Selection of reference genes for quantitative real-time polymerase chain reaction in porcine embryos
Won-Jae Lee A B , Si-Jung Jang A , Seung-Chan Lee A , Ji-Sung Park A , Ryoung-Hoon Jeon A , Raghavendra Baregundi Subbarao A , Dinesh Bharti A , Jeong-Kyu Shin C , Bong-Wook Park D and Gyu-Jin Rho A E FA Department of Theriogenology and Biotechnology, College of Veterinary Medicine, Gyeongsang National University, 501, Jinju-daero, Jinju 660-701, Republic of Korea.
B PWG Genetics Pvt. Ltd, 15 Tech Park Crescent, 638117, Singapore.
C Department of Obstetrics and Gynecology, School of Medicine and Institute of Health Science, Gyeongsang National University, Jinju 660-701, Republic of Korea.
D Department of Oral and Maxillofacial Surgery, Institute of Health Science, School of Medicine, Gyeongsang National University, Jinju 660-701, Republic of Korea.
E Research Institute of Life Sciences, Gyeongsang National University, 501, Jinju-daero, Jinju 660-701, Republic of Korea.
F Corresponding author. Email: jinrho@gnu.ac.kr
*These authors contributed equally to this work.
Reproduction, Fertility and Development 29(2) 357-367 https://doi.org/10.1071/RD14393
Submitted: 15 October 2014 Accepted: 14 July 2015 Published: 21 August 2015
Journal Compilation © CSIRO Publishing 2017 Open Access CC BY-NC-ND
Abstract
To study gene expression and to determine distinctive characteristics of embryos produced by different methods, normalisation of the gene(s) of interest against reference gene(s) has commonly been employed. Therefore, the present study aimed to assess which reference genes tend to express more stably in single porcine blastocysts produced in vivo (IVO) or by parthenogenetic activation (PA), in vitro fertilisation (IVF) and somatic cell nuclear transfer (SCNT) using different analysis programs, namely geNorm, Normfinder and Bestkeeper. Commonly used reference genes including 18S rRNA (18S), H2A histone family, member Z (H2A), hypoxanthine phosphoribosyltransferase1 (HPRT1), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ribosomal protein 4 (RPL4), peptidylprolyl isomerase A (PPIA), beta actin (ACTB), succinate dehydrogenase complex, subunit A (SDHA) and hydroxymethylbilane synthase (HMBS2) were analysed; most of them resulted in significantly (P < 0.05) different cycle threshold (CT) values in porcine embryos except for SDHA and H2A. In evaluation of stable reference genes across in vivo and in vitro porcine blastocysts, three kinds of programs showed slightly different results; however, there were similar patterns about the rankings of more or less stability overall. In conclusion, SDHA and H2A were determined as the most appropriate reference genes for reliable normalisation in order to find the comparative gene expression in porcine blastocysts produced by different methods, whereas 18S was regarded as a less-stable reference gene. The present study has evaluated the stability of commonly used reference genes for accurate normalisation in porcine embryos to obtain reliable results.
Additional keywords: single blastocyst, normalisation, qRT-PCR.
Introduction
Quantitative analysis of gene expression is an indispensable part of biology. The exploration of the change of gene expression is expected to provide understanding about the gene regulatory network (Nailis et al. 2006). The quantitative reverse transcription polymerase chain reaction (qRT-PCR) is a valuable and commonly used technique for quantitative analysis because of its sensitivity, specificity and reproducibility (Radonić et al. 2004; Davoren et al. 2008). The sensitivity of this technique allows working with a minimal amount of starting material from only a small number of cells (Fink et al. 1998), hence qRT-PCR has been widely used in embryo research (Goossens et al. 2005; Kuijk et al. 2007; Mamo et al. 2007; Llobat et al. 2012; Gu et al. 2014; Luchsinger et al. 2014).
Embryos can be generated not only by in vivo (IVO) development in the female uterus after natural mating or artificial insemination but also by in vitro culture in the incubator after embryo manipulation such as parthenogenetic activation (PA), in vitro fertilisation (IVF) and somatic cell nuclear transfer (SCNT). These embryos are important models for understanding embryology, reproduction, innate disease and for the generation of transgenic animals. Differences between embryos produced by different in vivo and in vitro methods have revealed their distinctive characteristics including developmental efficiency, total cell number and gene expression (Kumar et al. 2007, 2012; Mamo et al. 2007; Magnani and Cabot 2008; McElroy et al. 2008; du Puy et al. 2011). In particular, compared with others, SCNT embryos have revealed extensively different characteristics caused by the process of manipulation attributable to incomplete and abnormal epigenetic reprogramming (Somers et al. 2006; Beyhan et al. 2007; Suzuki et al. 2008; Zhao et al. 2010; Deshmukh et al. 2011; Kumar et al. 2013). Differences in gene expression were also caused by in vitro culture conditions (Niemann and Wrenzycki 2000; Lazzari et al. 2002; Rizos et al. 2002; Rinaudo and Schultz 2004; McElroy et al. 2008) and the methodological procedures used to generate the embryos (Zhou et al. 2008; Ross et al. 2010).
Although qRT-PCR is commonly employed for analysis of mRNA expression levels, there are several factors causing variation in overall transcriptional activity, such as amount of starting material, enzymatic efficiencies, diversities in protocols and instruments used (Bustin 2002). Furthermore, another distinct variable in embryo study that has to be considered is extremely low transcript mRNA yield from a single embryo for reliable quantification as starting material (Robert et al. 2002; Luchsinger et al. 2014). One way to minimise these variables for a well-conducted study in qRT-PCR assays is by normalisation of the gene of interest against reference genes (housekeeping genes), which should be expressed consistently and stably in all the samples regardless of experimental conditions and treatments (Bustin 2002; Vandesompele et al. 2002; Dheda et al. 2004; Huggett et al. 2005). The expression of reference genes plays a vital role in maintenance of cellular function and production, which are minimal essential necessities for normal physiology (Butte et al. 2001).
Gene expression studies rely on the use of reference genes, yet there has not been any attempt to identify the suitability of any reference gene under various experimental or culture conditions (Radonić et al. 2004; Huggett et al. 2005). The possible effects of such variation on commonly used reference genes such as beta actin (ACTB), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and 18S rRNA (18S) are, consequently, mostly unknown (Uddin et al. 2011). The use of these genes was evaluated in different studies and criticised due to observation of variation between treatment groups (Thellin et al. 1999; Tricarico et al. 2002; Radonić et al. 2004). Moreover, the use of inappropriate reference genes for normalisation could result in false conclusions and misleading interpretation of gene expression (Bas et al. 2004; Dheda et al. 2004; Haller et al. 2004). Evaluation for stability of reference genes, therefore, is required in each experimental condition to guarantee more reliable results of biological significance.
Several studies have focussed on evaluation of reference genes from pre-implantation embryos in various species including mouse (Mamo et al. 2007; Llobat et al. 2012; Gu et al. 2014), rabbit (Mamo et al. 2008), horse (Smits et al. 2009; Paris et al. 2011) and cattle (Goossens et al. 2005; Luchsinger et al. 2014). Although several studies of porcine reference genes have been investigated under various conditions such as different tissues, age and treatments (Nygard et al. 2007; Uddin et al. 2011; McCulloch et al. 2012), information on suitable reference genes from porcine embryos is available only from IVF embryos (Kuijk et al. 2007). However, evaluation of stable reference genes is a necessity for accurate analysis of porcine embryos, because the stability of reference genes can also vary depending on the method used to produce the embryos. Therefore, the aim of the present study was to assess the stable expression of nine widely used reference genes in several kinds of porcine pre-implantation embryos that were produced by IVO, PA, IVF and SCNT to select the most-stable ones by analysis of different algorithms including geNorm, Normfinder and Bestkeeper.
Material and methods
Chemicals and media
All chemicals and media, unless stated otherwise, were purchased from Sigma-Aldrich Chemical Inc. (St. Louis, MO, USA) and Gibco (Grand Island, NY, USA).
Preparation of donor cells for SCNT
All animal samples were collected and handled following the approval of the research ethical committee of the animal centre for biomedical experimentation, Gyeongsang National University, under set guidelines (GNU-140305-P0016). Porcine ear skin tissue from ~1-year-old pigs was collected, minced and explanted onto dishes. The isolated cells were cultured in Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin (pen-strep; 10 000 IU and 10 000 μg mL–1, respectively) at 38.5°C in a humidified incubator at 5% CO2 in air. Cells were further passaged to obtain a proliferating homogenous mixture under aseptic conditions. Fibroblasts at Passage 3 were used as SCNT donor cells.
In vitro maturation of oocytes
Cumulus–oocyte complexes (COCs) were collected from prepubertal pig ovaries obtained from a local slaughterhouse and in vitro maturation (IVM) performed as previously described (Kumar et al. 2007, 2013; Lee et al. 2014). Briefly, COCs were aspirated from ovarian follicles of 3–5 mm in diameter using a 19-guage needle attached to a 10-mL syringe. Sets of 100 COCs with even cytoplasm and multilayered cumulus cells were matured in 500 μL tissue culture medium (TCM)-199 medium supplemented with 5% FBS, 1% pen-strep, 0.57 mM cysteine, 10 ng mL–1 epidermal growth factor (EGF), 2.5 mM sodium pyruvate, 1 mM L-glutamine, 0.5 μg mL–1 luteinising hormone (LH) and 0.5 μg mL–1 follicle-stimulating hormone (FSH) for 22 h and then further cultured for an additional 20 h in the same medium without LH and FSH at 38.5°C in a humidified atmosphere of 5% CO2 in air. After 42 h of IVM, cumulus cells were denuded from oocytes by pipetting in Dulbecco’s phosphate-buffered saline (DPBS) with 0.1% hyaluronidase for 1 min. Oocytes having the first polar body (PB1), even cytoplasm and intact membranes were selected for the production of PA, IVF and SCNT embryos.
Preparation of pre-implantation embryos
For the production of in vivo and in vitro embryos, IVO and embryo manipulations for PA, IVF and SCNT were performed as previously described (Kumar et al. 2007, 2012, 2013; Lee et al. 2014). For preparation of IVO embryos, oestrous sows were fertilised by artificial insemination twice with a 12-h interval between inseminations. On Day 7 after the first insemination, the sows were shipped to a local slaughterhouse and withdrawn genital tracts were transported to the laboratory at optimal temperature conditions of 35–39°C within 1 h. The uterine horns and uteri were flushed with DPBS supplemented with 0.1% bovine serum albumin (BSA) for embryo recovery of IVO blastocysts (Kumar et al. 2007).
For production of PA embryos, MII oocytes were pulsed twice with 2.0 kV cm–1 DC for 30 μs using a BTX Electro Square Porator and a BTX Electro chamber (ECM 830; BTX Inc., San Diego, CA, USA) filled with 0.28 M mannitol solution containing 0.5 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), 0.1 mM MgSO4, 0.05 mM CaCl2 and 0.01% BSA. This was followed by culture in porcine zygote medium 5 (PZM5) containing 7.5 μg mL–1 cytochalasin B (CCB) for 3 h at 38.5°C in a humidified atmosphere of 5% CO2 in air to activate PA embryos (Kumar et al. 2012).
For SCNT embryo production, MII oocytes containing polar body1 (PB1) with small amount of cytoplasm was enucleated in HEPES-buffered TCM-199 supplemented with 7.5 µg mL–1 CCB, 0.3% BSA and 12 mM sorbitol and replaced with fibroblast. The couplets were fused and activated using the same activation parameters as were used for the PA embryos.
To generate IVF embryos, sets of 20 MII oocytes were fertilised using Percoll (Pharmacia, Uppsala, Sweden) density-gradient-treated spermatozoa (5 × 103 spermatozoa mL–1 final concentration) in 50-μL droplets of modified Tris-buffered medium (mTBM) consisting of 113.1 mM NaCl, 3.0 mM KCl, 7.5 mM CaCl2·2H2O, 20.0 mM Tris crystallised free base, 11.0 mM glucose and 5.0 mM sodium pyruvate, supplemented with 2 mM caffeine and 4 mg mL–1 BSA for 5 h at 38.5°C in a humidified atmosphere of 5% CO2 in air (Kumar et al. 2013). All embryos produced in vitro, including IVF, PA and SCNT embryos, were then cultured in sets of 30 embryos per 30-μL droplet of PZM5 medium supplemented with 3 mg mL–1 BSA, 20 μg mL–1 Eagle’s amino acids in basal medium (BME), 10 μg mL–1 non-essential amino acids (NEAA) for 4 days at 38.5°C in a humidified atmosphere of 5% CO2 in air and then further cultured in the same medium supplemented with 10% FBS for an additional 2 days, in the same way as previously described (Kumar et al. 2012; Lee et al. 2014). Day-7 blastocysts were frozen immediately by liquid nitrogen with small volume of DPBS and stored at –80°C until RNA extraction.
RNA extraction, cDNA synthesis and qRT-PCR
Blastocysts produced in vivo (n = 11) and in vitro (each n = 20) were employed for all molecular work. Total RNA was extracted from single embryos using the RNeasy Micro Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions and residual genomic DNA was removed by using RNase Free DNase (Qiagen) treatment for 15 min. Due to the low amounts of total RNA extracted from a single blastocyst, quantification of total mRNA yield could not be measured by UV spectrophotometer. First-strand cDNA was synthesised from the 12 µL of total RNA extracted using 8 µL of mixture with Sensiscript Reverse Transcription Kit (Qiagen), 10 units of RNaseOUT Recombinant Ribonuclease Inhibitor (Invitrogen, Grand Island, NY, USA) and 1 µM oligo dT primer (Invitrogen) at 37°C for 1 h. Quantitative RT-PCR was performed using a Rotor Gene Q qRT-PCR machine (Qiagen) with Rotor-Gene 2X SYBR Green mix (Qiagen) including 2 µL cDNA per reaction and 0.5 µM forward and reverse primers. All experiments were executed one reaction for each single embryo because of limited amounts of cDNA. Quantitative RT-PCR program settings were comprised of pre-denaturation at 95°C for 10 min, 45 PCR cycles at 95°C for 10 s, 60°C for 6 s and 72°C for 4 s, melting curve from 60°C to 95°C by 1°C s–1 and cooling at 40°C for 30 s with minor modifications in the manufacturer’s qRT-PCR program protocol. Rotor-Gene Q Series Software (Qiagen, Hilden, Germany) was used for analysis of amplification curves, melting curves and cycle threshold values (CT values). All amplicons from qRT-PCR were verified by electrophoresis using 1% agarose gel with 0.1 mg mL–1 ethidium bromide for checking non-specific amplification. Images were analysed using zoom browser EX5.7 software (Canon, Tokyo, Japan).
Candidate reference genes and primer efficiency
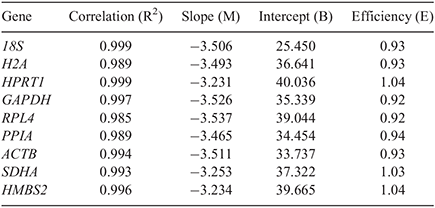
Nine commonly used reference genes were selected based on different intracellular functional activity. Candidates of reference genes were selected on the basis of previously described articles examining the stability of porcine reference genes under several experimental conditions (Nygard et al. 2007; Uddin et al. 2011; McCulloch et al. 2012; Lee et al. 2015) and evaluated according to cases where they were applied as embryo reference genes (Goossens et al. 2005; Bettegowda et al. 2006; Mamo et al. 2007; Smits et al. 2009; Gu et al. 2014; Luchsinger et al. 2014). Consequently, nine widely used reference genes including 18S, H2A histone family, member Z (H2A), hypoxanthine phosphoribosyltransferase1 (HPRT1), GAPDH, ribosomal protein 4 (RPL4), peptidylprolyl isomerase A (PPIA), ACTB, succinate dehydrogenase complex, subunit A (SDHA) and hydroxymethylbilane synthase (HMBS2), were selected and sequences of all reference genes were taken from Nygard et al. (2007) or Lee et al. (2015). Their annealing temperatures were confirmed as 60°C by a primer searching program (Primer 3 Plus). The complete information is presented in Table 1. To check PCR efficiencies of reference genes for embryos, a standard curve of each primer of reference gene was generated from the CT values by a four-fold serial dilution of cDNA from PA blastocysts. Standard curve parameters including slope (M), intercept (B), PCR efficiencies (E) and correlation (R2) calculated by Rotor-Gene Q Series Software (Qiagen) are presented in Table 2.

|
|
Evaluation of stable reference gene expression
CT values of each reference gene were log-transformed and analysed by three kinds of widely used algorithmic tools – geNorm, Normfinder and Bestkeeper – to assess the stability of the selected reference genes. By using these methods, we ranked the stability of the reference genes and confirmed the most-stable reference genes in porcine blastocysts produced by different methods. The geNorm program is based on a Visual Basic application (VBA) tool for Microsoft Excel and provides a measure of gene expression stability (M value). A gene presenting the highest M value is excluded from tested genes and the new M values from the remaining genes are calculated continuously until the last two genes presenting the lowest M value are left. The gene with the highest M value corresponds to the least-stable gene expression, whereas the two genes with the lowest M value determine the most-stable ones. In geNorm, the geometric average of expression of the most-stable reference genes generates normalisation factors (NF) and pairwise variation for investigating the optimal number of reference genes was calculated and presented Vn/n+1 by stepwise inclusion between two sequential NFs (Vandesompele et al. 2002). Normfinder is also a VBA program and is based on an ANOVA-based model to estimate intra- and inter-group variation to deduce the single most-stable reference gene. The values of stability and standard errors are calculated by the transcriptional variation of the reference genes. This program exhibits the most-stable gene, presenting the low stability values and best combination of two genes (Andersen et al. 2004). Bestkeeper, an Excel-based tool, analyses variability in expression of reference genes by calculating comparative analysis based on numerous pairwise correlations of all reference genes against each other (Pfaffl et al. 2004).
The application of normalisation to reference genes with different stability
The effect of most- or least-stable reference genes was examined by applying different reference genes against relative gene expression for the gene of interest. OCT4 (octamer-binding transcription factor 4) expression in each of 20 blastocysts from PA and IVF embryos was normalised against several reference genes to evaluate most-, moderately and least-stable reference genes by using three different types of programs. The primer sequence of OCT4 has been presented in Table 1.
Statistical analysis
The CT values of reference genes from embryos produced by different in vivo and in vitro methods were presented as mean ± s.e.m. and analysed using one-way ANOVA with Tukey’s post hoc test (PASW Statistics 18; SPSS Inc., IBM SPSS, Armonk, NY, USA) and Student’s t-test was applied to assess the application of normalisation. Significant differences were considered at P < 0.05. Moreover, correlation of the normalisation factor (NFn) between the optimal number of reference genes (NFopt) from pairwise variation of geNorm and the three most-stable reference genes (NF3) was analysed by Pearson’s correlation in PASW Statistics 18, in accordance with a previous article (Lee et al. 2015).
Results
Examination of primer specificity and amplicon size
For the nine reference genes used in this study, melting curve analysis was performed at the end of the PCR reactions. All reference genes showed specifically high amplifications of a single peak without non-specific dimer bindings in melting curve analysis. In addition, all amplicons were confirmed by expected product sizes presenting a single band in all samples by agarose gel electrophoresis. Agarose gel images are displayed at the bottom of each melting curve graph (Fig. 1).
To check primer efficiency, the standard curves were generated and measured by using four-fold serial dilutions of cDNA preparations from a single PA embryo. The assays for the candidate reference genes were displayed in a linear form and correlation coefficient (R2) indicated from 0.985 to 0.999. PCR efficiencies were found to be between 0.92 and 1.04 (Table 2). Based on the correlation coefficient and PCR efficiency results , the assays can be regarded as acceptable and valid for the quantification of transcripts and further experiments.
Average of CT values of reference genes in blastocysts produced by different methods
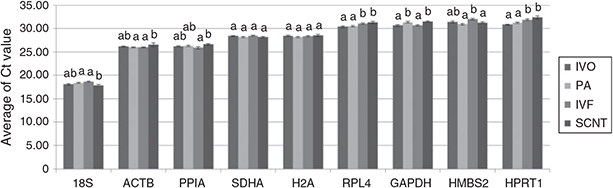
To investigate the RNA transcription levels across blastocysts produced by different in vitro and in vivo methods, the CT values from each reference gene were compared and presented in Fig. 2. Cycle threshold values of H2A and SDHA did not differ across different classes of blastocysts. However, the values of the other seven reference genes were significantly (P < 0.05) different. In particular, of the traditionally used reference genes such as ACTB, GAPDH and 18S, ACTB showed significantly (P < 0.05) higher CT values in IVO than IVF and SCNT embryos, while 18S showed significantly (P < 0.05) lower CT values under the same comparison. Further, significantly (P < 0.05) higher CT values of GAPDH were revealed in IVO and IVF than PA and SCNT embryos.
|
Examination of stable reference genes
geNorm analysis
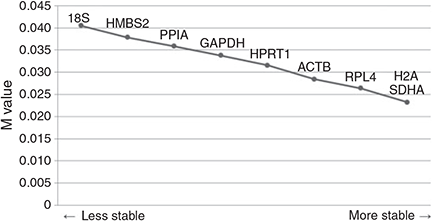
Using this program, we ranked the nine candidate reference genes following M values in blastocysts produced by different in vivo and in vitro methods according to their expression stability (Fig. 3). The gradual stepwise exclusion of the least-stable genes revealed that H2A, SDHA and RPL4 were the three most-stable genes; however, expression of 18S showed the least stability and GAPDH revealed moderately stable expression among the nine candidate reference genes. Moreover, pairwise variations showed that the use of seven reference genes (V7/8) was regarded as the optimal number of reference genes (Fig. 4a). However, because it is hard to apply seven reference genes to study embryo gene expression, correlation of normalisation factors between the optimal number (NF7) and three of the most-stable reference genes (NF3) was analysed to remove excessive usage of reference genes (Fig. 4b). High correlation (r = 0.905, P < 0.01) was determined between NF7 and NF3 by Pearson’s correlation.
|
Normfinder analysis
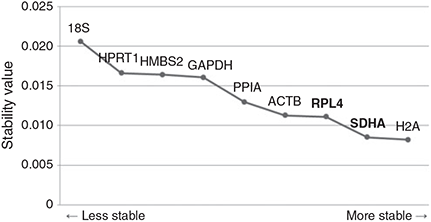
In Normfinder analysis, the most-stable reference gene in embryos produced by different in vivo and in vitro methods was determined as H2A and the best combination of two genes from the nine candidate reference genes was revealed as being RPL4 and SDHA (Fig. 5). This analysis displayed similar results to the geNorm analysis on 18S and GAPDH, which presented as the least-stable reference gene and having moderately stable expression, respectively.
|
Bestkeeper analysis
The result of reference gene evaluation by Bestkeeper is presented in Table 3. The most-stable reference gene can be identified as presenting the lowest coefficient of variance (CV ± s.e.m.). Several reports have mentioned that presenting a s.e.m. higher than 1 should be considered unacceptable (Pfaffl et al. 2004). In this analysis, all of the nine candidate genes were available to be accepted as reliable reference genes because the estimation of the s.e.m. (± CT) of the CV (% CT) values for all reference genes was lower than cut-off. SDHA was determined to be the most-stable reference gene in embryos produced by different in vivo and in vitro methods, whereas 18S, GAPDH and ACTB showed moderately stable expression.
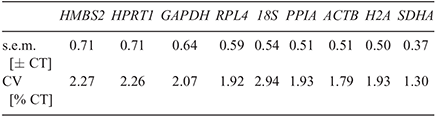
|
The application for normalisation of evaluated reference genes
Taken together, both SDHA and H2A were evaluated as the most-stable reference genes, whereas GAPDH and 18S showed moderate or the least stability, respectively, as analysed by three kinds of programs. Therefore, their effect as reference genes for normalisation purpose was examined and the levels of relative quantification are presented in Fig. 6. OCT4 expression in PA and IVF blastocysts was normalised against SDHA and H2A as the most-stable reference genes and those results showed significantly (P < 0.05) upregulated OCT4 expression in IVF blastocysts compared to PA blastocysts. In contrast, downregulation of OCT4 expression with no significant differences were observed when GAPDH or 18S were applied in normalisation, respectively.
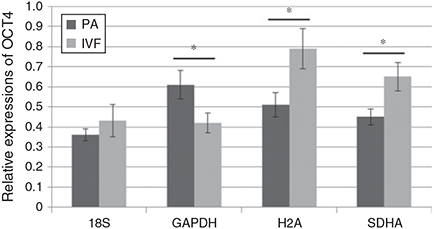
|
Discussion
Given the dynamic changes in pre-implantation embryos, it is hard to figure out embryonic development clearly because of limited amounts of starting material and due to little information about gene expression and species differences (Bettegowda et al. 2006; Kuijk et al. 2007; Mamo et al. 2008; Luchsinger et al. 2014). Prior to the fertilisation of embryos, the first meiotic prophase division of oocytes characterises transcription and translation of numerous genes, resulting in storage of mRNA and proteins for preparing early embryonic development. In contrast, transcription is halted and translation of mRNA is reduced at the second meiotic metaphase division of oocytes (Arya et al. 2005). Further, pre-implantation embryos attain distinct shapes due to morphological changes, including cleavage division, compact morula and blastocyst with activation of the embryonic genome after the fertilisation. During these processes, differential expression of genes related to embryonic development is regulated in a stage- and time-dependent manner (Niemann and Wrenzycki 2000).
Likewise, different methods of embryo production also result in drastic changes including gene expression, developmental rate, total cell number and apoptosis (Kumar et al. 2007; Magnani and Cabot 2008; McElroy et al. 2008; du Puy et al. 2011). Compared with embryos produced in vitro, in vivo embryos show increased patterns of pluripotent gene expression and total cell number at the blastocyst stage, lower levels of chromosomal abnormalities, decreased trends of imprinting and apoptosis (Kumar et al. 2007). Thus, embryos produced by different methods show a similar pattern, but not identical expression of pluripotent genes. The highest abundance of Nanog expression from in vivo embryos was observed at the four-cell and blastocyst stage (Magnani and Cabot 2008). Furthermore, an investigation focussed on the inefficiency of porcine SCNT embryos has revealed that abnormal patterns of developmental efficiency and gene expression were caused by incomplete reprogramming of the donor cell genome as well as aberrant epigenetic mechanisms related to DNA methylation and histone de-acetylation (Kumar et al. 2007, 2013; Zhao et al. 2010). It has been reported that reference genes in early embryonic developmental stages were differentially expressed and hence the accurate assessment of gene expression could not be performed (Robert et al. 2002; Mamo et al. 2007, 2008; Gu et al. 2014). Gene expression of different embryos reflects their own characteristics, therefore, investigations should be focussed on increasing the accuracy of reliable reference genes for appropriate comparative gene expression studies.
The validation of candidate reference genes is critical for accurate analysis of embryo gene expression to prevent false conclusions from invalidated reference genes (Bas et al. 2004; Dheda et al. 2004; Haller et al. 2004). Differences in relative quantification of IL-4 mRNA expression after normalisation against different reference genes, GAPDH and human acidic ribosomal protein (HuPO), were present in blood sample of patients with pulmonary tuberculosis and in healthy individuals. In case of normalisation against HuPO, there was a significant (P < 0.0001) difference between groups but it was not the same when data was normalised against GAPDH (Dheda et al. 2004). In agreement with these results, Fig. 6 showed that evaluation of reference genes was a highly essential step for a particular experiment. Relative quantification of OCT4 after normalisation against SDHA and H2A (evaluated as stable reference genes) presented significant (P < 0.05) upregulation of OCT4 in IVF blastocysts; however, GAPDH and 18S were assessed as less-stable genes and showed different results from those of SDHA and H2A. Therefore, these differences by normalisation against non-evaluated or less-stable reference genes can lead to false conclusions.
Several previous studies have provided evidence that transcription levels of reference genes from embryos did not express constantly in different developmental stages and under different production methods (Mamo et al. 2007, 2008; Smits et al. 2009; Gu et al. 2014). In the present study, the differences in CT values of reference gene expression were observed in blastocysts produced by different in vivo and in vitro methods (Fig. 2). The CT values were inversely proportional to the amount of template present in the PCR amplification (Walker 2002). The majority of reference genes showed CT values in the range of 26 to 31 but 18S showed a lower value of 18, which means more-abundant transcripts in any kind of blastocyst. However, most of the reference genes presented significant differences between different types of embryo, except for expression of H2A and SDHA, and there were no constant patterns that can be explained for all blastocysts produced by different in vivo and in vitro methods. Mouse and rabbit embryos also showed drastic changes at blastocyst stages presenting higher transcript copy numbers in the case of in vivo embryos compared with in vitro ones, and the variations were different in most of the reference genes (Mamo et al. 2007).
There are clear differences in morphological and molecular biological diversities within different mammalian embryos. Accordingly, even if some reference genes could be evaluated as having stable expression in certain embryos, there have been different results depending upon different species. Several studies have been performed to answer this issue of validating stable reference genes in embryos of different species, including mouse (Jeong et al. 2005; Mamo et al. 2007; Llobat et al. 2012; Gu et al. 2014), cow (Robert et al. 2002; Goossens et al. 2005; Luchsinger et al. 2014), rabbit (Mamo et al. 2008), horse (Smits et al. 2009; Paris et al. 2011) and chicken (Yang et al. 2013). PPIA, H2A and HPRT1 were determined as the most-stable reference genes in mouse embryos produced in vivo and in vitro by geNorm analysis (Mamo et al. 2007; Gu et al. 2014). In addition, the H2A gene was also suggested as a stable reference gene in single mouse eggs and pre-implantation embryos with the use of a simple freezing–thawing method for RNA collection and economic SYBR-green dye system for quantitative RT-PCR (Jeong et al. 2005). In rabbit pre-implantation embryos, the use of H2A, YWHAZ (tyrosine 3-monooxygenase/ Tryptophan 5-monooxygenase activation protein, zeta) and HPRT1 was preferable for normalisation (Mamo et al. 2008). While working with chicken embryo fibroblasts infected with avian leucosis virus, commonly used ACTB and GAPDH were found to be unsuitable reference genes with varied expression as compared with the most-stably expressed RPL30 (ribosomal protein L30) and SDHA with the lowest M value by geNorm analysis (Yang et al. 2013). In addition, SDHA, PGK1 (phosphoglycerate kinase 1) and TBP (TATA box binding protein) have been shown as the most suitable genes among the analysis of 14 internal control genes in case of embryonic (ES), trophectoderm (TS) and extra-embryonic endoderm (XEN; Veazey and Golding 2011). These results support our findings that H2A and SDHA can be a better choice for porcine embryology to get the most-stable expression in quantitative real-time PCR for normalisation of interest genes (Fig. 3 and 5; Table 3).
Earlier, in one porcine embryo study, only IVF embryos were tested for selection of reference genes by using a single-analysis program. The study determined that GAPDH was the most-stable reference gene in porcine IVF embryos. Furthermore, H2A and 18S were revealed as moderately stably expressed genes (Kuijk et al. 2007). In the last two previous articles about bovine embryos, GAPDH was investigated as a stable reference gene in bovine embryos regardless of not only different production procedures such as IVF, SCNT and intra-cytoplasmic sperm injection (ICSI) but also different pre-implantation stages (Goossens et al. 2005; Luchsinger et al. 2014); our results are not in agreement with those findings. We concluded that H2A expression is the most stable, whereas 18S or GAPDH were found to be expressed as least- or moderately stable genes, respectively. These differences in our results compared with other reported studies may be due to different species, composition of candidate reference genes (Table 1) and different experimental groups including IVO, PA, IVF and SCNT embryos.
The expression of reference genes in embryos produced by different in vivo and in vitro methods was compared using three different commonly used analysis programs for investigating stable expression. geNorm, Normfinder and Bestkeeper were programmed by different algorithms and analytical principles (Vandesompele et al. 2002; Andersen et al. 2004; Pfaffl et al. 2004). In agreement with other studies resulting in an optimal number of reference genes from geNorm analysis (Uddin et al. 2011; Lee et al. 2015), it is not easy to use many reference genes for normalisation in embryo studies because of insufficient starting material and unnecessarily excessive experiments. For this reason, the correlation of normalisation factors by Pearson’s correlation was analysed between the optimal number (NF7) from pairwise variation and three of the most-stable reference genes (NF3; Fig. 4). The analysis showed high correlation between NF7 and NF3 (r = 0.905) and this result demonstrated that three of the most-stable reference genes by geNorm analysis should be satisfactory to use for normalisation. Although rankings from analysis by the three programs presented slightly different results, there were similar patterns about the stability orders of more- or less-stable reference genes by each program despite the variations of embryo sources. SDHA, H2A and RPL4 were found to be three of the most-stable reference genes by geNorm and Normfinder analysis, whereas SDHA, H2A and ACTB presented stable gene expression by Bestkeeper analysis (Fig. 3 and 5; Table 3). Taken together, all analysis programs comprehensively concluded that SDHA and H2A were expressed stably across porcine blastocysts produced by different in vivo and in vitro methods. On the other hand, 18S and GAPDH were determined as the least-stable or moderately stable reference genes, respectively. Our results using various analysis programs are in agreement with other published studies. To verify appropriate reference genes to investigate both diploid and polyploid mouse embryos, the stability of twelve reference genes was compared and UBC (ubiquitin C), PPIA and PGK1 were determined to be the most-stable reference genes by four different programs (Gu et al. 2014). By using same sets of analysis programs as the present study, RPL4, PPIA and YWHAZ were determined as the most appropriate combination of reference genes for normalisation of different porcine tissues at different ages (Uddin et al. 2011) and the use of PPIA, ACTB, GAPDH and SDHA was recommended as appropriate reference genes for the study of porcine articular cartilage (McCulloch et al. 2012).
In conclusion, genomic DNA-free total RNA extracted from single blastocysts produced by different in vivo and in vitro methods was evaluated for stable expression of reference genes for reliable normalisation by qRT-PCR analysis. Three kinds of analysis programs displayed similar results that the SDHA and H2A genes were suggested as the appropriate reference genes across various porcine blastocysts and 18S was revealed as the least-stable reference gene. The present study suggests that appropriate evaluation of reference genes for normalisation guarantees more-reliable results of gene expression studies in porcine embryos.
Acknowledgement
This research was supported by a grant from the Next-Generation BioGreen 21 Program (No. PJ009021), Rural Development Administration, Republic of Korea.
References
Andersen, C. L., Jensen, J. L., and Ørntoft, T. F. (2004). Normalisation of real-time quantitative reverse-transcription PCR data: a model-based variance estimation approach to identify genes suited for normalisation, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250.| Normalisation of real-time quantitative reverse-transcription PCR data: a model-based variance estimation approach to identify genes suited for normalisation, applied to bladder and colon cancer data sets.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXmtF2mtbg%3D&md5=21f0e7dbbe80b169de8977b03b8c2a3bCAS | 15289330PubMed |
Arya, M., Shergill, I. S., Williamson, M., Gommersall, L., Arya, N., and Patel, H. R. (2005). Basic principles of real-time quantitative PCR. Expert Rev. Mol. Diagn. 5, 209–219.
| Basic principles of real-time quantitative PCR.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjt1Kmu7c%3D&md5=04eebd9ff636567650e73f340f2d29d6CAS | 15833050PubMed |
Bas, A., Forsberg, G., Hammarström, S., and Hammarström, M. L. (2004). Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalisation in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand. J. Immunol. 59, 566–573.
| Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalisation in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXlsF2qsb0%3D&md5=614b5ac9a16565be83d12bc0ac990f83CAS | 15182252PubMed |
Bettegowda, A., Patel, O. V., Ireland, J. J., and Smith, G. W. (2006). Quantitative analysis of messenger RNA abundance for ribosomal protein L-15, cyclophilin-A, phosphoglycerokinase, beta-glucuronidase, glyceraldehyde 3-phosphate dehydrogenase, beta-actin and histone H2A during bovine oocyte maturation and early embryogenesis in vitro. Mol. Reprod. Dev. 73, 267–278.
| Quantitative analysis of messenger RNA abundance for ribosomal protein L-15, cyclophilin-A, phosphoglycerokinase, beta-glucuronidase, glyceraldehyde 3-phosphate dehydrogenase, beta-actin and histone H2A during bovine oocyte maturation and early embryogenesis in vitro.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XhsVKht78%3D&md5=dd6d97f2ba2aeeedeb76fc5db1a6a994CAS | 16261607PubMed |
Beyhan, Z., Forsberg, E. J., Eilertsen, K. J., Kent-First, M., and First, N. L. (2007). Gene expression in bovine nuclear transfer embryos in relation to donor cell efficiency in producing live offspring. Mol. Reprod. Dev. 74, 18–27.
| Gene expression in bovine nuclear transfer embryos in relation to donor cell efficiency in producing live offspring.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xht1yjsb%2FJ&md5=c49b5641b5338021310a0ffb2247c384CAS | 16941691PubMed |
Bustin, S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29, 23–39.
| Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XmvFOhu7s%3D&md5=18aa0a1d85b90082336be1eaf096aa50CAS | 12200227PubMed |
Butte, A. J., Dzau, V. J., and Glueck, S. B. (2001). Further defining housekeeping, or “maintenance” genes. Focus on “a compendium of gene expression in normal human tissues”. Physiol. Genomics 7, 95–96.
| 1:CAS:528:DC%2BD38XhtFagsLo%3D&md5=892c395e979a77bfc8964fa7bc8417b2CAS | 11773595PubMed |
Davoren, P. A., McNeill, R. E., Lowery, A. J., Kerin, M. J., and Miller, N. (2008). Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer. BMC Mol. Biol. 9, 76.
| Identification of suitable endogenous control genes for microRNA gene expression analysis in human breast cancer.Crossref | GoogleScholarGoogle Scholar | 18718003PubMed |
Deshmukh, R. S., Østrup, O., Østrup, E., Vejlsted, M., Niemann, H., Lucas-Hahn, A., Petersen, B., Li, J., Callesen, H., and Hyttel, P. (2011). DNA methylation in porcine pre-implantation embryos developed in vivo and produced by in vitro fertilisation, parthenogenetic activation and somatic cell nuclear transfer. Epigenetics 6, 177–187.
| DNA methylation in porcine pre-implantation embryos developed in vivo and produced by in vitro fertilisation, parthenogenetic activation and somatic cell nuclear transfer.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjs12ksLY%3D&md5=7f6332ef229ed49e4350ab26c9fc021dCAS | 20935454PubMed |
Dheda, K., Huggett, J. F., Bustin, S. A., Johnson, M. A., Rook, G., and Zumla, A. (2004). Validation of housekeeping genes for normalising RNA expression in real-time PCR. Biotechniques 37, 112–114, 116, 118–119.
| 1:CAS:528:DC%2BD2cXlvVagtrg%3D&md5=00a8dddbec28456169416f1210469a2fCAS | 15283208PubMed |
du Puy, L., Lopes, S. M., Haagsman, H. P., and Roelen, B. A. (2011). Analysis of co-expression of OCT4, NANOG and SOX2 in pluripotent cells of the porcine embryo, in vivo and in vitro. Theriogenology 75, 513–526.
| Analysis of co-expression of OCT4, NANOG and SOX2 in pluripotent cells of the porcine embryo, in vivo and in vitro.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXjs1Kmsw%3D%3D&md5=a0939d8f4bf2b8afc0994b1bde2837f4CAS | 21074831PubMed |
Fink, L., Seeger, W., Ermert, L., Hanze, J., Stahl, U., Grimminger, F., Kummer, W., and Bohle, R. M. (1998). Real-time quantitative RT-PCR after laser-assisted cell picking. Nat. Med. 4, 1329–1333.
| Real-time quantitative RT-PCR after laser-assisted cell picking.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXntlaru7s%3D&md5=b3c00dd6c9658f5f4aa0532b144351d7CAS | 9809560PubMed |
Goossens, K., Van Poucke, M., Van Soom, A., Vandesompele, J., Van Zeveren, A., and Peelman, L. J. (2005). Selection of reference genes for quantitative real-time PCR in bovine pre-implantation embryos. BMC Dev. Biol. 5, 27.
| Selection of reference genes for quantitative real-time PCR in bovine pre-implantation embryos.Crossref | GoogleScholarGoogle Scholar | 16324220PubMed |
Gu, Y., Shen, X., Zhou, D., Wang, Z., Zhang, N., Shan, Z., Jin, L., and Lei, L. (2014). Selection and expression profiles of reference genes in mouse pre-implantation embryos of different ploidies at various developmental stages. PLoS One 9, e98956.
| Selection and expression profiles of reference genes in mouse pre-implantation embryos of different ploidies at various developmental stages.Crossref | GoogleScholarGoogle Scholar | 24927500PubMed |
Haller, F., Kulle, B., Schwager, S., Gunawan, B., von Heydebreck, A., Sültmann, H., and Füzesi, L. (2004). Equivalence test in quantitative reverse transcription polymerase chain reaction: confirmation of reference genes suitable for normalisation. Anal. Biochem. 335, 1–9.
| Equivalence test in quantitative reverse transcription polymerase chain reaction: confirmation of reference genes suitable for normalisation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXpt1Sqt74%3D&md5=327daf08decbc0e47dee3cb630d2987cCAS | 15519565PubMed |
Huggett, J., Dheda, K., Bustin, S., and Zumla, A. (2005). Real-time RT-PCR normalisation: strategies and considerations. Genes Immun. 6, 279–284.
| Real-time RT-PCR normalisation: strategies and considerations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXks1Gmtrs%3D&md5=f4bb44b857e6f231b18f69ca22ef5253CAS | 15815687PubMed |
Jeong, Y. J., Choi, H. W., Shin, H. S., Cui, X. S., Kim, N. H., Gerton, G. L., and Jun, J. H. (2005). Optimisation of real time RT-PCR methods for the analysis of gene expression in mouse eggs and pre-implantation embryos. Mol. Reprod. Dev. 71, 284–289.
| Optimisation of real time RT-PCR methods for the analysis of gene expression in mouse eggs and pre-implantation embryos.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXltFyiu7c%3D&md5=16032da79708595290d9f5105db0a92cCAS | 15806558PubMed |
Kuijk, E. W., du Puy, L., van Tol, H. T., Haagsman, H. P., Colenbrander, B., and Roelen, B. A. (2007). Validation of reference genes for quantitative RT-PCR studies in porcine oocytes and pre-implantation embryos. BMC Dev. Biol. 7, 58.
| Validation of reference genes for quantitative RT-PCR studies in porcine oocytes and pre-implantation embryos.Crossref | GoogleScholarGoogle Scholar | 17540017PubMed |
Kumar, B. M., Jin, H. F., Kim, J. G., Ock, S. A., Hong, Y., Balasubramanian, S., Choe, S. Y., and Rho, G. J. (2007). Differential gene expression patterns in porcine nuclear transfer embryos reconstructed with fetal fibroblasts and mesenchymal stem cells. Dev. Dyn. 236, 435–446.
| Differential gene expression patterns in porcine nuclear transfer embryos reconstructed with fetal fibroblasts and mesenchymal stem cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXisFamt70%3D&md5=77b9122e6729998163d970ead529659aCAS | 17191234PubMed |
Kumar, B. M., Maeng, G. H., Jeon, R. H., Lee, Y. M., Lee, W. J., Jeon, B. G., Ock, S. A., and Rho, G. J. (2012). Developmental expression of lineage-specific genes in porcine embryos of different origins. J. Assist. Reprod. Genet. 29, 723–733.
| Developmental expression of lineage-specific genes in porcine embryos of different origins.Crossref | GoogleScholarGoogle Scholar | 22639061PubMed |
Kumar, B. M., Maeng, G. H., Lee, Y. M., Lee, J. H., Jeon, B. G., Ock, S. A., Kang, T., and Rho, G. J. (2013). Epigenetic modification of fetal fibroblasts improves developmental competency and gene expression in porcine cloned embryos. Vet. Res. Commun. 37, 19–28.
| Epigenetic modification of fetal fibroblasts improves developmental competency and gene expression in porcine cloned embryos.Crossref | GoogleScholarGoogle Scholar | 23065456PubMed |
Lazzari, G., Wrenzycki, C., Herrmann, D., Duchi, R., Kruip, T., Niemann, H., and Galli, C. (2002). Cellular and molecular deviations in bovine in vitro-produced embryos are related to the large offspring syndrome. Biol. Reprod. 67, 767–775.
| Cellular and molecular deviations in bovine in vitro-produced embryos are related to the large offspring syndrome.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XmsV2js7Y%3D&md5=596f0bd100ff38f31be18a66075c17bfCAS | 12193383PubMed |
Lee, J. H., Lee, W. J., Jeon, R. H., Lee, Y. M., Jang, S. J., Lee, S. L., Jeon, B. G., Ock, S. A., King, W. A., and Rho, G. J. (2014). Development and gene expression of porcine cloned embryos derived from bone marrow stem cells with overexpressing Oct4 and Sox2. Cell. Reprogram. 16, 428–438.
| Development and gene expression of porcine cloned embryos derived from bone marrow stem cells with overexpressing Oct4 and Sox2.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhvF2lur3K&md5=7156ec9562a9be588891f451ce1778cbCAS | 25437870PubMed |
Lee, W. J., Jeon, R. H., Jang, S. J., Park, J. S., Lee, S. C., Baregundi Subbarao, R., Lee, S. L., Park, B. W., King, W. A., and Rho, G. J. (2015). Selection of reference genes for quantitative gene expression in porcine mesenchymal stem cells derived from various sources along with differentiation into multi-lineages. Stem Cells Int. 2015, 235192.
| Selection of reference genes for quantitative gene expression in porcine mesenchymal stem cells derived from various sources along with differentiation into multi-lineages.Crossref | GoogleScholarGoogle Scholar | 25972899PubMed |
Llobat, L., Marco-Jiménez, F., Peñaranda, D. S., Saenz-de-Juano, M. D., and Vicente, J. S. (2012). Effect of embryonic genotype on reference gene selection for RT-qPCR normalisation. Reprod. Domest. Anim. 47, 629–634.
| Effect of embryonic genotype on reference gene selection for RT-qPCR normalisation.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC38XhsVKjurvO&md5=3dc8a1386b1f9d00dfb6f8210b6bee50CAS | 22044783PubMed |
Luchsinger, C., Arias, M. E., Vargas, T., Paredes, M., Sánchez, R., and Felmer, R. (2014). Stability of reference genes for normalisation of reverse transcription quantitative real-time PCR (RT-qPCR) data in bovine blastocysts produced by IVF, ICSI and SCNT. Zygote 22, 505–512.
| Stability of reference genes for normalisation of reverse transcription quantitative real-time PCR (RT-qPCR) data in bovine blastocysts produced by IVF, ICSI and SCNT.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC2cXhs1WisbvE&md5=7a906d2da8dd69f9a3c90eebce1ed1cbCAS | 23731783PubMed |
Magnani, L., and Cabot, R. A. (2008). In vitro- and in vivo-derived porcine embryos possess similar, but not identical, patterns of Oct4, Nanog and Sox2 mRNA expression during cleavage development. Mol. Reprod. Dev. 75, 1726–1735.
| In vitro- and in vivo-derived porcine embryos possess similar, but not identical, patterns of Oct4, Nanog and Sox2 mRNA expression during cleavage development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtlyqsrzN&md5=69be01962ccbaed83f949ec7e47dee5aCAS | 18425776PubMed |
Mamo, S., Gal, A. B., Bodo, S., and Dinnyes, A. (2007). Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev. Biol. 7, 14.
| Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro.Crossref | GoogleScholarGoogle Scholar | 17341302PubMed |
Mamo, S., Gal, A. B., Polgar, Z., and Dinnyes, A. (2008). Expression profiles of the pluripotency marker gene POU5F1 and validation of reference genes in rabbit oocytes and pre-implantation stage embryos. BMC Mol. Biol. 9, 67.
| Expression profiles of the pluripotency marker gene POU5F1 and validation of reference genes in rabbit oocytes and pre-implantation stage embryos.Crossref | GoogleScholarGoogle Scholar | 18662377PubMed |
McCulloch, R. S., Ashwell, M. S., O’Nan, A. T., and Mente, P. L. (2012). Identification of stable normalisation genes for quantitative real-time PCR in porcine articular cartilage. J. Anim. Sci. Biotechnol. 3, 36.
| Identification of stable normalisation genes for quantitative real-time PCR in porcine articular cartilage.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3sXivFGks7Y%3D&md5=d194020fc8dcd8140adc9530032c70c6CAS | 23146128PubMed |
McElroy, S. L., Kim, J. H., Kim, S., Jeong, Y. W., Lee, E. G., Park, S. M., Hossein, M. S., Koo, O. J., Abul Hashem, M. D., Jang, G., Kang, S. K., Lee, B. C., and Hwang, W. S. (2008). Effects of culture conditions and nuclear transfer protocols on blastocyst formation and mRNA expression in pre-implantation porcine embryos. Theriogenology 69, 416–425.
| Effects of culture conditions and nuclear transfer protocols on blastocyst formation and mRNA expression in pre-implantation porcine embryos.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhsVSmsro%3D&md5=ba46e4eddaf30c0425b215575aac1babCAS | 18055008PubMed |
Nailis, H., Coenye, T., Van Nieuwerburgh, F., Deforce, D., and Nelis, H. J. (2006). Development and evaluation of different normalisation strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol. Biol. 7, 25.
| Development and evaluation of different normalisation strategies for gene expression studies in Candida albicans biofilms by real-time PCR.Crossref | GoogleScholarGoogle Scholar | 16889665PubMed |
Niemann, H., and Wrenzycki, C. (2000). Alterations of expression of developmentally important genes in pre-implantation bovine embryos by in vitro culture conditions: implications for subsequent development. Theriogenology 53, 21–34.
| Alterations of expression of developmentally important genes in pre-implantation bovine embryos by in vitro culture conditions: implications for subsequent development.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXkvVyqug%3D%3D&md5=e4b2eb879d9d29d3cb3c378096ea5b3fCAS | 10735059PubMed |
Nygard, A. B., Jørgensen, C. B., Cirera, S., and Fredholm, M. (2007). Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR. BMC Mol. Biol. 8, 67.
| Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR.Crossref | GoogleScholarGoogle Scholar | 17697375PubMed |
Paris, D. B., Kuijk, E. W., Roelen, B. A., and Stout, T. A. (2011). Establishing reference genes for use in real-time quantitative PCR analysis of early equine embryos. Reprod. Fertil. Dev. 23, 353–363.
| Establishing reference genes for use in real-time quantitative PCR analysis of early equine embryos.Crossref | GoogleScholarGoogle Scholar | 21211469PubMed |
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515.
| Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXhvF2mtLk%3D&md5=2dca329043b816f75670dae0aa22729dCAS | 15127793PubMed |
Radonić, A., Thulke, S., Mackay, I. M., Landt, O., Siegert, W., and Nitsche, A. (2004). Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862.
| Guideline to reference gene selection for quantitative real-time PCR.Crossref | GoogleScholarGoogle Scholar | 14706621PubMed |
Rinaudo, P., and Schultz, R. M. (2004). Effects of embryo culture on global pattern of gene expression in pre-implantation mouse embryos. Reproduction 128, 301–311.
| Effects of embryo culture on global pattern of gene expression in pre-implantation mouse embryos.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXot1KitLg%3D&md5=fef5c0d364691265319f364b0b5e7516CAS | 15333781PubMed |
Rizos, D., Lonergan, P., Boland, M. P., Arroyo-García, R., Pintado, B., de la Fuente, J., and Gutiérrez-Adán, A. (2002). Analysis of differential messenger RNA expression between bovine blastocysts produced in different culture systems: implications for blastocyst quality. Biol. Reprod. 66, 589–595.
| Analysis of differential messenger RNA expression between bovine blastocysts produced in different culture systems: implications for blastocyst quality.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XhvVeitLo%3D&md5=f1e8ff9c4886ecc0210ffa3997126194CAS | 11870062PubMed |
Robert, C., McGraw, S., Massicotte, L., Pravetoni, M., Gandolfi, F., and Sirard, M. A. (2002). Quantification of housekeeping transcript levels during the development of bovine pre-implantation embryos. Biol. Reprod. 67, 1465–1472.
| Quantification of housekeeping transcript levels during the development of bovine pre-implantation embryos.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xot1Kjs78%3D&md5=343e500481c0ae49f0b3250f2ecb6034CAS | 12390877PubMed |
Ross, P. J., Wang, K., Kocabas, A., and Cibelli, J. B. (2010). Housekeeping gene transcript abundance in bovine fertilised and cloned embryos. Cell. Reprogram. 12, 709–717.
| Housekeeping gene transcript abundance in bovine fertilised and cloned embryos.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXhsFygsLrK&md5=0396fcf84cc45b093a81ca7572bf9b12CAS | 20973679PubMed |
Smits, K., Goosens, K., Soom, A. V., Govaere, J., Hoogevijs, M., Vanhaesebrouck, E., Galli, C., Colleoni, S., Vandesompele, M., and Peelman, L. (2009). Selection of reference genes for quantitative real-time PCR in equine in vivo and fresh and frozen–thawed in vitro blastocysts. BMC Res. Notes 2, 246.
| Selection of reference genes for quantitative real-time PCR in equine in vivo and fresh and frozen–thawed in vitro blastocysts.Crossref | GoogleScholarGoogle Scholar | 20003356PubMed |
Somers, J., Smith, C., Donnison, M., Wells, D. N., Henderson, H., McLeay, L., and Pfeffer, P. L. (2006). Gene expression profiling of individual bovine nuclear transfer blastocysts. Reproduction 131, 1073–1084.
| Gene expression profiling of individual bovine nuclear transfer blastocysts.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xpt1ehur0%3D&md5=0df3091d630af199e6f6c4f84715165aCAS | 16735546PubMed |
Suzuki, T., Kondo, S., Wakayama, T., Cizdziel, P. E., and Hayashizaki, Y. (2008). Genome-wide analysis of abnormal H3K9 acetylation in cloned mice. PLoS One 3, e1905.
| Genome-wide analysis of abnormal H3K9 acetylation in cloned mice.Crossref | GoogleScholarGoogle Scholar | 18398451PubMed |
Thellin, O., Zorzi, W., Lakaye, B., De Borman, B., Coumans, B., Hennen, G., Grisar, T., Igout, A., and Heinen, E. (1999). Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75, 291–295.
| Housekeeping genes as internal standards: use and limits.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXmsFajt74%3D&md5=f0f997f5c32cdcbca0e2d0a1cc651eb0CAS | 10617337PubMed |
Tricarico, C., Pinzani, P., Bianchi, S., Paglierani, M., Distante, V., Pazzagli, M., Bustin, S. A., and Orlando, C. (2002). Quantitative real-time reverse transcription polymerase chain reaction: normalisation to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 309, 293–300.
| Quantitative real-time reverse transcription polymerase chain reaction: normalisation to rRNA or single housekeeping genes is inappropriate for human tissue biopsies.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38Xot1Kmu7s%3D&md5=12d1086a35a3a539ce32aaed2dc8a0f5CAS | 12413463PubMed |
Uddin, M. J., Cinar, M. U., Tesfaye, D., Looft, C., Tholen, E., and Schellander, K. (2011). Age-related changes in relative expression stability of commonly used housekeeping genes in selected porcine tissues. BMC Res. Notes 4, 441.
| Age-related changes in relative expression stability of commonly used housekeeping genes in selected porcine tissues.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhsV2ltb7N&md5=ad1c628cd5c985535c716212f6338cdaCAS | 22023805PubMed |
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., and Speleman, F. (2002). Accurate normalisation of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034–research0034.11.
| Accurate normalisation of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes.Crossref | GoogleScholarGoogle Scholar | 12184808PubMed |
Veazey, K. J., and Golding, M. C. (2011). Selection of stable reference genes for quantitative RT-PCR comparison of mouse embryonic and extra-embryonic stem cells. PLoS One 6, e27592.
| Selection of stable reference genes for quantitative RT-PCR comparison of mouse embryonic and extra-embryonic stem cells.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhsFyntLbO&md5=b75919c682a02aa7a73f0e7cf5665905CAS | 22102912PubMed |
Walker, N. J. (2002). Tech.Sight. A technique whose time has come. Science 296, 557–559.
| Tech.Sight. A technique whose time has come.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XjtFCjt7s%3D&md5=b116e13ec59c3a34de047141509935fdCAS | 11964485PubMed |
Yang, F., Lei, X., Palacios, R. R., Tang, C., and Yue, H. (2013). Selection of reference gene for quantitative real-time PCR analysis in chicken embryo fibroblasts infected with avian leucosis virus subgroup J. BMC Res. Notes 6, 402.
| Selection of reference gene for quantitative real-time PCR analysis in chicken embryo fibroblasts infected with avian leucosis virus subgroup J.Crossref | GoogleScholarGoogle Scholar | 24099561PubMed |
Zhao, J., Whyte, J., and Prather, R. S. (2010). Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer. Cell Tissue Res. 341, 13–21.
| Effect of epigenetic regulation during swine embryogenesis and on cloning by nuclear transfer.Crossref | GoogleScholarGoogle Scholar | 20563602PubMed |
Zhou, W., Xiang, T., Walker, S., Farrar, V., Hwang, E., Findeisen, B., Sadeghieh, S., Arenivas, F., Abruzzese, R. V., and Polejaeva, I. (2008). Global gene expression analysis of bovine blastocysts produced by multiple methods. Mol. Reprod. Dev. 75, 744–758.
| Global gene expression analysis of bovine blastocysts produced by multiple methods.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXktFGhsb8%3D&md5=c5360ccf94d334f84489096bbe432ca8CAS | 17886272PubMed |




