Organic matter and metal loadings influence the spatial gradient of the benthic bacterial community in a temperate estuary
Eric J. Raes A B * , Bronwyn H. Holmes A , Kristen Karsh A , Katie E. Hillyer C , Mark Green A , Jodie van de Kamp A , Levente Bodrossy A , Sam Whitehead D , Bernadette Proemse D , Ursula Taylor D , Akira Weller-Wong D , Andrew T. Revill A , Elizabeth A. Brewer A and Andrew Bissett A
A B * , Bronwyn H. Holmes A , Kristen Karsh A , Katie E. Hillyer C , Mark Green A , Jodie van de Kamp A , Levente Bodrossy A , Sam Whitehead D , Bernadette Proemse D , Ursula Taylor D , Akira Weller-Wong D , Andrew T. Revill A , Elizabeth A. Brewer A and Andrew Bissett A
A Oceans and Atmosphere, CSIRO, Hobart, Tas. 7004, Australia.
B Flourishing Oceans, Minderoo Foundation, PO Box 3155, Broadway, Nedlands, WA 6009, Australia.
C Land and Water, CSIRO, Ecoscience Precinct, Dutton Park, Brisbane, Qld 4102, Australia.
D Derwent Estuary Program, Hobart, Tas. 7000, Australia.
Marine and Freshwater Research 73(4) 428-440 https://doi.org/10.1071/MF21225
Submitted: 2 August 2021 Accepted: 6 December 2021 Published: 18 January 2022
© 2022 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Omics-based monitoring using bacterial marker genes can provide valuable mechanistic insights into the functioning of ecosystems. Here, we present a 2.5-year dataset with monthly sampling of sediment genomic bacterial DNA (n = 160) in a temperate, urbanised estuary in Tasmania, Australia. Molecular data were collected with physical and biochemical bottom water data, sediment organic matter and metal concentrations. Our study supports evidence that sediment-specific variables (organic matter composition) have a larger influence over the sediment bacterial community than do large-scale environmental conditions (seasonal water changes). The observed spatial and temporal differences are interesting, given the significant seasonal variation in bottom water data (e.g. temperature differences of up to 10°C and 3-fold increases for NOx concentrations in the bottom water between summer and winter months). Whereas bottom water parameters changed seasonally, metal concentrations in the sediments did not show seasonal variations. Metal concentrations explained a larger variance in the bacterial community among sites but not on an estuary-wide scale. The disconnect between environmental bottom water conditions and the sediment bacterial communities has important ramifications, because it indicates that seasonal changes have little effect on the compositional dynamics of sediment microbes and may, therefore, be difficult to trace with marker-gene surveys.
Keywords: 16S rRNA gene, bacteria, bacterial eDNA, environmental monitoring, functional diversity, metals, organic matter, temporal change, temperate estuary, urban.
Introduction
Estuaries provide valuable ecological, economic and cultural services (Gillanders et al. 2011). These services include, among others, coastal protection, nursery habitats for commercially and ecologically important fish species and cultural, spiritual experiences (Martin et al. 2020). Estuaries are spatially and temporally dynamic systems, and although estuaries vary in size, geomorphology and energy exchange between the land and oceans, many of them have one key thing in common, namely, a growing human population (Kennish 2002). Since 2000, the global human population has increased by 21%, and the expansion of urbanisation along coast lines throughout the world is leading to an array of negative environmental impacts (United Nations 2019). Urbanisation is directly and indirectly affecting all the aforementioned services that estuaries provide by habitat loss and fragmentation, a decrease in biodiversity, harmful algal blooms, over-exploitation of resources, hypoxia, increases in water temperatures, contamination by sewage (both biological and chemical), pesticides, heavy metals and other organic and inorganic pollutants (Diaz and Rosenberg 2008; Greaver et al. 2016; Scanes et al. 2020). Building a baseline understanding of natural variation and seasonal patterns is, therefore, paramount if we, as a society, want to disentangle natural from anthropogenic pressures in urbanised estuaries.
Management strategies, protection and restoration plans are crucial to mitigate current and future impacts on estuaries and coastal zones in general. In Australia, indicators of water-column quality are typically measured and managed at a local (council) and state government level, with little data centralisation at a national level (Raes et al. 2021). To indicate water quality and toxicity, local monitoring programs generally survey abiotic and biotic variables, such as concentrations of nitrate, nitrite and phosphate, turbidity, total suspended and dissolved solids, O2 concentrations, pH values, and chlorophyll-a (Chl-a) concentrations (Coughanowr et al. 2009; Visby and Weller-Wong 2020). However, there are opportunities in using molecular approaches such as genomic bacteria DNA from environmental samples to improve ecosystem surveillance strategies, monitoring programs and, ultimately, environmental management decisions (Thomsen and Willerslev 2015; Clarke et al. 2020; Raes et al. 2021). Studies utilising metabarcoding and quantitative PCR to investigate sediment and water column bacterial communities (bacterial eDNA) provide both qualitative and quantitative data. For example, bacterial eDNA can be used to investigate bacterial removal and retention of nitrogen (Francis et al. 2013; Raes et al. 2020) to identify the footprint of aquaculture pens (Li et al. 2011; Stoeck et al. 2018), reveal global trends in anti-microbial resistance (Hendriksen et al. 2019) and even the evolutionary responses of bacteria to long-term mercury exposure (Ruuskanen et al. 2020). However, routinely integrating metabarcoding data in environmental monitoring programs remains challenging.
Here, we present the findings from a 2.5-year monitoring survey from sediment samples collected monthly and analysed for genomic bacterial DNA, and physical and biochemical metadata, including sediment total nitrogen (TN), total organic carbon (TOC), analyses of the δ15N and δ13C and metal concentrations in the Derwent estuary in Tasmania, Australia. The Derwent estuary is a microtidal-influenced system connected to the Southern Ocean. Non-biological monitoring programs covering the past 15 years have shown a strong metal pollution gradient in the Derwent estuary, with zinc (Zn), arsenic (As), cadmium (Cd), copper (Cu) and lead (Pb) all exceeding the Australian quality guideline values at sites close to the zinc refinery (Simpson et al. 2007; Coughanowr et al. 2009). Earlier work on sediment bacterial communities in the Derwent estuary showed strong spatial patterns of the bacterial community composition (Abell et al. 2013). More specifically, bacterial nitrifiers, archaeal ammonia oxidisers, denitrifiers and total bacterial and archaeal biomass were positively correlated with C:N ratios and the isotope ratios of N (δ15N) and C (δ13C) of the organic matter (OM) in the sediment.
On the basis of the previous work from Abell et al. (2013) and the results from several studies that have highlighted that bacteria exhibit substrate preferences for the size and age of OM (Findlay 2003; Ding et al. 2015; Wang et al. 2015), we hypothesise that the variability of the bacterial community would be best explained by the source (indicated by the isotopic δ13C values; see Peterson (1999)) and quantity of OM. Second, we hypothesise that, although the quantity and source of the OM would be the main explanatory variable for the bacterial community composition on an estuary scale, the sediment metal concentrations would significantly determine the bacterial community composition at the most metal-affected sites near the zinc refinery, but not on an estuary scale. Last, we selected several independent environmental parameters that may have some influence over the bacterial community composition, and thus help determine its variability, including the inferred metabolic potentials of these communities. As a result of this research, we provide a baseline from which we hope to further increase our understanding and the relationships between the sediment geo-chemistry, bacterial assemblages and the ecosystem functions that the latter perform.
Materials and methods
Water chemistry measurements
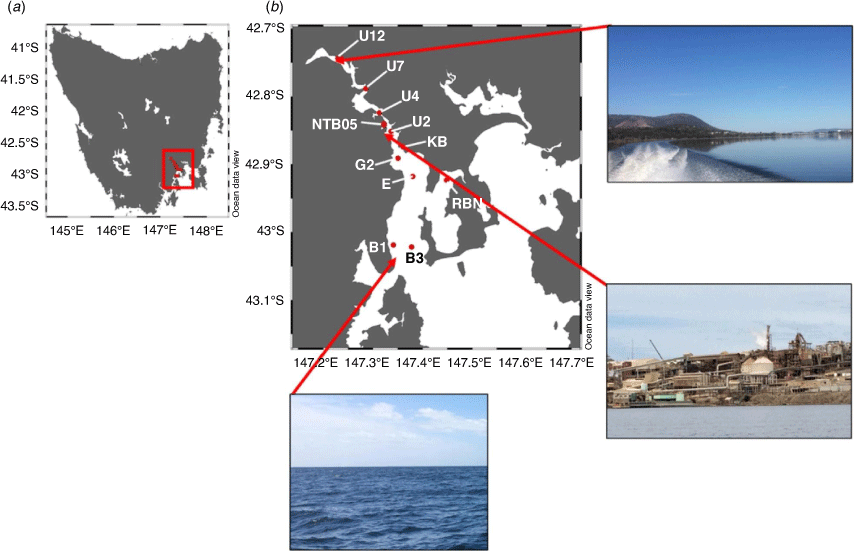
We present trends in physico-biochemical water column characteristics at 11 sites in the Derwent estuary, Tasmania, since 2010 (Fig. 1, 2). Monthly water profiles (every metre from the surface to 0.5 m from the bottom) for temperature, salinity and dissolved oxygen were collected (using a YSI EXO 3 multiparameter sonde and a Hydrolab 4a DataSonde) by the following three organisations: The Derwent Estuary Program (DEP), Norske Skog Boyer, and Nyrstar Hobart. Dissolved measurements for inorganic nitrate (NO3–), nitrite (NO2–) and Chl-a concentrations were collected using Niskin bottles at the surface and bottom by these agencies. Samples were collected 0.5 m from the bottom and the depths ranged from 3 to 26 m (see also Supplementary Table S1). All samples were analysed at Analytical Services Tasmania (AST; https://analyticalservices.tas.gov.au/). All biochemical and physical water column data were requested through the DEP (https://www.derwentestuary.org.au/contact/); the data were received on 20 August 2020. The data and detailed methodologies are available in the Supplementary material and the State of the Derwent Update 2020 (Visby and Weller-Wong 2020); https://www.derwentestuary.org.au/state-of-the-derwent/.
|
Sediment sampling
Monthly sediment surveys were conducted between May 2018 and February 2020 at the same 11 sites where the DEP collected physico-biochemical water-column data. Over the span of 2.5 years, 160 sediment samples were collected for abiotic and genomic bacterial DNA analyses from the lower to the upper basin of the estuary. At each site, three sediment cores were collected using a triangular configured sediment corer with polyethylene sample tubes (4.5-cm internal diameter) located at each corner. The top 1 cm of three sediment cores were homogenously mixed in glass jars, and subsamples were taken for genomic DNA (gDNA) extractions, TN, TOC, including analyses of the δ15N and δ13C, total phosphorus (P) as phosphate from sediment digests (referred to as total PO43– dry weight) and metals, which included As, Cd, Cu, iron (Fe), Pb, Zn and mercury (Hg).
Elemental, organic matter and stable isotope analyses
Samples for elemental, organic matter and stable isotope analyses in the sediments were collected between May 2018 and February 2020. Sediment metal concentrations and total phosphorus were analysed by inductively coupled plasma–atomic emission spectroscopy (ICP-AES; Method 2301). Sediment Hg analyses were performed by cold-vapour atomic fluorescence spectroscopy (CV-AFS; Method 2304; please see the ‘Metals and stable isotope analyses’ section in the Supplementary material for more details). Metal concentrations and total P are expressed in milligrams per kilogram on a dry matter basis (DMB). Stable isotope analysis was used to determine the fractionation and potential source of C and N in the estuary. The isotopic composition of the total N and organic C in the sediments (δ15N and δ13C) were analysed at the CSIRO laboratories in Hobart, Tasmania. A Carlo Erba NA1500 CNS analyser was interfaced with a Conflo IV to a Thermo Scientific Delta V Plus isotope ratio mass spectrometer and operated in the continuous-flow mode during sample analyses. Results are presented in the standard δ notation (please see Supplementary material for more details).
gDNA extractions
Sediment samples for genomics were collected between May 2018 and February 2020 and stored at –80°C until gDNA extraction. Approximately 0.25 g of wet sediment was weighed into powerbead tubes and gDNA was extracted using the QIAGEN DNeasy PowerSoil Total DNA Kits (QIAGEN; Cat. No. 12888-100) according to the manufacturer’s instructions. Nucleic acids were quantified on a QuBIT 2.0 fluorometer (Supplementary Table S1).
16S rRNA amplicon sequencing
Bacterial diversity was investigated by tag sequencing targeting the V1–V3 region of the 16S rRNA gene with the bacterial forward 27F and reverse 519R primer sets (Lane et al. 1985; Weisburg et al. 1991), by using the Illumina MiSeq platform. Amplicons targeting the 16S rRNA gene were amplified from environmental sediment gDNA extracts and sequenced at the Ramaciotti Centre for Genomics (UNSW, Sydney, NSW, Australia). Nextera XT barcode incorporation, purification, library generation and sequencing using the Illumina MiSeq platform (Illumina, Inc., San Diego, CA, USA), with 300-bp paired reads, were performed according to the manufacturer’s directions.
Amplicon sequence variant (ASV) tables were prepared after Bissett et al. (2016) (see also; https://www.australianmicrobiome.com/protocols/16sanalysisworkflow/). Briefly, FLASH (ver. 1.2.11, see https://ccb.jhu.edu/software/FLASH/; Magoč and Salzberg 2011) was used to merge paired-end reads and unique sequences were denoised into ASVs with the UNOISE3 algorithm (Edgar and Flyvbjerg 2015), using USEARCH 64 bit (ver. 8.0.1517, see https://www.drive5.com/usearch/download.html; Edgar 2010). Abundance profiles per sample were constructed by mapping all the reads to the unique ASVs by using the USEARCH ‘otutab’ command. The SILVA database (ver. 138, see https://www.arb-silva.de/; Quast et al. 2012; Yilmaz et al. 2014) was used to derive taxonomy by using the naive Bayesian classifier method of Wang et al. (2007) with the classify.seqs command in Mothur (settings: cutoff = 60, probs = TRUE; Schloss et al. 2009). ASVs were classified as follows: Domains: ‘unknown’, ‘Archaea’ and ‘Eukaryota’; Phylum: ‘Bacteria_unclassified’; Family: ‘Mitochondria’; Order: ‘Chloroplast’, and likely erroneous sequences were removed before the analyses. The final 16S rRNA gene library had 13 803 ASVs and the sequencing depth ranged between 4121 and 40 253 reads per sample. Genomic data are available at the NCBI under the Bioproject accession number PRJNA611638.
Functional marker gene predictions using PICRUSt2
PICRUSt2 (Langille et al. 2013; Douglas et al. 2020) was used to infer an approximate phylogenetically inferred metabolic potential of the bacterial communities in the Derwent estuary. To reduce run time, and because PICRUSt2 estimates function from the nearest ‘ancestor’, we clustered the ASVs at the 97% similarity threshold (97% of operational taxonomic units (OTUs) were generated using the USEARCH ‘cluster_fast’ function with -id 0.97; see Supplementary material for the full workflow). The PICRUSt2 pipeline (v. picrust/2.3.0b) was run with default settings, and sequences with Nearest Sequenced Taxon Index (NSTI) scores of >2 were removed. The average NSTI score, based on 163 samples, was 0.52 ± 0.28 (±s.d.). Only 3 of the 13 803 OTUs were above the maximum NSTI cut-off of 2 (these ASVs had less than 31 reads). Pathways with fewer than 10 reads were removed. MetaCyc pathway abundances are the main high-level prediction output by PICRUSt2. MetaCyc is a curated database (Caspi et al. 2016) of experimentally deduced metabolic pathways from all domains of life and an open-source alternative to the Kyoto Encyclopaedia of Genes and Genomes (KEGG; Kanehisa et al. 2007). A superclass or parental class contains a set of pathways that accomplish roughly the same biological function, such as degradation of a given starting material, or biosynthesis of an end product.
Statistical analysis
The Phyloseq package (ver. 1.32.0, see https://joey711.github.io/phyloseq/; McMurdie and Holmes 2013) was used to analyse, visualise and plot the microbiome and physical and bio-chemical metadata using R (ver. 4.0.2, R Foundation for Statistical Computing, Vienna, Austria) in RStudio (ver. 1.1.442, RStudio Inc., Boston, MA, USA, see https://www.rstudio.com/). Statistical tests were conducted using the Vegan package (ver. 2.5-6, J. Oksanen, R. Kindt, P. Legendre, B. O’Hara, M. H. H. Stevens, M. J. Oksanen and M. Suggests, see https://cran.r-project.org/web/packages/vegan/). Analyses of similarities (ANOSIM) were used to test whether we could identify statistical differences among the 11 estuarine sites and the 4 estuarine zones using the PRIMER-e (ver. 7.0.17, see https://www.primer-e.com/; Clarke and Gorley 2006). The plyr package (ver. 1.8.4, see https://cran.r-project.org/web/packages/plyr/index.html) was used to calculate means and summarise the data (Wickham 2011).
Differential abundance analyses were used to identify changes in taxa and functions between the four different estuarine zones, by using ANOVA-like differential expression (ALDEx2 ver. 1.20.0, see https://bioconductor.org/packages/release/bioc/html/ALDEx2.html; Gloor et al. 2017). Prior to the analyses, ASVs were agglomerated at the family level (on average and across all sites <5% of the ASVs at the phyla were assigned as ‘uncultured’). To account for the compositionality of the data, both the ASV count data and the inferred metabolic pathway (PICRUSt2) data were transformed using the centre log ratio (CLR) for all statistical analyses. The CLR transformations were performed using the microbiome package (ver. 1.16.0, see https://microbiome.github.io/) with a pseudo-count of min(relative abundance) ÷ 2 to exact zero relative abundance entries. The differential abundances were generated by 128 Monte Carlo samples sourced from a Dirichlet distribution (Fernandes et al. 2014).
Unconstrained (principal component analyses; PCA) and constrained (redundancy analyses; RDA) were constructed from Aitchison (Euclidean) distance matrices generated from CLR-transformed ASVs. Environmental parameters were standardised with the ‘standardise’ function (variables were scaled to zero mean and unit variance) using decostand from the Vegan package (J. Oksanen et al., see https://cran.r-project.org/web/packages/vegan/) and significant (P < 0.05) environmental parameters were derived using the ‘envfit’ function in Vegan and overlaid as vectors to identify multiple explanatory variables among the estuarine zones (J. Oksanen et al., see https://cran.r-project.org/web/packages/vegan/).
Results
Seasonal bottom water changes
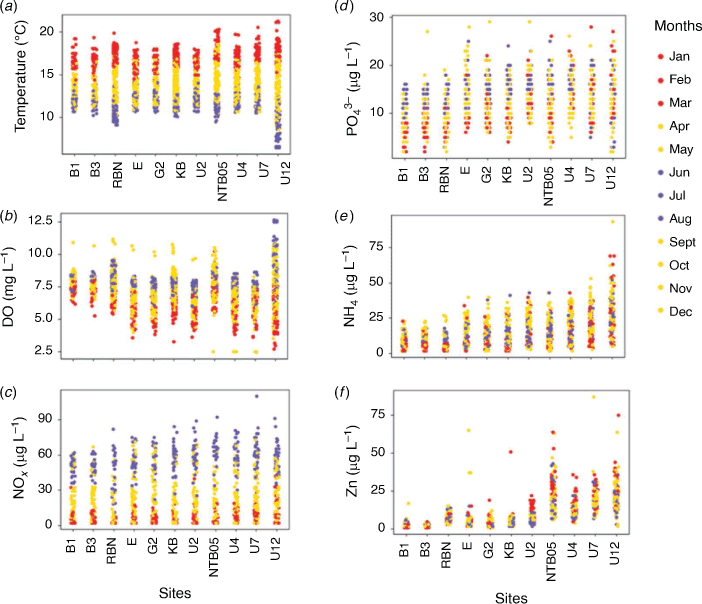
During the past decade, annual bottom water temperatures ranged between 6.5 and 21.3°C. Bottom water temperatures were significantly different among the winter, transition and summer months (Fig. 2a; Wilcoxon test, P < 0.05). Dissolved oxygen (DO) concentrations displayed similar significant changes among the winter, transition and summer months, with highest DO concentrations being recorded in the colder months (>7.5 mg L–1), and significantly lower concentrations in summer (Wilcoxon test, P < 0.05; Fig. 2b). Dissolved inorganic NO3– + NO2– (NOx) concentrations showed a clear annual trend, with significant differences among the winter, transition and summer months, with highest concentrations in the colder and lowest concentrations in the warmer months (Wilcoxon test, P < 0.05; Fig. 2c). Dissolved inorganic PO43–, NH4+ and Zn concentrations showed a general increase from the lower to the upper estuary (Fig. 2d–f). Dissolved inorganic PO43– was significantly higher in winter, whereas NH4+ concentrations were significantly higher in the transition months (Fig. 2e; Wilcoxon test, P < 0.05). Bottom water Zn concentrations were significantly higher in the warmer months (Wilcoxon test, P < 0.05). Salinity measurements from the bottom waters showed an increasing trend from fresh water at the upper estuary (i.e. site U12), up to 32 PSU in the lower estuary (i.e. B1 and B3 sites; Supplementary Fig. S2). Across the estuary, all bottom water measurements showed clear seasonal cycles across the entire estuary (Supplementary Fig. S2a–f).
Sediment chemistry
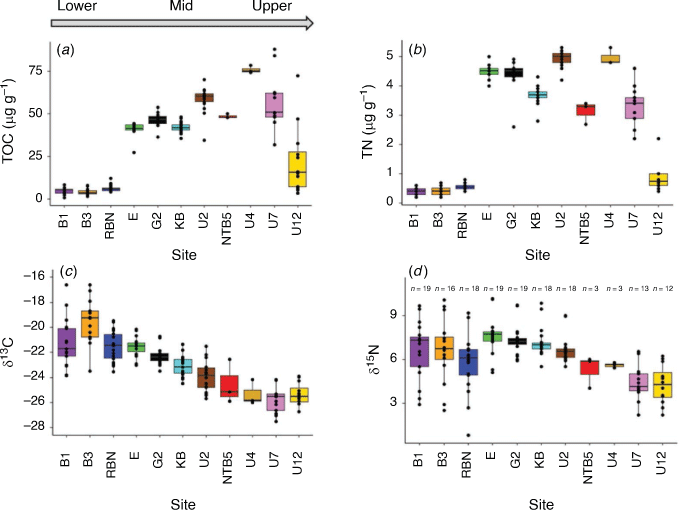
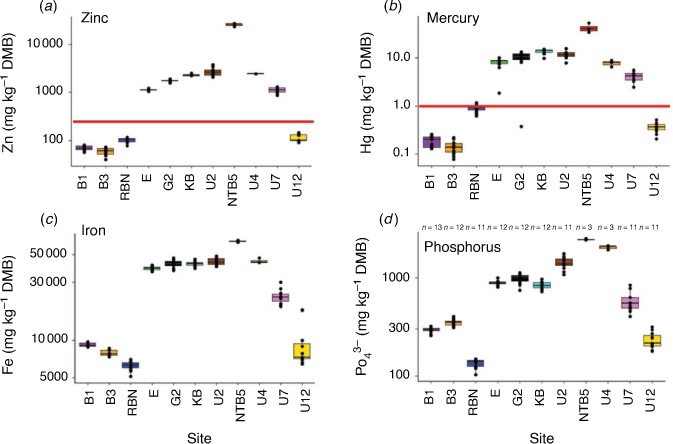
Concentrations of TOC and TN in the first centimetres of the sediments showed similar, and significant, trends across the estuary, with the lowest concentrations occurring at the lower estuarine sites (i.e. B1, B3 and RBN sites; ~5 µg C g–1 and ~0.5 µg N g–1), an increase at the mid-estuary sites (i.e. E, G2 and KB sites; ~50 µg C g–1 and ~4 µg N g–1), and a declining trend for the upper estuary sites (i.e. U7 and U12 sites; ~20 µg C g–1 and ~1 µg N g–1; Fig. 1b, 3a, b; Wilcoxon test, P < 0.05). The isotopic composition of the TOC (δ13C) showed a significant trend to more negative values from the lower to the upper estuary (Wilcoxon test, P < 0.05). The highest variability for δ15N was measured at the sites with the lowest TOC and TN concentrations (Fig. 3c, d). Besides at Site U12 (where seasonal differences were noted for TOC, TN and δ15N between the transition and warm months; Wilcoxon tests, P > 0.05), no seasonal effect was noted at the other sites for the TOC, TN and isotopic measurements (Wilcoxon tests, P > 0.05). At most sites, metal (As, Cd, Cu, Hg, Pb and Zn) concentrations in the sediments exceeded the Australian and New Zealand upper sediment quality guideline values (SQGV), with the highest concentrations at the sites near the zinc refinery (Sites U2 and NTB5; Supplementary Table S2). Overall highest metal concentrations were always recorded at Site NTB5 (Fig. 4, Supplementary Table S2). Similar trends along the estuary were noted for all other metal concentrations (Supplementary Table S2), and no seasonal differences were found at each site (Wilcoxon tests, P > 0.05).
Patterns in bacterial diversity
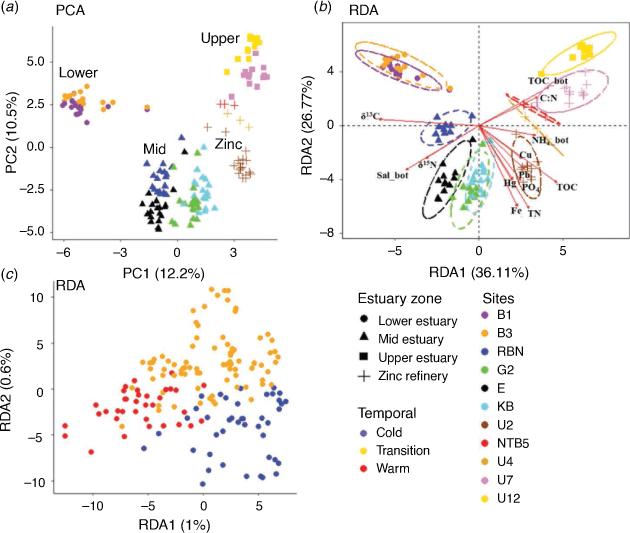
An unconstrained PCA analysis showed that, similar to the bio-geochemical sediment data, the bacterial communities displayed a clear spatial pattern along the Derwent estuary, with sites at the lower estuary, sites in the mid-estuary, sites close to the zinc refinery and sites in the upper estuary clustering closer to each other respectively (Fig. 5a). Analysis of similarities (ANOSIM) complemented these findings and showed a high and significant separation among the 11 sites and the different estuary zones (ANOSIM statistic R: 0.893 and 0.840 respectively; significance: 0.001; pairwise combinations are shown in Supplementary Tables S3, S4).
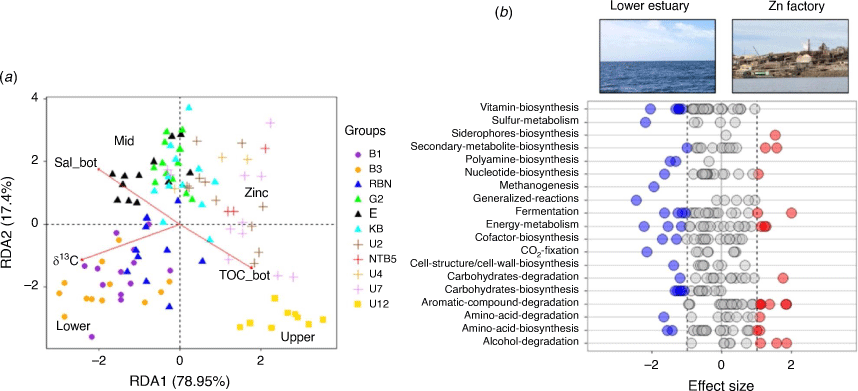
Redundancy analyses (RDAs) showed that a variety of sediment and bottom water parameters were significantly correlated with the bacterial community composition (Supplementary Table S5). More specifically, regression fitting of single explanatory variables including, TN, δ13C, TOC, Fe, bottom water salinity, dissolved organic C, sediment PO43–, Hg, the C:N ratio of the organic matter in the sediment, Pb concentrations and changes in the δ15N each explained more than 20% of the variance in the prokaryotic community composition (R2: 0.65, 0.60, 0.58, 0.58, 0.47, 0.41, 0.31, 0.29, 0.28, 0.27, 0.26 respectively, P < 0.001 for all; Supplementary Table S5). Bottom water dissolved O2, NH4+, Cu, PO43–, As, Cd and surface Chl-a concentrations also showed significant correlations with the bacterial community composition, but each parameter contributed <20% of the variance (R2: 0.19, 0.19, 0.19, 0.12, 0.10, 0.06 and 0.12 respectively, P < 0.001 for all; Supplementary Table S5). All metals showed significant correlations with the bacterial community composition, but Fe (mg kg–1 DMB, which showed a trend similar to the TOC and TN concentrations; Fig. 4c, d, 5b) showed the strongest effect on the community composition (R2: 0.58 Supplementary Table S5). A corresponding redundancy analysis constrained by seasonality showed that a very low percentage of the variance (~1%) could be explained by seasonal changes; Fig. 5c). Most of the measured sediment and bottom water measurements significantly (P < 0.001) correlated with the bacterial community data, except for bottom water temperatures, bottom water TN, bottom water NOx and sediment Cd concentrations (Supplementary Table S5).
|
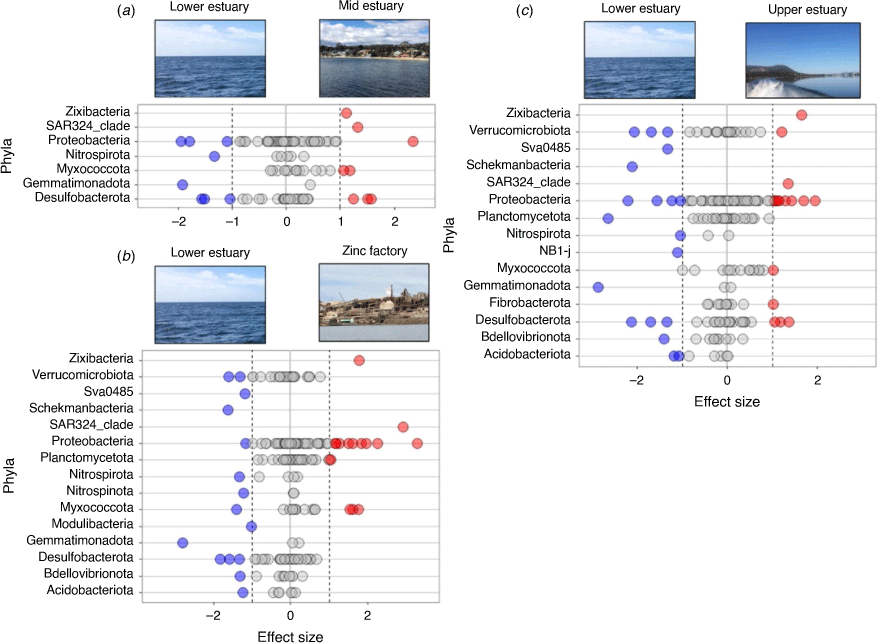
No significant differences were found within each site between the sediment bacterial communities in the warmer, transition and colder months (PERMANOVA P > 0.05). These results confirmed the strong differences in the bacterial communities between the sites and estuary zones (see ANOSIM analyses; Supplementary Tables S3, S4), along with the low percentage of the variance (1.6%) explained by seasonal changes in bottom water (RDA results; Fig. 5c). Differential relative abundance analyses were then used to identify the main phyla that contributed to the variations in the sediment communities between the estuary zones (as observed in the PCA analysis, Fig. 5). Results showed that the lower estuarine sediments were enriched with the phyla Proteobacteria, Nitrospinota, Gemmatimonadota and Desulfobacterota when compared with mid-estuarine samples. The zinc refinery sites were shown to have higher relative abundances of the phyla Zixibacteria, SAR324 clade, Proteobacteria, Planctomycetota and Myxococcota than in sediments in the lower estuary. The upper sediments could mostly be characterised by an increase in the phyla Zixibacteria, Verrucomicrobiota, SAR324 clade, Myxococcota and Firbobacterota, compared with the lower estuary (Fig. 6a, c).
Patterns in inferred bacterial functional diversity
The 16S rRNA gene-based metabolic reconstructions resulted in 369 inferred MetaCyc pathways and could be collapsed into 40 superclasses. Once again, site was a highly explanatory variable for bacterial functional community composition (ANOSIM; R: 0.430; significance: 0.001; with significant differences for all pairwise combinations except between B1–B3 and U7–U4 sites; see Supplementary Table S6). Estuarine zone showed a statistically significant effect albeit with a lower explanatory R value than for the site comparisons (ANOSIM; R: 0.406; significance: 0.001; see Supplementary Table S7 for pairwise combinations and Fig. 7a). RDAs showed that again a variety of sediment and bottom water parameters were significantly correlated with the phylogenetically inferred functional community composition, but only three parameters (δ13C, TOC and salinity in bottom waters) contributed >20% of the variance (Fig. 7a, Supplementary Table S8). Differential relative abundance analysis identified a higher number of metabolic superclasses related to biosynthesis and methanogenesis pathways in the lower estuary. The sediments near the zinc refinery were associated with a higher relative abundance or more diverse and complex degradation pathways and siderophore biosynthesis pathways (Fig. 7b).
Discussion
Our study complements the previous results from Abell et al. (2013) in the Derwent estuary, who found that both the abundance and community structure of bacterial nitrifiers, archaeal ammonia oxidisers, and denitrifiers are largely influenced by the sediment organic matter composition. However, the results from the current study extend these findings from those bacteria involved in the N-cycle to the whole bacterial community. Abell et al. (2013) already noted that the differences in OM sources are potentially affecting the rate of mineralisation by heterotrophic microorganisms. The trend to more negative δ13C values of the OM, from the lower to the upper estuary, indicates an increase of more terrestrial C sources (Stevens et al. 2021). By contrast, the low OM concentrations (both TN and TOC), the low C:N ratios, and the less negative δ13C values in the sediments of the lower estuary indicate a greater contribution of marine phytoplankton (Peterson 1999). The study from Abell et al. (2013) suggested that the OM in the lower estuary is more labile and, hence, more easily remineralised to NH4+, which was indirectly confirmed by observing a higher biomass of heterotrophic organisms (the authors reported higher 16S rRNA gene copy numbers for both bacteria and archaea in the lower estuary). In this study, we have also provided evidence that sites near the zinc refinery, with higher organic matter quantity, had a greater diversity of degradation pathways, including the presence of more complex compound degradation pathways. These results suggest that although the organic matter is less labile (from a C:N ratio perspective) in the mid-estuary and zinc refinery sites, the larger abundance of different organic substrates also seems to provide niche diversification for the bacterial community.
Although metal concentrations in the sediments exceeded the Australian and New Zealand upper sediment quality guideline values at most sites, and always at the sites near the zinc refinery, they did not show a large effect on the bacterial community composition on an estuary-wide basis (Supplementary Table S5). These results suggest that metal-tolerant species have adapted to the high metal concentrations in the estuary, and highlight the need to have a baseline from which we can untangle the natural variation owing to past and future pressures. These findings are also supported by the recent study from Hillyer et al. (2022) who showed that changes in the metabolite profiles for three macroinvertebrate groups were not significantly correlated with Zn concentrations on an estuary basis in the Derwent. However, zinc sediment concentrations were significantly and positively correlated with changes in metabolite profiles at the most metal-affected sites near the zinc refinery. These results, including those from our study, confirmed our second hypothesis that metal concentrations will influence site-specific spatial variation but not on a whole of estuary scale, because metals did not exceed the upper sediment quality guideline values at every site. In general, these results also align with those presented by Chen et al. (2018), who showed that the microbiome is able to adapt to long-term heavy-metal pollution exposure, and that sediment bacterial diversity, therefore, can be best examined among similar affected sites and not on an estuary-wide scale.
Water column data are the most commonly collected metadata, and we believed it would be a reasonable proxy to explain variability in the sediment bacterial communities. However, our study, and the earlier results from Abell et al. (2013) showed that the bacterial communities had a high site-specific community structure with distinct communities over time. These results are interesting, given the significant changes in environmental conditions in the estuary between seasons (e.g. temperature differences of 8°C, 3-fold increases for NOx and NH4+ and doubling of PO43– concentrations in the bottom waters between summer and winter months). Neither bottom water temperature or dissolved NOx concentrations showed significant correlations with the sediment microbiomes, suggesting that bacterial groups are more tightly controlled by local sediment abiotic and biotic parameters rather than large fluctuations in the overlying bottom water. The disconnect between the bottom water and the sediment has important ramifications, because it has been suggested that dynamic water column changes, including nutrient bursts, may have little effect on the compositional dynamics of sediment microbes (Orland et al. 2020). These findings suggest that rather than an indication of current state, perhaps our data give some indication of trajectory or temporal stability. Sediment microbes may instead respond stronger to fine‐scale variations in the composition (quantity, quality and source, e.g. marine and terrestrial) of organic matter (Fagervold et al. 2014; Ding et al. 2015; Wang et al. 2015), and, consequently, the physicochemical gradients and geochemistry of their surroundings. These findings are consistent with observational evidence that the abundance rather than presence of bacterial taxa is limited by resource availability in sediments (Louca et al. 2018; Orland et al. 2019). Our study, therefore, supports the necessity to sample multiple sites and consider sediments differently from the water column when bacterial processes are upscaled to entire estuary basins (Orland et al. 2020).
Monitoring and reporting on estuary health in Australia
The cumulative effects of climate change and increasing pressures from urbanisation are affecting coastal ecosystems worldwide (United Nations 2019). Over the past 12 years in Australia, estuarine temperatures have increased on average by 2.16°C, with waters acidifying at a rate of 0.09 pH units and freshening at 0.086 PSU year–1 (Scanes et al. 2020). The benefits of protecting and managing estuaries is directly related to the fact that these systems provide valuable ecological, cultural and economic services, including aquaculture industry, nursery grounds for commercial and wild fisheries, N removal, C storage, and recreational tourism (Evans et al. 2017; Fulford et al. 2020). Identifying reliable and inexpensive methods to track the status of estuaries and to predict the effects of specific stressors or management remains a major challenge (United Nations 2019). The scientific community are increasingly incorporating genomic surveys into their methodologies for biomonitoring because of its high accuracy, taxonomically holistic lens and ease of deployment (Berry et al. 2021). Although the integration of genomic data comes with its own challenges (Porter and Hajibabaei 2018; Quinn et al. 2018), it is becoming a core technology for ecosystem and water-quality assessment (Glasl et al. 2017; Berry et al. 2021). However, the potential to include genomic surveys into environmental monitoring programs can be achieved only once a baseline (the aim of this study) is established; from there, change can be estimated and quantified.
Omics-based monitoring technologies using bacterial marker genes have been shown to reveal valuable mechanistic insights into the functioning of ecosystems and the metabolic potential of bio-geochemical cycles (Ruuskanen et al. 2020). The underlying reason is that microbes are seen as useful, reliable and responsive indicators of ecosystem health because of their high abundance, the diversity of nutrient cycling functions they perform, and because their short life-cycle makes them able to respond quickly to environmental change. Integrating bacterial eDNA data in environmental monitoring programs remains a challenge. Our study did provide insights into the inferred metabolic potential of the sediment bacterial community, by showing relatively larger proportions of pathways associated with the biosynthesis of organic molecules in the sites from the lower estuary (referred to as the more ‘pristine’ sites compared with those close to the zinc refinery in terms of metal concentrations). These results also highlighted significant differences in the metabolic potential between these pristine and metal-polluted sites and open the possibility to get insights into the functional bacterial diversity. The opportunity to infer and measure this functional bacterial diversity, or metabolic potential, may provide new meeting points for microbial ecologists, biogeochemists and ecosystem modellers. However, one major gap in many omics surveys including ours is the lack of rate or metabolite data to truly allow extrapolation of marker gene predictions to metabolic processes. The congruent data collection for marker gene data, metabolic rate data (in situ or from experiments) and metabolomic data (e.g. Hillyer et al. 2022) are highly needed to fully unravel the interactions among different bacterial taxa within a community and their influence on the environment. These insights will be key to appreciate the pivotal role microbes have in an ecosystem.
Conclusions
How ecosystem functioning changes with bacterial communities remains an open question in many natural ecosystems, and merits investigation; hence, the main purpose of this study was to provide a descriptive analysis of the sediment bacterial communities of the Derwent estuary. Overall, our study supports evidence that small-scale environmental conditions (such as organic sediment composition) have a larger influence over the bacterial community assemblages in the Derwent estuary than do large‐scale environmental conditions (such as seasonal bottom water changes). We hope that these data will increase our understanding and ability to predict or infer the metabolic state of coastal ecosystems in a holistic way, and one day may improve environmental management decisions.
Data availability
All biochemical and physical water-column data are available through the DEP (https://www.derwentestuary.org.au/contact/). Genomic data are available at the NCBI under the Bioproject accession number PRJNA611638.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Declaration of funding
This research was funded by CSIRO Environomics Future Science Platform.
Author contributions
E. J. Raes, K. Karsh, B. H. Holmes, K. E. Hillyer, M. Green and A. Bissett collected monthly sediment data for genomic DNA, metal and organic matter analyses. A. T. Revill and E. A. Brewer analysed and quality controlled the organic matter analyses. S. Whitehead, B. Proemse, U. Taylor and A. Weller-Wong collected, analysed and provided long-term metadata. L. Bodrossy designed the study. A. Bissett, J. van de Kamp and E. J. Raes analysed the sequence data. E. J. Raes provided ecological interpretation and wrote the manuscript. All authors contributed to the editing of the paper.
Supplementary material
Supplementary material is available online.
Acknowledgements
We pay our respects to the Traditional Owners of this land, the Muwinina people, and pay respect to those that have passed before us and to acknowledge today’s and future’s Tasmanian Aboriginal people who are the custodians of the land and waters where we conducted our fieldwork. The Derwent estuary is named timtumili minanya in the indigenous dialect. We thank Mina Brock for processing sediment samples for stable isotope analysis.
References
Abell, GCJ, Ross, DJ, Keane, JP, Oakes, JM, Eyre, BD, Robert, SS, and Volkman, JK (2013). Nitrifying and denitrifying microbial communities and their relationship to nutrient fluxes and sediment geochemistry in the Derwent Estuary, Tasmania. Aquatic Microbial Ecology 70, 63–75.| Nitrifying and denitrifying microbial communities and their relationship to nutrient fluxes and sediment geochemistry in the Derwent Estuary, Tasmania.Crossref | GoogleScholarGoogle Scholar |
Berry, O, Jarman, S, Bissett, A, Hope, M, Paeper, C, Bessey, C, Schwartz, M, Hale, J, and Bunce, M (2021). Making environmental DNA (eDNA) biodiversity records globally accessible. Environmental DNA 3, 699–705.
| Making environmental DNA (eDNA) biodiversity records globally accessible.Crossref | GoogleScholarGoogle Scholar |
Bissett, A, Fitzgerald, A, Meintjes, T, Mele, PM, Reith, F, Dennis, PG, Breed, MF, Brown, B, Brown, MV, and Brugger, J (2016). Introducing BASE: the biomes of Australian soil environments soil microbial diversity database. GigaScience 5, 21.
| Introducing BASE: the biomes of Australian soil environments soil microbial diversity database.Crossref | GoogleScholarGoogle Scholar | 27195106PubMed |
Caspi, R, Billington, R, Ferrer, L, Foerster, H, Fulcher, CA, Keseler, IM, Kothari, A, Krummenacker, M, Latendresse, M, and Mueller, LA (2016). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Research 44, D471–D480.
| The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases.Crossref | GoogleScholarGoogle Scholar | 26527732PubMed |
Chen, Y, Jiang, Y, Huang, H, Mou, L, Ru, J, Zhao, J, and Xiao, S (2018). Long‐term and high‐concentration heavy‐metal contamination strongly influences the microbiome and functional genes in Yellow River sediments. Science of the Total Environment 637, 1400–1412.
Clarke KR, Gorley RN (2006) ‘Primer.’ (PRIMER-e: Plymouth, UK)
Clarke, LJ, Jones, PJ, Ammitzboll, H, Barmuta, LA, Breed, MF, Chariton, A, Charleston, M, Dakwa, V, Dewi, F, and Eri, R (2020). Mainstreaming microbes across biomes. Bioscience 70, 589–596.
| Mainstreaming microbes across biomes.Crossref | GoogleScholarGoogle Scholar |
Coughanowr CA, Whitehead J, Whitehead S, Einoder LE, Taylor U, Weeding B (2009) The state of the Derwent estuary 2015. A review of environmental data from 2009 to 2014. (Derwent Estuary Program) Available at https://www.derwentestuary.org.au/assets/State_of_the_Derwent_Estuary_2015.pdf
Diaz, RJ, and Rosenberg, R (2008). Spreading dead zones and consequences for marine ecosystems. Science 321, 926–929.
| Spreading dead zones and consequences for marine ecosystems.Crossref | GoogleScholarGoogle Scholar | 18703733PubMed |
Ding, J, Zhang, Y, Wang, M, Sun, X, Cong, J, Deng, YE, Lu, H, Yuan, T, Van Nostrand, JD, and Li, D (2015). Soil organic matter quantity and quality shape microbial community compositions of subtropical broadleaved forests. Molecular Ecology 24, 5175–5185.
| Soil organic matter quantity and quality shape microbial community compositions of subtropical broadleaved forests.Crossref | GoogleScholarGoogle Scholar | 26363284PubMed |
Douglas, GM, Maffei, VJ, Zaneveld, JR, Yurgel, SN, Brown, JR, Taylor, CM, Huttenhower, C, and Langille, MGI (2020). PICRUSt2 for prediction of metagenome functions. Nature Biotechnology 38, 685–688.
| PICRUSt2 for prediction of metagenome functions.Crossref | GoogleScholarGoogle Scholar | 32483366PubMed |
Edgar, RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461.
| Search and clustering orders of magnitude faster than BLAST.Crossref | GoogleScholarGoogle Scholar | 20709691PubMed |
Edgar, RC, and Flyvbjerg, H (2015). Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482.
| Error filtering, pair assembly and error correction for next-generation sequencing reads.Crossref | GoogleScholarGoogle Scholar | 26139637PubMed |
Evans K, Bax N, Smith DC (2017) Australia state of the environment 2016: marine environment. Independent report to the Australian Government Minister for the Environment and Energy. Australian Government Department of the Environment and Energy, Canberra, ACT, Australia.
Fagervold, SK, Bourgeois, S, Pruski, AM, Charles, F, Kerherve, P, Vetion, G, and Galand, PE (2014). River organic matter shapes microbial communities in the sediment of the Rhone prodelta. The ISME Journal 8, 2327–2338.
| River organic matter shapes microbial communities in the sediment of the Rhone prodelta.Crossref | GoogleScholarGoogle Scholar | 24858780PubMed |
Fernandes, AD, Reid, JN, Macklaim, JM, McMurrough, TA, Edgell, DR, and Gloor, GB (2014). Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis. Microbiome 2, 15.
| Unifying the analysis of high-throughput sequencing datasets: characterizing RNA-seq, 16S rRNA gene sequencing and selective growth experiments by compositional data analysis.Crossref | GoogleScholarGoogle Scholar | 24910773PubMed |
Findlay S (2003) Bacterial response to variation in dissolved organic matter. In ‘Aquatic Ecosystems’. (Eds SEG Findlay, RL Sinsabaugh) pp. 363–379. (Elsevier)
Francis, CA, O’Mullan, GD, Cornwell, JC, and Ward, BB (2013). Transitions in nirS-type denitrifier diversity, community composition, and biogeochemical activity along the Chesapeake Bay estuary. Frontiers in Microbiology 4, 237.
| Transitions in nirS-type denitrifier diversity, community composition, and biogeochemical activity along the Chesapeake Bay estuary.Crossref | GoogleScholarGoogle Scholar | 24009603PubMed |
Fulford, RS, Russell, M, Hagy, JD, and Breitburg, D (2020). Managing estuaries for ecosystem function. Global Ecology and Conservation 21, e00892.
| Managing estuaries for ecosystem function.Crossref | GoogleScholarGoogle Scholar | 33365365PubMed |
Gillanders, BM, Elsdon, TS, Halliday, IA, Jenkins, GP, Robins, JB, and Valesini, FJ (2011). Potential effects of climate change on Australian estuaries and fish utilising estuaries: a review. Marine and Freshwater Research 62, 1115–1131.
| Potential effects of climate change on Australian estuaries and fish utilising estuaries: a review.Crossref | GoogleScholarGoogle Scholar |
Glasl, B, Webster, NS, and Bourne, DG (2017). Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems. Marine Biology 164, 91.
| Microbial indicators as a diagnostic tool for assessing water quality and climate stress in coral reef ecosystems.Crossref | GoogleScholarGoogle Scholar |
Gloor, GB, Macklaim, JM, Pawlowsky-Glahn, V, and Egozcue, JJ (2017). Microbiome datasets are compositional: and this is not optional. Frontiers in Microbiology 8, 2224.
| Microbiome datasets are compositional: and this is not optional.Crossref | GoogleScholarGoogle Scholar | 29187837PubMed |
Greaver, TL, Clark, CM, Compton, JE, Vallano, D, Talhelm, AF, Weaver, CP, Band, LE, Baron, JS, Davidson, EA, and Tague, CL (2016). Key ecological responses to nitrogen are altered by climate change. Nature Climate Change 6, 836–843.
| Key ecological responses to nitrogen are altered by climate change.Crossref | GoogleScholarGoogle Scholar |
Hendriksen, RS, Bortolaia, V, Tate, H, Tyson, GH, Aarestrup, FM, and McDermott, PF (2019). Using genomics to track global antimicrobial resistance. Frontiers in Public Health 7, 242.
| Using genomics to track global antimicrobial resistance.Crossref | GoogleScholarGoogle Scholar | 31552211PubMed |
Hillyer, KE, Raes, E, Karsh, K, Holmes, B, Bissett, A, and Beale, DJ (2022). Metabolomics as a tool for in situ study of chronic metal exposure in estuarine invertebrates. Environmental Pollution 292, 118408.
| Metabolomics as a tool for in situ study of chronic metal exposure in estuarine invertebrates.Crossref | GoogleScholarGoogle Scholar | 34718088PubMed |
Kanehisa, M, Araki, M, Goto, S, Hattori, M, Hirakawa, M, Itoh, M, Katayama, T, Kawashima, S, Okuda, S, and Tokimatsu, T (2007). KEGG for linking genomes to life and the environment. Nucleic Acids Research 36, D480–D484.
| KEGG for linking genomes to life and the environment.Crossref | GoogleScholarGoogle Scholar | 18077471PubMed |
Kennish, MJ (2002). Environmental threats and environmental future of estuaries. Environmental Conservation 29, 78–107.
| Environmental threats and environmental future of estuaries.Crossref | GoogleScholarGoogle Scholar |
Lane, DJ, Pace, B, Olsen, GJ, Stahl, DA, Sogin, ML, and Pace, NR (1985). Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proceedings of the National Academy of Sciences of the United States of America 82, 6955–6959.
| Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses.Crossref | GoogleScholarGoogle Scholar | 2413450PubMed |
Langille, MGI, Zaneveld, J, Caporaso, JG, McDonald, D, Knights, D, Reyes, JA, Clemente, JC, Burkepile, DE, Thurber, RLV, and Knight, R (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology 31, 814–821.
| Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences.Crossref | GoogleScholarGoogle Scholar |
Li, M, Cao, H, Hong, Y-G, and Gu, J-D (2011). Seasonal dynamics of anammox bacteria in estuarial sediment of the Mai Po Nature Reserve revealed by analyzing the 16S rRNA and hydrazine oxidoreductase (hzo) genes. Microbes and Environments 26, 15–22.
| Seasonal dynamics of anammox bacteria in estuarial sediment of the Mai Po Nature Reserve revealed by analyzing the 16S rRNA and hydrazine oxidoreductase (hzo) genes.Crossref | GoogleScholarGoogle Scholar | 21487198PubMed |
Louca, S, Polz, MF, Mazel, F, Albright, MBN, Huber, JA, O’Connor, MI, Ackermann, M, Hahn, AS, Srivastava, DS, and Crowe, SA (2018). Function and functional redundancy in microbial systems. Nature Ecology & Evolution 2, 936–943.
| Function and functional redundancy in microbial systems.Crossref | GoogleScholarGoogle Scholar |
Magoč, T, and Salzberg, SL (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963.
| FLASH: fast length adjustment of short reads to improve genome assemblies.Crossref | GoogleScholarGoogle Scholar | 21903629PubMed |
Martin, CL, Momtaz, S, Gaston, T, and Moltschaniwskyj, NA (2020). Estuarine cultural ecosystem services valued by local people in New South Wales, Australia, and attributes important for continued supply. Ocean and Coastal Management 190, 105160.
| Estuarine cultural ecosystem services valued by local people in New South Wales, Australia, and attributes important for continued supply.Crossref | GoogleScholarGoogle Scholar |
McMurdie, PJ, and Holmes, S (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8, e61217.
| phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data.Crossref | GoogleScholarGoogle Scholar | 23630581PubMed |
Orland, C, Emilson, EJS, Basiliko, N, Mykytczuk, NCS, Gunn, JM, and Tanentzap, AJ (2019). Microbiome functioning depends on individual and interactive effects of the environment and community structure. The ISME Journal 13, 1–11.
| Microbiome functioning depends on individual and interactive effects of the environment and community structure.Crossref | GoogleScholarGoogle Scholar | 30042502PubMed |
Orland, C, Yakimovich, KM, Mykytczuk, NCS, Basiliko, N, and Tanentzap, AJ (2020). Think global, act local: the small‐scale environment mainly influences microbial community development and function in lake sediment. Limnology and Oceanography 65, S88–S100.
| Think global, act local: the small‐scale environment mainly influences microbial community development and function in lake sediment.Crossref | GoogleScholarGoogle Scholar |
Peterson, BJ (1999). Stable isotopes as tracers of organic matter input and transfer in benthic food webs: a review. Acta Oecologica 20, 479–487.
| Stable isotopes as tracers of organic matter input and transfer in benthic food webs: a review.Crossref | GoogleScholarGoogle Scholar |
Porter, TM, and Hajibabaei, M (2018). Scaling up: a guide to high‐throughput genomic approaches for biodiversity analysis. Molecular Ecology 27, 313–338.
| Scaling up: a guide to high‐throughput genomic approaches for biodiversity analysis.Crossref | GoogleScholarGoogle Scholar | 29292539PubMed |
Quast, C, Pruesse, E, Yilmaz, P, Gerken, J, Schweer, T, Yarza, P, Peplies, J, and Glöckner, FO (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research 41, D590–D596.
| The SILVA ribosomal RNA gene database project: improved data processing and web-based tools.Crossref | GoogleScholarGoogle Scholar | 23193283PubMed |
Quinn, TP, Erb, I, Richardson, MF, and Crowley, TM (2018). Understanding sequencing data as compositions: an outlook and review. Bioinformatics 34, 2870–2878.
| Understanding sequencing data as compositions: an outlook and review.Crossref | GoogleScholarGoogle Scholar | 29608657PubMed |
Raes, EJ, Karsh, K, Kessler, AJ, Cook, PLM, Holmes, BH, van de Kamp, J, Bodrossy, L, and Bissett, A (2020). Can we use functional genetics to predict the fate of nitrogen in estuaries? Frontiers in Microbiology 11, 1261.
| Can we use functional genetics to predict the fate of nitrogen in estuaries?Crossref | GoogleScholarGoogle Scholar | 32655525PubMed |
Raes, EJ, Karsh, K, Sow, SLS, Ostrowski, M, Brown, MV, van de Kamp, J, Franco-Santos, RM, Bodrossy, L, and Waite, AM (2021). Metabolic pathways inferred from a bacterial marker gene illuminate ecological changes across South Pacific frontal boundaries. Nature Communications 12, 2213.
| Metabolic pathways inferred from a bacterial marker gene illuminate ecological changes across South Pacific frontal boundaries.Crossref | GoogleScholarGoogle Scholar | 33850115PubMed |
Raes EJ, Participants of the CSIRO Oceans, Atmosphere ECR workshop Life Below Water (2022) Measuring success of SDG 14: an Australian perspective. In ‘Life below water: Encyclopedia of the UN Sustainable Development Goals’. (Eds W Leal Filho, AM Azul, L Brandli, A Lange Salvia, T Wall) pp. 1–14. (Springer International Publishing).
| Crossref |
Ruuskanen, MO, Aris-Brosou, S, and Poulain, AJ (2020). Swift evolutionary response of microbes to a rise in anthropogenic mercury in the Northern Hemisphere. The ISME Journal 14, 788–800.
| Swift evolutionary response of microbes to a rise in anthropogenic mercury in the Northern Hemisphere.Crossref | GoogleScholarGoogle Scholar | 31831837PubMed |
Scanes, E, Scanes, PR, and Ross, PM (2020). Climate change rapidly warms and acidifies Australian estuaries. Nature Communications 11, 1803.
| Climate change rapidly warms and acidifies Australian estuaries.Crossref | GoogleScholarGoogle Scholar | 32286277PubMed |
Schloss, PD, Westcott, SL, Ryabin, T, Hall, JR, Hartmann, M, Hollister, EB, Lesniewski, RA, Oakley, BB, Parks, DH, and Robinson, CJ (2009). Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology 75, 7537–7541.
| Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities.Crossref | GoogleScholarGoogle Scholar | 19801464PubMed |
Simpson SL, Batley GE, Chariton AA (2007) Revision of the ANZECC/ARMCANZ sediment quality guidelines. CSIRO Land and Water science report 08/07. Prepared for the Department of Sustainability, Environment, water, population and communities. CSIRO Land and Water.
Stevens, H, Chase, Z, Zawadzki, A, Wong, H, and Proemse, BC (2021). Reconstructing the history of nutrient loads and sources in the Derwent Estuary, Tasmania, Australia, using isotopic fingerprinting techniques. Estuaries and Coasts 44, 2236–2249.
| Reconstructing the history of nutrient loads and sources in the Derwent Estuary, Tasmania, Australia, using isotopic fingerprinting techniques.Crossref | GoogleScholarGoogle Scholar |
Stoeck, T, Frühe, L, Forster, D, Cordier, T, Martins, CIM, and Pawlowski, J (2018). Environmental DNA metabarcoding of benthic bacterial communities indicates the benthic footprint of salmon aquaculture. Marine Pollution Bulletin 127, 139–149.
| Environmental DNA metabarcoding of benthic bacterial communities indicates the benthic footprint of salmon aquaculture.Crossref | GoogleScholarGoogle Scholar | 29475645PubMed |
Thomsen, PF, and Willerslev, E (2015). Environmental DNA – an emerging tool in conservation for monitoring past and present biodiversity. Biological Conservation 183, 4–18.
| Environmental DNA – an emerging tool in conservation for monitoring past and present biodiversity.Crossref | GoogleScholarGoogle Scholar |
United Nations (2019) Global indicator framework for the sustainable development goals and targets of the 2030 Agenda for sustainable development. Available at https://unstats.un.org/sdgs/indicators/Global%20Indicator%20Framework%20after%202021%20refinement_Eng.pdf
Visby I, Weller-Wong A (2020) Derwent Estuary recreational water quality program. Annual report 2019–2020. (Derwent Estuary Program) Available at https://www.derwentestuary.org.au/assets/2019-20_End_of_season_RWQ_report_2.pdf
Wang, Q, Garrity, GM, Tiedje, JM, and Cole, JR (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology 73, 5261–5267.
| Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy.Crossref | GoogleScholarGoogle Scholar | 17586664PubMed |
Wang, H, Boutton, TW, Xu, W, Hu, G, Jiang, P, and Bai, E (2015). Quality of fresh organic matter affects priming of soil organic matter and substrate utilization patterns of microbes. Scientific Reports 5, 10102.
| Quality of fresh organic matter affects priming of soil organic matter and substrate utilization patterns of microbes.Crossref | GoogleScholarGoogle Scholar | 25960162PubMed |
Weisburg, WG, Barns, SM, Pelletier, DA, and Lane, DJ (1991). 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology 173, 697–703.
| 16S ribosomal DNA amplification for phylogenetic study.Crossref | GoogleScholarGoogle Scholar | 1987160PubMed |
Wickham, H (2011). ggplot2. Wiley Interdisciplinary Reviews: Computational Statistics 3, 180–185.
| ggplot2.Crossref | GoogleScholarGoogle Scholar |
Yilmaz, P, Parfrey, LW, Yarza, P, Gerken, J, Pruesse, E, Quast, C, Schweer, T, Peplies, J, Ludwig, W, and Glöckner, FO (2014). The SILVA and ‘all-species living tree project (LTP)’ taxonomic frameworks. Nucleic Acids Research 42, D643–D648.
| The SILVA and ‘all-species living tree project (LTP)’ taxonomic frameworks.Crossref | GoogleScholarGoogle Scholar | 24293649PubMed |