. 2011). Thus, even if the molecular mechanism of mouse and human PXT1 is different, both seem to be involved in apoptosis regulation. In addition, in the testis of Fxra−/− knockout mice (knockout of the Nrlh4 gene) the expression of Pxt1 was significantly lower as compared to control animals. Targeted disruption of Nrlh4 was accompanied by reduced apoptosis rates of male germ cells in mice at the age of 15 days postpartum (dpp) to 3 months, besides other phenotypic features (Martinot et al. 2017). Notably, Pxt1 expression in mouse testis starts at 15 dpp (Grzmil et al. 2007). The DFI was analysed in spermatozoa isolated from cauda epididymidis of Pxt1−/− mice, and we raise the question whether PXT1 acts only in testis or also in epididymis. No Pxt1 transcript could be detected in epididymis using Northern blot analysis (Grzmil et al. 2007). However, the sensitivity of Northern blot analysis is lower compared to PCR-based techniques. Therefore, we used RT-PCR to analyse the expression of Pxt1 in caput, corpus and cauda epididymidis and confirmed that this gene is not expressed in any of the analysed regions of the epididymis. All above-mentioned observations suggest that PXT1 acts exclusively in testis and its activity starts with the spermatocyte stage of spermatogenesis.
From the pioneering works of Evenson and co-workers (Evenson et al. 1978, 1980a, 1980b), it is widely accepted that an increased DFI correlates with a negative impact on male fertility (reviewed in Evenson 2016). More specifically, in humans and other species a correlation between DFI and the outcome of natural or artificial fertilisation has been reported (Bochenek et al. 2001; Guérin and Benchaïb 2004; Miciński et al. 2009; Jurewicz et al. 2015; Gupta et al. 2021). Spermatozoa of Pxt1−/− mice demonstrate an increased DFI, particularly a highly increased proportion of spermatozoa with high red fluorescence value (termed DFI high). If sperm DNA damage is minor, it could be repaired in the oocyte (Evenson et al. 2002), but an increased population of spermatozoa indexed as DFI high may suggest that Pxt1-deficient males face fertility problems. Another population of spermatozoa detected through SCSA analysis are HDS spermatozoa. The population of spermatozoa with HDS reflects immature spermatozoa (Evenson 2013) and was calculated on the basis of the percentage of spermatozoa with high levels of green fluorescence (Evenson et al. 2002). Reduced populations of HDS spermatozoa in Pxt1−/− mice can be explained by a high proportion of spermatozoa with DFI high that demonstrate a fluorescence signal shifted into red. Therefore, only a minor population of spermatozoa from Pxt1-deficient mice is measured as ‘highly green’.
In spite of the high DFI, Pxt1−/− males are fertile. Because in most mammals females mate promiscuously with multiple males, male competition and sexual selection are important aspects of reproductive success (Birkhead 2001). Sexual selection could act against Pxt1−/− mutants before copulation (e.g. male dominance, female choice) or after copulation (e.g. spermatozoa with high DFI could be impaired in their competition with wild type spermatozoa in the female reproductive tract) (Kleene 2005). In a wild population of Mus domesticus, nearly 20% of females produced progeny derived from mating with multiple males (Dean et al. 2006). Therefore, we performed the competition breeding test. To our surprise, Pxt1-deficient males were able to mate when wild type competitors were present. As no promiscuous matings were observed in this competition breeding test, it can only be concluded that precopulatory selection against Pxt1−/− males does not occur.
Our observation that Pxt1-deficient males are fertile despite a high sperm DFI does not mean that the DFI has no impact on fertility in mice. DFI does not constitute a constant parameter throughout the lifespan (e.g. in boar DFI changes occur seasonally, Ausejo et al. 2021), and in mice this parameter varies even within the day (Ni et al. 2019). To avoid any inconsistency resulting from this phenomenon, we have always sacrificed mice for spermatozoa isolation at the same time. In humans, DFI changes with age as reported previously (Deenadayal Mettler et al. 2020; Evenson et al. 2020), an observation not noticed in rodents and rabbits (Gogol et al. 2002; Taylor et al. 2019). In humans, DNA damage in spermatozoa related to paternal age could be a reflection of lifestyle (Fraga et al. 1996; Lane et al. 2014) or a wide range of environmental pollutants (Grizard et al. 2007; Khan et al. 2015; Barbonetti et al. 2016; Lu et al. 2017). It is also known that chromatin damage (e.g. increased DFI) has an impact on the next generation (reviewed in Aitken 2018). The increased proportion of spermatozoa with high DFI in Pxt1 mice could impact offspring, leading to a decrease of fertility in the next generations. In the current study, we utilised mice from the F1–F2 generation of heterozygous parents but not beyond due to the long timeframe necessary. Therefore, this aspect will be a matter of future studies.
In conclusion, targeted disruption of the Pxt1 gene resulted in increased sperm DFI. Because Pxt1 induces cell death (Kaczmarek et al. 2011), we suggest that the function of this gene may be related to elimination of male germ cells with DNA damage; however, further analyses are needed to provide more evidence confirming this suggestion. The expression of Pxt1 was not detected in epididymal samples containing epididymal spermatozoa. Earlier studies have demonstrated the expression of Pxt1 in spermatocytes (Grzmil et al. 2007); therefore, we hypothesise that this gene acts in spermatocytes and/or some later stages rather than in spermatozoa. In humans, a homologue Pxt1 gene is located on chromosome 6 and shares 74% similarity with the mouse gene (Grzmil et al. 2007). Thus, human PXT1 can be considered for mutation screening in patients with increased DFI.
Data availability
The data that support this study will be shared upon request to the corresponding author.
Declaration of funding
This work was supported by the Polish National Science Centre grant no. 2015/19/B/NZ4/00576 to PG. The open access publication of this article was funded by the program ‘Excellence Initiative – Research University’ at the Faculty of Biology of the Jagiellonian University in Kraków, Poland.
Acknowledgements
We thank A. Herwig and J. Manz for excellent technical support and S. Wolf for help in breeding of knockout mice. We also thank M. Pabijan for critical reading of the manuscript.
References
Adham IM, Eck TJ, Mierau K, Müller N, Sallam MA, Paprotta I, Schubert S, Hoyer-Fender S, Engel W (2005) Reduction of spermatogenesis but not fertility in Creb3l4-deficient mice. Molecular and Cellular Biology 25(17), 7657-7664.
| Crossref | Google Scholar |
Agarwal A, Barbăroșie C, Ambar R, Finelli R (2020) The impact of single- and double-strand DNA breaks in human spermatozoa on assisted reproduction. International Journal of Molecular Sciences 21(11), 3882.
| Crossref | Google Scholar |
Aguilar F, Yu S, Grant RA, Swanson S, Ghose D, Su BG, Sarosiek KA, Keating AE (2023) Peptides from human BNIP5 and PXT1 and non-native binders of pro-apoptotic BAK can directly activate or inhibit BAK-mediated membrane permeabilization. Structure 31(3), 265-281.e7.
| Crossref | Google Scholar |
Aitken RJ (2018) Not every sperm is sacred; a perspective on male infertility. Molecular Human Reproduction 24(6), 287-298.
| Crossref | Google Scholar |
Aouacheria A, Combet C, Tompa P, Hardwick JM (2015) Redefining the BH3 death domain as a ‘Short Linear Motif’. Trends in Biochemical Sciences 40(12), 736-748.
| Crossref | Google Scholar |
Auger J, Mesbah M, Huber C, Dadoune JP (1990) Aniline blue staining as a marker of sperm chromatin defects associated with different semen characteristics discriminates between proven fertile and suspected infertile men. International Journal of Andrology 13(6), 452-462.
| Crossref | Google Scholar |
Ausejo R, Martínez JM, Soler-Llorens P, Bolarín A, Tejedor T, Falceto MV (2021) Seasonal changes of nuclear DNA fragmentation in boar spermatozoa in Spain. Animals 11(2), 465.
| Crossref | Google Scholar |
Barbonetti A, Castellini C, Di Giammarco N, Santilli G, Francavilla S, Francavilla F (2016) In vitro exposure of human spermatozoa to bisphenol A induces pro-oxidative/apoptotic mitochondrial dysfunction. Reproductive Toxicology 66, 61-67.
| Crossref | Google Scholar |
Bochenek M, Smorąg Z, Pilch J (2001) Sperm chromatin structure assay of bulls qualified for artificial insemination. Theriogenology 56(4), 557-567.
| Crossref | Google Scholar |
Casciola-Rosen L, Rosen A, Petri M, Schlissel M (1996) Surface blebs on apoptotic cells are sites of enhanced procoagulant activity: implications for coagulation events and antigenic spread in systemic lupus erythematosus. Proceedings of the National Academy of Sciences 93(4), 1624-1629.
| Crossref | Google Scholar |
Dean MD, Ardlie KG, Nachman MW (2006) The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus). Molecular Ecology 15(13), 4141-4151.
| Crossref | Google Scholar |
Deenadayal Mettler A, Govindarajan M, Srinivas S, Mithraprabhu S, Evenson D, Mahendran T (2020) Male age is associated with sperm DNA/chromatin integrity. The Aging Male 23(5), 822-829.
| Crossref | Google Scholar |
De Felici M, Klinger FG (2015) Programmed cell death in mouse primordial germ cells. The International Journal of Developmental Biology 59(1-3), 41-49.
| Crossref | Google Scholar |
Du L, Chen W, Cheng Z, Wu S, He J, Han L, He Z, Qin W (2021) Novel gene regulation in normal and abnormal spermatogenesis. Cells 10(3), 666.
| Crossref | Google Scholar |
Evenson DP (2016) The Sperm Chromatin Structure Assay (SCSA®) and other sperm DNA fragmentation tests for evaluation of sperm nuclear DNA integrity as related to fertility. Animal Reproduction Science 169, 56-75.
| Crossref | Google Scholar |
Evenson DP (2017) Evaluation of sperm chromatin structure and DNA strand breaks is an important part of clinical male fertility assessment. Translational Andrology and Urology 6(Suppl 4), S495-S500.
| Crossref | Google Scholar |
Evenson DP (2018) Sperm Chromatin Structure Assay (SCSAVR): evolution from origin to clinical utility. In ‘A clinician’s guide to sperm DNA and chromatin damage’. (Eds A Zini, A Agarwal) pp. 65–89. (Springer International: Cham, Switzerland) doi:10.1007/978-3-319-71815-6
Evenson DP, Witkin SS, de Harven E, Bendich A (1978) Ultrastructure of partially decondensed human spermatozoal chromatin. Journal of Ultrastructure Research 63(2), 178-187.
| Crossref | Google Scholar |
Evenson DP, Darzynkiewicz Z, Melamed MR (1980a) Comparison of human and mouse sperm chromatin structure by flow cytometry. Chromosoma 78(2), 225-238.
| Crossref | Google Scholar |
Evenson DP, Darzynkiewicz Z, Melamed MR (1980b) Relation of mammalian sperm chromatin heterogeneity to fertility. Science 210(4474), 1131-1133.
| Crossref | Google Scholar |
Evenson DP, Larson KL, Jost LK (2002) Sperm chromatin structure assay: its clinical use for detecting sperm DNA fragmentation in male infertility and comparisons with other techniques. Journal of Andrology 23(1), 25-43.
| Crossref | Google Scholar |
Evenson DP, Djira G, Kasperson K, Christianson J (2020) Relationships between the age of 25,445 men attending infertility clinics and sperm chromatin structure assay (SCSA®) defined sperm DNA and chromatin integrity. Fertility and Sterility 114(2), 311-320.
| Crossref | Google Scholar |
Fraga CG, Motchnik PA, Wyrobek AJ, Rempel DM, Ames BN (1996) Smoking and low antioxidant levels increase oxidative damage to sperm DNA. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 351(2), 199-203.
| Crossref | Google Scholar |
Gogol P, Bochenek M, Smorag Z (2002) Effect of rabbit age on sperm chromatin structure. Reproduction in Domestic Animals 37(2), 92-95.
| Crossref | Google Scholar |
Grizard G, Ouchchane L, Roddier H, Artonne C, Sion B, Vasson M-P, Janny L (2007) In vitro alachlor effects on reactive oxygen species generation, motility patterns and apoptosis markers in human spermatozoa. Reproductive Toxicology 23(1), 55-62.
| Crossref | Google Scholar |
Grzmil P, Burfeind C, Preuss T, Dixkens C, Wolf S, Engel W, Burfeind P (2007) The putative peroxisomal gene Pxt1 is exclusively expressed in the testis. Cytogenetic and Genome Research 119(1–2), 74-82.
| Crossref | Google Scholar |
Grzmil P, Boinska D, Kleene KC, Adham I, Schlüter G, Kämper M, Buyandelger B, Meinhardt A, Wolf S, Engel W (2008) Prm3, the fourth gene in the mouse protamine gene cluster, encodes a conserved acidic protein that affects sperm motility. Biology of Reproduction 78(6), 958-967.
| Crossref | Google Scholar |
Guérin J-F, Benchaïb M (2004) Tests d’exploration de la qualité nucléaire du spermatozoïde: relations avec la fertilité et la qualité du conceptus [Assays for assessment of sperm DNA integrity: relationships with fertility and conceptus quality]. Gynécologie Obstétrique & Fertilité 32(9), 799-802.
| Crossref | Google Scholar |
Gupta S, Finelli R, Agarwal A, Henkel R (2021) Total antioxidant capacity – relevance, methods and clinical implications. Andrologia 53(2), e13624.
| Crossref | Google Scholar |
Januskauskas A, Söderquist L, Håård MG, Ch M, Lundeheim N, Rodriguez-Martinez H (1996) Influence of sperm number per straw on the post-thaw sperm viability and fertility of Swedish red and white A.I. bulls. Acta Veterinaria Scandinavica 37(4), 461-470.
| Crossref | Google Scholar |
Jurewicz J, Radwan M, Wielgomas B, Sobala W, Piskunowicz M, Radwan P, Bochenek M, Hanke W (2015) The effect of environmental exposure to pyrethroids and DNA damage in human sperm. Systems Biology in Reproductive Medicine 61(1), 37-43.
| Crossref | Google Scholar |
Kaczmarek K, Studencka M, Meinhardt A, Wieczerzak K, Thoms S, Engel W, Grzmil P (2011) Overexpression of peroxisomal testis-specific 1 protein induces germ cell apoptosis and leads to infertility in male mice. Molecular Biology of the Cell 22(10), 1766-1779.
| Crossref | Google Scholar |
Khan S, Jan MH, Kumar D, Telang AG (2015) Firpronil induced spermotoxicity is associated with oxidative stress, DNA damage and apoptosis in male rats. Pesticide Biochemistry and Physiology 124, 8-14.
| Crossref | Google Scholar |
Kikukawa Y, Minami R, Shimada M, Kobayashi M, Tanaka K, Yokosawa H, Kawahara H (2005) Unique proteasome subunit Xrpn10c is a specific receptor for the antiapoptotic ubiquitin-like protein Scythe. FEBS Journal 272(24), 6373-6386.
| Crossref | Google Scholar |
Kleene KC (2005) Sexual selection, genetic conflict, selfish genes, and the atypical patterns of gene expression in spermatogenic cells. Developmental Biology 277(1), 16-26.
| Crossref | Google Scholar |
Lane M, McPherson NO, Fullston T, Spillane M, Sandeman L, Kang WX, Zander-Fox DL (2014) Oxidative stress in mouse sperm impairs embryo development, fetal growth and alters adiposity and glucose regulation in female offspring. PLoS ONE 9(7), e100832.
| Crossref | Google Scholar |
Lim D, Jin S, Shin H-C, Kim W, Choi JS, Oh D-B, Kim SJ, Seo J, Ku B (2022) Structural and biochemical analyses of Bcl-xL in complex with the BH3 domain of peroxisomal testis-specific 1. Biochemical and Biophysical Research Communications 625, 174-180.
| Crossref | Google Scholar |
Lolis D, Georgiou I, Syrrou M, Zikopoulos K, Konstantelli M, Messinis I (1996) Chromomycin A3-staining as an indicator of protamine deficiency and fertilization. International Journal of Andrology 19(1), 23-27.
| Crossref | Google Scholar |
Lomonosova E, Chinnadurai G (2008) BH3-only proteins in apoptosis and beyond: an overview. Oncogene 27(Suppl 1), S2-S19.
| Crossref | Google Scholar |
Lu Y, Lin M, Aitken RJ (2017) Exposure of spermatozoa to dibutyl phthalate induces abnormal embryonic development in a marine invertebrate Galeolaria caespitosa (Polychaeta: Serpulidae). Aquatic Toxicology 191, 189-200.
| Crossref | Google Scholar |
Lyon MF, Searle AG (1989) ‘Genetic variants and strains of the laboratory mouse.’ 2nd edn. pp. XIV, 876. (Oxford University Press: UK) doi:10.1002/jobm.3620300716
Mannan AU, Nayernia K, Mueller C, Burfeind P, Adham IM, Engel W (2003) Male mice lacking the Theg (testicular haploid expressed gene) protein undergo normal spermatogenesis and are fertile. Biology of Reproduction 69(3), 788-796.
| Crossref | Google Scholar |
Martinot E, Sèdes L, Baptissart M, Holota H, Rouaisnel B, Damon-Soubeyrand C, De Haze A, Saru J-P, Thibault-Carpentier C, Keime C, Lobaccaro J-MA, Baron S, Benoit G, Caira F, Beaudoin C, Volle DH (2017) The bile acid nuclear receptor FXRα is a critical regulator of mouse germ cell fate. Stem Cell Reports 9(1), 315-328.
| Crossref | Google Scholar |
Matzuk MM, Lamb DJ (2002) Genetic dissection of mammalian fertility pathways. Nature Cell Biology 8(Suppl 10), s41-s49.
| Crossref | Google Scholar |
Matzuk MM, Lamb DJ (2008) The biology of infertility: research advances and clinical challenges. Nature Medicine 14(11), 1197-1213.
| Crossref | Google Scholar |
Miciński P, Pawlicki K, Wielgus E, Bochenek M, Tworkowska I (2009) The sperm chromatin structure assay (SCSA) as prognostic factor in IVF/ICSI program. Reproductive Biology 9(1), 65-70.
| Crossref | Google Scholar |
Mund T, Gewies A, Schoenfeld N, Bauer MKA, Grimm S (2003) Spike, a novel BH3-only protein, regulates apoptosis at the endoplasmic reticulum. The FASEB Journal 17(6), 696-698.
| Crossref | Google Scholar |
Nayernia K, Adham IM, Shamsadin R, Müller C, Sancken U, Engel W (2002) Proacrosin-deficient mice and zona pellucida modifications in an experimental model of multifactorial infertility. Molecular Human Reproduction 8(5), 434-440.
| Crossref | Google Scholar |
Nayernia K, Drabent B, Adham IM, Möschner M, Wolf S, Meinhardt A, Engel W (2003) Male mice lacking three germ cell expressed genes are fertile. Biology of Reproduction 69(6), 1973-1978.
| Crossref | Google Scholar |
Ni W, Liu K, Hou G, Pan C, Wu S, Zheng J, Cao J, Chen Q, Huang X (2019) Diurnal variation in sperm DNA fragmentation: analysis of 11,382 semen samples from two populations and in vivo animal experiments. Chronobiology International 36(11), 1455-1463.
| Crossref | Google Scholar |
Nozawa K, Fritzler MJ, Takasaki Y, Wood MR, Chan EKL (2009) Co-clustering of Golgi complex and other cytoplasmic organelles to crescentic region of half-moon nuclei during apoptosis. Cell Biology International 33(2), 148-157.
| Crossref | Google Scholar |
Oleszczuk K, Giwercman A, Bungum M (2016) Sperm chromatin structure assay in prediction of in vitro fertilization outcome. Andrology 4(2), 290-296.
| Crossref | Google Scholar |
Ozaki T, Hanaoka E, Naka M, Nakagawara A, Sakiyama S (1999) Cloning and characterization of rat BAT3 cDNA. DNA and Cell Biology 18(6), 503-512.
| Crossref | Google Scholar |
Rosahl TW, Spillane D, Missler M, Herz J, Selig DK, Wolff JR, Hammer RE, Malenka RC, Südhof TC (1995) Essential functions of synapsins I and II in synaptic vesicle regulation. Nature 375(6531), 488-493.
| Crossref | Google Scholar |
Shukla KK, Mahdi AA, Rajender S (2012) Apoptosis, spermatogenesis and male infertility. Frontiers in Bioscience 4(2), 746-754.
| Crossref | Google Scholar |
Sinha Hikim AP, Wang C, Lue Y, Johnson L, Wang X-H, Swerdloff RS (1998) Spontaneous germ cell apoptosis in humans: evidence for ethnic differences in the susceptibility of germ cells to programmed cell death. The Journal of Clinical Endocrinology & Metabolism 83(1), 152-156.
| Crossref | Google Scholar |
Taylor JD, Baumgartner A, Schmid TE, Brinkworth MH (2019) Responses to genotoxicity in mouse testicular germ cells and epididymal spermatozoa are affected by increased age. Toxicology Letters 310, 1-6.
| Crossref | Google Scholar |
Tripathi R, Mishra DP, Shaha C (2009) Male germ cell development: turning on the apoptotic pathways. Journal of Reproductive Immunology 83(1–2), 31-35.
| Crossref | Google Scholar |
Wang R, Liew C-C (1994) The human BAT3 ortholog in rodents is predominantly and developmentally expressed in testis. Molecular and Cellular Biochemistry 136(1), 49-57.
| Crossref | Google Scholar |
Westerman R (2020) Biomarkers for demographic research: sperm counts and other male infertility biomarkers. Biodemography and Social Biology 65(1), 73-87.
| Crossref | Google Scholar |
Wu Y-H, Shih S-F, Lin J-Y (2004) Ricin triggers apoptotic morphological changes through caspase-3 cleavage of BAT3. Journal of Biological Chemistry 279(18), 19264-19275.
| Crossref | Google Scholar |
Yatsenko AN, Iwamori N, Iwamori T, Matzuk MM (2010) The power of mouse genetics to study spermatogenesis. Journal of Andrology 31(1), 34-44.
| Crossref | Google Scholar |


