Molecular cloning and characterisation of an acyl carrier protein thioesterase gene (CocoFatB1) expressed in the endosperm of coconut (Cocos nucifera) and its heterologous expression in Nicotiana tabacum to engineer the accumulation of different fatty acids
Yijun Yuan A B , Yinhua Chen B , Shan Yan A , Yuanxue Liang A , Yusheng Zheng A C and Li Dongdong A CA Department of Biotechnology, Hainan University, Haikou, Hainan 570228, China.
B Hainan Key Laboratory for Sustainable Utilisation of Tropical Bioresource, Hainan University, Haikou, Hainan 570228, China.
C Corresponding authors. Emails: liddfym@hotmail.com; hainanzyh@yahoo.com.cn
Functional Plant Biology 41(1) 80-86 https://doi.org/10.1071/FP13050
Submitted: 6 March 2013 Accepted: 31 May 2013 Published: 8 July 2013
Abstract
Coconut (Cocos nucifera L.) contains large amounts of medium chain fatty acids, which mostly recognise acyl-acyl carrier protein (ACP) thioesterases that hydrolyse acyl-ACP into free fatty acids to terminate acyl chain elongation during fatty acid biosynthesis. A full-length cDNA of an acyl-ACP thioesterase, designated CocoFatB1, was isolated from cDNA libraries prepared from coconut endosperm during fruit development. The gene contained an open reading frame of 1254 bp, encoding a 417-amino acid protein. The amino acid sequence of the CocoFatB1 protein showed 100% and 95% sequence similarity to CnFatB1 and oil palm (Elaeis guineensis Jacq.) acyl-ACP thioesterases, respectively. Real-time fluorescent quantitative PCR analysis indicated that the CocoFatB1 transcript was most abundant in the endosperm from 8-month-old coconuts; the leaves and endosperm from 15-month-old coconuts had ~80% and ~10% of this level. The CocoFatB1 coding region was overexpressed in tobacco (Nicotiana tabacum L.) under the control of the seed-specific napin promoter following Agrobacterium tumefaciens-mediated transformation. CocoFatB1 transcript expression varied 20-fold between different transgenic plants, with 21 plants exhibiting detectable levels of CocoFatB1 expression. Analysis of the fatty acid composition of transgenic tobacco seeds showed that the levels of myristic acid (14 : 0), palmitic acid (16 : 0) and stearic acid (18 : 0) were increased by 25%, 34% and 17%, respectively, compared with untransformed plants. These results indicated that CocoFatB1 acts specifically on 14 : 0-ACP, 16 : 0-ACP and 18 : 0-ACP, and can increase medium chain saturated fatty acids. The gene may valuable for engineering fatty acid metabolism in crop improvement programmes.
Additional keywords: medium chain fatty acids, myristic acid, palmitic acid, stearic acid, transgenic tobacco.
Introduction
Coconut (Cocos nucifera L.), a member of the monocotyledonous family Aracaceae (Palmaceace), is an important oil-yielding plantation crop that is of considerable economic and social importance in the tropics (Rivera et al. 1999; Samsudeen et al. 2006). The endosperm tissue of coconut stores a substantial amount of oil, which has been used extensively for human consumption and other purposes all over the world. In physicochemical terms, coconut oil differs from other vegetable oils in that it is rich in saturated oil (≈93%), with a high percentage of medium chain fatty acids (MCFA) (≈60%), especially lauric acid (12 : 0) (≈50%) (Ceniza et al. 1991; Bhatnagar et al. 2009). Compared with long-chain triacylglycerols, medium-chain triacylglycerols that contain caprylic acid (8 : 0), capric acid (10 : 0), lauric acid (12 : 0) and myristic acid (14 : 0) are more soluble in water and have a lower melting point, enabling them to be absorbed and metabolised faster (Jeukendrup and Sarah 2004; Beermann et al. 2007). In addition, the unique antiviral, antibacterial and antiprotozoal properties of medium-chain triacylglycerols have found applications in the food industry (Enig 1998), and MCT oils have been used therapeutically since the 1950s. The use of MCT oils as part of a ketogenic diet to treat epilepsy, premature infants and patients infected with human immunodeficiency virus, as well as to prevent fat malabsorption in cystic fibrosis patients, is widely accepted (Ramírez et al. 2001; Beermann et al. 2007). The potential industrial and medical applications of uncommon seed oils have resulted in rapid advances in efforts to bioengineer their accumulation.
The mechanisms by which coconut endosperm accumulates unusual fatty acids are still unknown. Fatty acid biosynthesis in higher plants occurs predominantly in the plastids by a de novo iterative ‘polymerisation’ process, which is commonly primed with the acetyl moiety from acetyl-CoA and proceeds via iterative chain extension through reaction with malonyl- acyl carrier protein (ACP). The synthesis of 16- and 18-carbon (C16 and C18) fatty acids is terminated by the acyl-ACP thioesterase, which catalyses acyl-ACP thioester bond hydrolysis, the terminal reaction of fatty acid biosynthesis that releases a free fatty acid and ACP (Voelker 1996; Jing et al. 2011). Therefore, the specificities of thioesterases largely determine the chain lengths of most plant fatty acids (Stumpf 1987).
Acyl-ACP thioesterases are nuclear-encoded, plastid-targeted globular proteins (Voelker et al. 1992). Based on amino acid sequence alignments, these enzymes have been functionally characterised and classified into two general families, termed FatA and FatB (Jones et al. 1995). All FatAs are orthologous in different species, with the highest activities towards oleoyl–ACP (18 : 1 9-ACP) (Hitr and Yadav 1992; Sánchez-García et al. 2010). In contrast with the high level of conservation in the specificity of FatAs, FatBs primarily hydrolyse saturated acyl-ACPs with chain lengths that contain between 8 and 18 carbons (Voelker and Davies 1994; Jones et al. 1995; Jha et al. 2010). The first FatB gene was isolated from the developing seeds of California Bay Tree (Umbellularia californica), and the strong preference of the enzyme for 12 : 0-ACP was verified in Arabidopsis thaliana (L.) Heynh. (Davies et al. 1991; Voelker et al. 1992). This work demonstrated, for the first time, the role of FatB in determing the chain lengths of fatty acids, which spurred efforts to isolate similar enzymes from other plant species with unusual fatty acid phenotypes. Such enzymes included the MCFA-specific thioesterases from Cuphea (Dehesh et al. 1996; Leonard et al. 1997), American elm (Ulmus americana L.; Voelker et al. 1997) and coconut (Jing et al. 2011). Because of the potential applications of special seed oils, several studies have focussed on engineering plant thioesterases with medium-chain-specificities. Three acyl-ACP thioesterases (CnFatB1 (JF338903), CnFatB2 (JF338904), CnFatB3 (JF338905)) from coconut have been isolated and characterised (Jing et al. 2011). However, their in vivo activities and substrate specificities were only shown in Escherichia coli, with no function analyses performed using plants. In the present work, a full-length cDNA of an acyl-ACP thioesterase (CocoFatB1: JX275886) was isolated from cDNA libraries prepared from coconut endosperm during fruit development (Li and Fan 2009). The CocoFatB1 gene was heterologously expressed in transgenic tobacco (Nicotiana tabacum L.) under the control of the seed-specific napin promoter (Kridl et al. 1991; Zheng et al. 2007). Our results provide new insights into the function of CocoFatB1 and how it might be used to impact on the composition of plant oils.
9-ACP) (Hitr and Yadav 1992; Sánchez-García et al. 2010). In contrast with the high level of conservation in the specificity of FatAs, FatBs primarily hydrolyse saturated acyl-ACPs with chain lengths that contain between 8 and 18 carbons (Voelker and Davies 1994; Jones et al. 1995; Jha et al. 2010). The first FatB gene was isolated from the developing seeds of California Bay Tree (Umbellularia californica), and the strong preference of the enzyme for 12 : 0-ACP was verified in Arabidopsis thaliana (L.) Heynh. (Davies et al. 1991; Voelker et al. 1992). This work demonstrated, for the first time, the role of FatB in determing the chain lengths of fatty acids, which spurred efforts to isolate similar enzymes from other plant species with unusual fatty acid phenotypes. Such enzymes included the MCFA-specific thioesterases from Cuphea (Dehesh et al. 1996; Leonard et al. 1997), American elm (Ulmus americana L.; Voelker et al. 1997) and coconut (Jing et al. 2011). Because of the potential applications of special seed oils, several studies have focussed on engineering plant thioesterases with medium-chain-specificities. Three acyl-ACP thioesterases (CnFatB1 (JF338903), CnFatB2 (JF338904), CnFatB3 (JF338905)) from coconut have been isolated and characterised (Jing et al. 2011). However, their in vivo activities and substrate specificities were only shown in Escherichia coli, with no function analyses performed using plants. In the present work, a full-length cDNA of an acyl-ACP thioesterase (CocoFatB1: JX275886) was isolated from cDNA libraries prepared from coconut endosperm during fruit development (Li and Fan 2009). The CocoFatB1 gene was heterologously expressed in transgenic tobacco (Nicotiana tabacum L.) under the control of the seed-specific napin promoter (Kridl et al. 1991; Zheng et al. 2007). Our results provide new insights into the function of CocoFatB1 and how it might be used to impact on the composition of plant oils.
Materials and methods
Plant materials
Coconut (Cocos nucifera L.) fruits (i.e. coconuts), including immature coconuts (8 months old) and ripe coconuts (15 months old), and coconut leaves were obtained from the Coconut Research Institute, Chinese Agricultural Academy of Tropical Crops, Hainan, China. The fruits and leaves were harvested at random, and endosperm tissues were physically isolated and immediately frozen in liquid nitrogen to form three sample pools. The samples were then analysed in duplicate for RNA concentration, cDNA synthesis, gene amplification and differential expression analysis. Escherichia coli strain DH5α (Clontech Palo Alto, CA, USA), which was grown in Lysogeny broth (LB) medium (Sangon, Shanghai, China) supplemented with 50 mg L–1 ampicilin or kanamycin at 37°C,was used for bacterial cloning. The pCAMBIA1300S vector was donated by Yongjun Lin (Professor of Huazhong Agricultural University). All chemicals, endonucleases and other required enzymes were obtained from Sigma (St. Louis, MO, USA) or TaKaRa (Dalian, China), unless otherwise stated.
RNA extraction and cDNA library construction
Total RNA from the pulp of coconuts was extracted using the cetyltrimethylammonium bromide-based methods described by Li and Fan (2007). The quantity and quality of isolated total RNA was examined using spectrophotometry and gel electrophoresis, respectively. Construction of the cDNA library prepared from RNA isolated from coconut endosperm have been reported previously (Li and Fan 2009).
Cloning of the CocoFatB1 gene and bioinformatic analysis
EST sequence information was obtained for 1000 clones that were randomly selected from the cDNA library. All clones were sequenced by Oebiotech Co. (Shanghai, China). A homology search was conducted based on BLAST searches using the National Center for Biotechnology Information BLAST server (http://www.ncbi.nlm.nih.gov/BLAST). Among the EST sequences obtained from the library, a partial clone was discovered as being 91% identical to the palmitoyl-acyl carrier protein thioesterase gene from oil palm (GenBank accession: AF147879.2). Complete sequencing of the clone was performed using the primers M13+ (5′-GTAAAACGACGGCCAGT-3′) and M13– (5′-AACAGCTATGACCATGTTCA-3′) followed by primer-walking until complete overlapping sequence data were obtained from both sides.
Fluorescence quantitative reverse transcription–PCR analysis
Total RNA from coconut leaves and endosperm tissues were isolated separately from immature coconuts (8 months old) and ripe coconuts (15 months old). First-strand cDNA was synthesised from 2 µg of total RNA using the TIANScript OneStep RT-PCR Kit (TIANGEN, Beijing, China). Reverse transcription was performed at 42°C for 60 min, with a final denaturation at 70°C for 15 min. The cDNA was then subjected to real-time fluorescent quantitative reverse transcription–PCR (RT-PCR) using standard methods (Marone et al. 2001). The RT-PCR primers for CocoFatB1 were designed using the Primer3 program based on the cDNA sequence. The β-actin gene was used as an internal control for expression. The primers used in this study were:
-
RTActin-F: 5′-TTACTCTGAAATACCCCATTGAGC-3′,
-
RTActin-R: 5′-CTCTCTGTTAGCCTTGGGGTTG-3′,
-
RTFatBF: 5′-ACACTTCTTGATTGGAAACCACG-3′, and
-
RTFatBR: 5′-GCGTTTCTATAGAAGCCGTCC-3′.
The RT-PCR amplification step was performed using the SYBR Premix Ex Taq II (TaKaRa) and a RT-PCR detector (TaKaRa Smart Cycler II system) by using the SYBR Green I chimeric fluorescence method according to the manufacturer’s instructions. Expression was quantified in terms of comparative threshold cycle (Ct) using the 2–
 Ct method, and the results were expressed as the binary logarithm of the relative quantity of the transcript used to normalise gene expression (Livak and Schmittgen 2001). Reactions were performed in triplicate, including the ‘no template’ and ‘no reverse transcriptase’ controls, and were monitored using an Applied Biosystems (Foster City, CA, USA) 7500 RT-PCR instrumentation outfitted with SDS software ver. 1.3.1 (Applied Biosystems).
Ct method, and the results were expressed as the binary logarithm of the relative quantity of the transcript used to normalise gene expression (Livak and Schmittgen 2001). Reactions were performed in triplicate, including the ‘no template’ and ‘no reverse transcriptase’ controls, and were monitored using an Applied Biosystems (Foster City, CA, USA) 7500 RT-PCR instrumentation outfitted with SDS software ver. 1.3.1 (Applied Biosystems).
Construction of expression vectors
The CocoFatB1 coding sequence was cloned from cDNA prepared from coconut endosperm, and KpnI and BamHI sites were added at the 5′ and 3′ ends, respectively. The primers used for the PCR amplification were: FatBF 5′-TATGGTACCATGGTTGCTTCAGTTGCCGCTT-3′ (forward) and FatBR 5′-TATGGATCCTCAAGCACTTCCAGCTGAAGTGG-3′ (reverse). The conditions for PCR amplification were 94°C for 3 min, 30 cycles of 94°C for 45 s, 47°C for 45 s, 72°C for 1 min and extension at 72°C for 8 min. The PCR product was cloned into trhe pMD18-T vector (TaKaRa) and the recombinant plasmid was transformed into the Escherichia coli strain DH5α. To generate the plant overexpression construct, the coding region of CocoFatB1 was subcloned into the binary pCAMBIA1300S vector (KpnI or BamHI site) under the control of the seed-specific napin promoter (Kridl et al. 1991; Fig. 1).

|
Transformation, selection and regeneration of Nicotiana tabacum
The CocoFatB1 gene in the pCAMBIA1300S expression vector was transformed into Agrobacterium tumefaciens LBA4404/EHA105 by electroporation, as described by Hoekema et al. (1983). Young leaves from wild-type tobacco (Nicotiana tabacum L.) were cut into small square discs (0.5 × 0.5 cm2) and immersed in 10× diluted cultures of Agrobacterium tumefaciens for 15 min. The leaf discs were first transferred to sterile filter paper to remove excess Agrobacterium, and then transferred onto solidified cocultivation MS medium (Sigma; Murashige and Skoog 1962). After cocultivation, the transformants were selected on MS medium containing 500 mg L–1 carbenicillin (Sigma) and 10 mg L–1 hygromycin B (Sigma). Regenerated tobacco plants propagated in vitro were transferred to soil and grown to maturity in a greenhouse in a 16-h-light ; 8-h-darkness photoperiod with a PPFD of 200–900 μmol m–2 s–1. Positive transformants were identified by PCR screening of genomic DNA. Primary transformants were self-fertilised and the seeds were collected.
Analysis of CocoFatB1 expression in transgenic tobacco using fluorescence quantitative RT-PCR
To detect the introduced transgene with PCR in the putative regenerated transgenic plants, DNA samples were extracted from leaf tissue using the DNAeasy Mini-kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The FatBF–FatBR primer pair was used for PCR. The PCR conditions were identical to those described above, and the PCR products were then electrophoresed on a 1.0% (w/v) agarose gel and visualised under ultraviolet light. Positive transformants that yielded a single PCR product (~1200 bp) were selected for future investigation.
Total RNA from each of the mature tobacco seeds was isolated as described above. First-strand cDNA synthesis and fluorescent quantitative RT-PCR were carried out as described above using the RTFatB-F (5′-GTAGCCAAACCCACCTCT-3′) and RTFatB-R (5′-TTTCAGCCCAACCTTCG-3′) primers, which were used to detect the expression level of CocoFatB1 in the seeds of transgenic plants. Transcripts of the 18S gene, which was used as an internal control for expression, were amplified using the RT18S-F (5′-GCAACAAACCCCGACTTCT-3′) and RT18S-R (5′-GCGATCCGTCGAGTTATCAT-3′) primers.
Fatty acid methyl ester analysis by GC
Total lipids were extracted in triplicate using dichloromethane : methanol (2 : 1) from mature seeds from single plantlets of the transgenic and wild-type tobacco plants. The fatty acid methyl esters were recovered using N-hexane. Analysis of fatty acid methyl esters was performed using GC, with methyl heptadecanoate (17 : 0) (Sigma) as an internal standard. All GC analysis was performed using a HP5890 GC instrument equipped with a BPX-70 (30 m × 0.25 mm) chromatography column (SGE, Melbourne, Vic., Australia). The initial column temperature (90°C) was held for 10 min and then raised at 4°C min–1 until it reached 240°C, after which it was held at this temperature for another 10 min (Beermann et al. 2007; Chi et al. 2011).
Statistical analysis
All experiments were performed in triplicate, and the data provided are means ± s.d. Intergroup comparisons between the two tested groups were performed using a paired t-test, using Statview ver. 6.0 software (SAS Institute, Cary, NC, USA) and Microsoft Office Excel ver. 2007 (Microsoft Corporation, Richmond, WA, USA). A P-value of <0.05 was regarded as indicating a statistically significant difference.
Results
Complementary DNA cloning and conservation analysis of the CocoFatB1 sequence
The full-length cDNA sequence (1858 bp) of an acyl-ACP thioesterase was isolated (termed CocoFatB1; GenBank accession: JX275886) from total RNA from the coconut endosperm using EST sequences and RT-PCR. Sequence analysis revealed that the CocoFatB1 sequence was homologous to other acyl-ACP thioesterases and the predicted protein has similar properties to previously identified orthologues and is homologous across its entire length. CocoFatB1 has 73% amino acid identity with the rice enzyme (Os06 g0143400).
Expression of CocoFatB1 in different tissues and at different development stages
To reveal the expression of CocoFatB1 genes during the development of coconut pulp, fluorescence quantitative RT-PCR was used to analyse CocoFatB1 expression during two different developmental stages: 8-month-old and 15-month-old coconut fruits, with the abundance of β-actin transcripts providing an internal control. The CocoFatB1 transcript was most abundant in the endosperm from 8-month-old coconuts, whereas the leaves and endosperm from 15-month-old coconuts had ~80% and ~10% of this level.
Generation of transgenic tobacco plants expressing the CocoFatB1 gene under the control of the napin promoter
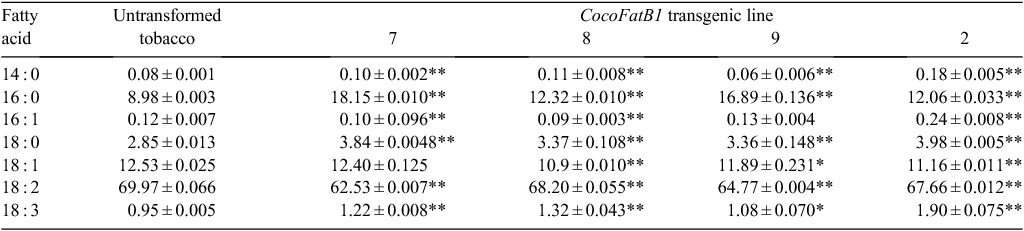
To further determine the function of CocoFatB1 and to establish whether its expression can change the fatty acid profile in plants, we investigated the effects of CocoFatB1 expression in transgenic tobacco. Analysis of the transgenic plants’ genomic DNA using PCR indicated the presence of the CocoFatB1 coding sequence in the tobacco genome. Following transformation and selection in the presence of hygromycin, 32 independent transgenic plants were obtained. CocoFatB1 transcript expression varied 20-fold between different transgenic plants, with 21 plants exhibiting detectable levels of CocoFatB1 expression (Fig. 2). Four transformant lines (7, 8, 9 and 2) that showed different levels of CocoFatB1 transcript were selected for further analysis.

|
Analysis of fatty acid composition
As an acyl-ACP thioesterase, CocoFatB1 was expected to increase the medium chain saturated fatty acid composition of tissues in which it is expressed. To further confirm its function in vivo, the fatty acid composition of transgenic and untransformed tobacco plants were determined and compared. Expression of transgenic CocoFatB1 in tobacco seed increased levels of myristic acid (14 : 0), palmitic acid (16 : 0) and stearic acid (18 : 0) by 10.2%, 3.9%, 8.4% and 4.3% in Plants 7, 8, 11 and 15 respectively. Meanwhile, there was no obvious difference in the levels of other fatty acids when the seeds of transgenic and untransformed tobacco plants were compared (Table 1).
Discussion
Plant fatty acids are synthesised in the stroma of the plastids of both leaves and developing seeds (Weaire and Kekwick 1975; Ohlrogge et al. 1979). Accordingly, CocoFatB1 transcripts are detected not only in the endosperm, but also in the leaves of coconut plants. Thioesterases play a pivotal role in fatty acid synthesis owing to their role in catalysing the terminal reaction of fatty acid biosynthesis, which regulates the fatty acid composition of storage lipids, especially in plant seeds (Brown et al. 2010; Jing et al. 2011). The expression of thioesterase genes displayed the highest levels in expanding tissues that are typically very active in lipid biosynthesis, such as developing seed endosperm and young expanding leaves (Oo and Stumpf 1979; Sánchez-García et al. 2010).
To confirm the activity and substrate specificity of CocoFatB1 in plants, we analysed the effects of its expression in transgenic tobacco. This result indicated that CocoFatB1 showed specificity towards 14 : 0-ACP, 16 : 0-ACP and 18 : 0-ACP. Compared with the results of function analysis of CnFatB1, CocoFatB1 is specific not only towards 14 : 0-ACP and 16 : 0-ACP, which have been demonstrated in E. coli by Jing et al. (2011), but also showed specificity to 18 : 0-ACP in plant. These results are similar to those previously reported for FatB thioesterases from other plants, such as Elaeis guineensis (Othman et al. 2000), Jatropha curcas L. (Wu et al. 2009), Cuphea hookeriana (Jones et al. 1995), Diploknema (Madhuca) butyracea (Jha et al. 2006) and Indian mustard (Brassica juncea L. Czern.; Jha et al. 2010). All of these enzymes displayed a high level of activity towards 16 : 0-ACP; BjFatBs from B. juncea were also specific to 18 : 0-ACP. Moreover, like other FatB thioesterases, which preferably hydrolyse acyl-ACPs with saturated fatty acid chains (Jones et al. 1995), CocoFatB1 showed a preference for saturated acyl-ACPs, especially palmitoyl-ACP.
However, compared with some FatBs from other plants that contain rich MCFAs, CocoFatB1 still displayed some unique characteristics. The FatBs from U. californica (Pollard et al. 1991; Voelker et al. 1992), A. thaliana (Dormann et al. 1995), U. americana (Voelker et al. 1997) and nutmeg (Myristica fragrans; Voelker et al. 1997) are specific for 12 : 0-ACP and play a critical role in MCFA production. Unlike these FatBs, CocoFatB1 shows a preference for 14 : 0-ACP, 16 : 0-ACP and 18 : 0-ACP. Recently, CnFatB3 has been demonstrated to be specific for 12 : 0-ACP, 14 : 0-ACP and 14 : 1-ACP in E. coli (Jing et al. 2011), which may make a great contribution to fatty acid profiles containing abundant MCFAs in coconut endosperm. More functional analysis is needed to confirm the characterisation of CnFatB3, especially in plants. Meanwhile, some crucial enzymes involved in fatty acid synthesis may also play an important role in determining the lengths of fatty acid chains.
Although the specificities of thioesterases determine the chain length of most plant fatty acids to a large extent, the action of specific β-ketoacyl-ACP synthases (KAS) and acyl-ACP acyltransferases shift the synthesis of fatty acids towards molecules with shorter chains (Davies et al. 1995; Leonard et al. 1998). Of three known classes of plant KAS enzymes, only KASI elongates substrates from 4 : 0-ACP to 14 : 0-ACP (Shimakata and Stumpf 1983). Seeds transformed with CwKASA and CwFatB2 thioesterases in comparison with the seeds transformed with thioesterases only had greatly increased concentrations of 10 : 0 (capric acid) and 12 : 0 (lauric acid). Coexpression of CwKASA with California bay FatB1 in transgenic canola (Brassica napus L.) increased amounts of 12 : 0 fatty acids when compared with expression of the FatB1 only (Leonard et al. 1998). Additionally, expression of the LPAAT gene of coconut endosperm in E. coli and canola indicates that the enzyme displays a marked preference for the transfer of medium-chain CoAs to 12 : 0-lysophosphatidic acid relative to unsaturated long-chain substrates(Wiberg et al. 1997). Co-expressed with thioesterase, lysophosphatidyl acyltransferase (LPAAT) expression generated triacylglycerol (TAGs) with high levels of lauric acid (12 : 0) at sn-2 (Davies et al. 1995; Knutzon et al. 1999; Wiberg et al. 2000).
The aim of this study was to explore why coconut endosperm is so rich in MCFAs and to identify one of the genes responsible for this phenotype. The CocoFatB1 gene we have isolated and characterised was highly effective in redirecting plant fatty acid synthases to palmitate and myristate production, and appears to be specific to 14 : 0-ACP, 16 : 0-ACP and 18 : 0-ACP. The ectopic expression of C. nucifera CocoFatB1 in N. tobaccum increased the levels of saturated acids, including myristic acid, palmitic acid and stearic acid, to provide a fatty acid profile distinct from that of other oil crops that produce large amounts of MCFAs, such as U. californica (Pollard et al. 1991; Voelker et al. 1992). Based on the previous studies, more detailed research about the actions of the thioesterases, KAS and LPAAT from coconut endosperm is needed to confirm the function of these crucial enzymes in fatty acid synthesis. Co-expression of special thioesterases with KASI or LPAAT in plants should enable the effects of the various enzymes on fatty acid composition to be separated. The ability to engineer the accumulation of these MCFAs, especially lauric acid (12 : 0), will be beneficial in efforts to improve crops by engineering fatty acid metabolism.
Acknowledgements
YY and YC contributed equally to the work. This research was supported by the National Natural Science Foundation of China (No.: 31160171, 31060259 and 31260193) and Youth Foundation of Hainan University (No.: QNJJ1001).
References
Beermann C, Winterling N, Green A, Mobius M, Schmitt JJ, Boehm G (2007) Comparison of the structures of triacylglycerols from native and transgenic medium-chain fatty acid-enriched rape seed oil by liquid chromatography-atmospheric pressure chemical ionization ion-trap mass spectrometry (LC-APCI-ITMS). Lipids 42, 383–394.| Comparison of the structures of triacylglycerols from native and transgenic medium-chain fatty acid-enriched rape seed oil by liquid chromatography-atmospheric pressure chemical ionization ion-trap mass spectrometry (LC-APCI-ITMS).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXkvFCmtLk%3D&md5=bb41d57eb4da05e2e50352990471f88dCAS | 17406932PubMed |
Bhatnagar AS, Prasanth Kumar PK, Hemavathy J, Gopala Krishna AG (2009) Fatty acid composition, oxidative stability, and radical scavenging activity of vegetable oil blends with coconut oil. Journal of the American Oil Chemists’ Society 86, 991–999.
| Fatty acid composition, oxidative stability, and radical scavenging activity of vegetable oil blends with coconut oil.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtFaqt77P&md5=9dd08726bf45c15354562ffab7396686CAS |
Brown AP, Slabas AR, Rafferty JB (2010) ‘Lipids in photosynthesis: essential and regulatory functions.’ (Springer:Dordrecht, The Netherlands)
Ceniza MS, Ueda S, Sugimura Y (1991) In vitro culture of coconut endosperm: callus induction and its fatty acids. Plant Cell Reports 11, 546–549.
| In vitro culture of coconut endosperm: callus induction and its fatty acids.Crossref | GoogleScholarGoogle Scholar |
Chi X, Yang Q, Pan L, Chen M, He Y, Yang Z, Yu S (2011) Isolation and characterization of fatty acid desaturase genes from peanut (Arachis hypogaea L.). Plant Cell Reports 30, 1393–1404.
| Isolation and characterization of fatty acid desaturase genes from peanut (Arachis hypogaea L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXovVWgtr4%3D&md5=3c8ba0b90d82357f316117a0fd5cc6f3CAS | 21409552PubMed |
Davies HM, Anderson L, Fan C, Hawkins DJ (1991) Developmental induction, purification, and further characterization of 12 : 0-ACP thioesterase from immature cotyledons of Umbellularia californica. Archives of Biochemistry and Biophysics 290, 37–45.
| Developmental induction, purification, and further characterization of 12 : 0-ACP thioesterase from immature cotyledons of Umbellularia californica.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK38Xis1Sh&md5=01f526da3737650c1bafb8dce2571ecdCAS | 1898097PubMed |
Davies HM, Hawkins DJ, Nelsen JS (1995) Lysophosphatidic acid acyltransferase from immature coconut endosperm having medium chain length substrate specificity. Phytochemistry 39, 989–996.
| Lysophosphatidic acid acyltransferase from immature coconut endosperm having medium chain length substrate specificity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXnsFSnsro%3D&md5=0aaddf4c5d01023299eb34cde042d8e3CAS |
Dehesh K, Jones A, Knutzon DS, Voelker TA (1996) Production of high levels of 8 : 0 and 10 : 0 fatty acids in transgenic canola by overexpression of ChFatB2, a thioesterase cDNA from Cuphea hookeriana. The Plant Journal 9, 167–172.
| Production of high levels of 8 : 0 and 10 : 0 fatty acids in transgenic canola by overexpression of ChFatB2, a thioesterase cDNA from Cuphea hookeriana.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK28XitVyjur8%3D&md5=d2ca325a8e2996e7db538fb00abb1236CAS | 8820604PubMed |
Dormann P, Voelker TA, Ohlrogge JB (1995) Cloning and expression in Escherichia coli of a novel thioesterase from Arabidopsis thaliana specific for long-chain acyl-acyl carrier proteins. Archives of Biochemistry and Biophysics 316, 612–618.
| Cloning and expression in Escherichia coli of a novel thioesterase from Arabidopsis thaliana specific for long-chain acyl-acyl carrier proteins.Crossref | GoogleScholarGoogle Scholar | 1:STN:280:DyaK2M7kt1KrtA%3D%3D&md5=f1f7f4a871309304102b79080f9dc484CAS | 7840673PubMed |
Enig MG (1998) Lauric oils as antimicrobial agents: theory of effect, scientific rationale, and dietary applications as adjunct nutritional support for HIV-infected individuals. In ‘Nutrients and foods in AIDS’. (Ed RR Watson) pp. 81–87. (CRC Press, Boca Raton)
Hitr WD, Yadav NS (1992) ‘Nucleotide sequences of soybean acyl-ACP thioesterase genes. International Patent WO 92/11373.’ (Wilmington Del)
Hoekema A, Hirsch P, Hooykaas PJ, Schilperoort R (1983) A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180.
| A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3sXktVWhsrY%3D&md5=353167e3076e43eef8a5aeb1994d330bCAS |
Jeukendrup AE, Sarah A (2004) Fat supplementation, health, and endurance performance. Nutrition 20, 678–688.
| Fat supplementation, health, and endurance performance.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXltVGitbk%3D&md5=71ed9ba15c7aa3d31723873bc626f579CAS | 15212751PubMed |
Jha JK, Maiti MK, Bhattacharjee A, Basu A, Sen PC, Sen SK (2006) Cloning and functional expression of an acyl-ACP thioesterase FatB type from Diploknema (Madhuca) butyracea seeds in Escherichia coli. Plant Physiology and Biochemistry 44, 645–655.
| Cloning and functional expression of an acyl-ACP thioesterase FatB type from Diploknema (Madhuca) butyracea seeds in Escherichia coli.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXhsl2jtQ%3D%3D&md5=4ec6e2b5b30caa24b730c86c833108a6CAS | 17092734PubMed |
Jha SS, Jha JK, Chattopadhyaya B, Basu A, Sen SK, Maiti MK (2010) Cloning and characterization of cDNAs encoding for long-chain saturated acyl-ACP thioesterases from the developing seeds of Brassica juncea. Plant Physiology and Biochemistry 48, 476–480.
| Cloning and characterization of cDNAs encoding for long-chain saturated acyl-ACP thioesterases from the developing seeds of Brassica juncea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3cXlvVeisrc%3D&md5=cdde5d262c7ae3be68236a3ab69a7c1dCAS | 20356753PubMed |
Jing F, Cantu DC, Tvaruzkova J, Chipman JP, Nikolau BJ, Yandeau-Nelson MD, Reilly PJ (2011) Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochemistry 12, 44–59.
| Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BC3MXhtFKms7rJ&md5=31103d41c4112054c3c7ef0116a15d89CAS | 21831316PubMed |
Jones A, Davies HM, Voelker TA (1995) Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases. The Plant Cell 7, 359–371.
| Palmitoyl-acyl carrier protein (ACP) thioesterase and the evolutionary origin of plant acyl-ACP thioesterases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2MXks1Chsb0%3D&md5=5d0633b302fc912cc7fa158f4bb9beeaCAS | 7734968PubMed |
Knutzon DS, Hayes TR, Wyrick A, Xiong H, Maelor Davies H, Voelker TA (1999) Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn-2 position of triacylglycerols in lauric rapeseed oil and can increase total laurate levels. Plant Physiology 120, 739–746.
| Lysophosphatidic acid acyltransferase from coconut endosperm mediates the insertion of laurate at the sn-2 position of triacylglycerols in lauric rapeseed oil and can increase total laurate levels.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1MXks1amtLg%3D&md5=433848a6085b5f1da6de9af65e0192cdCAS | 10398708PubMed |
Kridl JC, McCarter DW, Rose RE, Scherer DE, Knutzon DS, Radke SE, Knauf VC (1991) Isolation and characterization of an expressed napin gene from Brassica rapa. Seed Science Research 1, 202–219.
| Isolation and characterization of an expressed napin gene from Brassica rapa.Crossref | GoogleScholarGoogle Scholar |
Leonard JM, Slabaugh MB, Knapp SJ (1997) Cuphea wrightii thioesterases have unexpected broad specificities on saturated fatty acids. Plant Molecular Biology 34, 669–679.
| Cuphea wrightii thioesterases have unexpected broad specificities on saturated fatty acids.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXltFertLw%3D&md5=c27e7e09ecbf03b49d28ef092ffec478CAS | 9247548PubMed |
Leonard JM, Knapp SJ, Slabaugh MB (1998) A Cuphea beta-ketoacyl-ACP synthase shifts the synthesis of fatty acids towards shorter chains in Arabidopsis seeds expressing Cuphea FatB thioesterases. The Plant Journal 13, 621–628.
| A Cuphea beta-ketoacyl-ACP synthase shifts the synthesis of fatty acids towards shorter chains in Arabidopsis seeds expressing Cuphea FatB thioesterases.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK1cXisVKgtLg%3D&md5=9f271d0f99e27942a25692857a4b4405CAS | 9681004PubMed |
Li DD, Fan YM (2007) Extraction and quality analysis of total RNA from pulp of coconut (Cocos nucifera L.). Molecular Plant Breeding 5, 883–886. [In Chinese]
Li DD, Fan YM (2009) Cloning, characterization, and expression analysis of an oleosin gene in coconut (Cocos nucifera L.) pulp. The Journal of Horticultural Science & Biotechnology 84, 483–488.
Livak K, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2–

 Ct) method. Methods 25, 402–408.
Ct) method. Methods 25, 402–408.| Analysis of relative gene expression data using real-time quantitative PCR and the 2–

 Ct) method.Crossref |
Ct) method.Crossref |  Ct) method.&journal=Methods&volume=25&pages=402-408&publication_year=2001&author=K%20Livak&hl=en&doi=10.1006/meth.2001.1262" target="_blank" rel="nofollow noopener noreferrer" class="reftools">GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XhtFelt7s%3D&md5=cda22da73d6034ad2e4c74827f633baaCAS | 11846609PubMed |
Ct) method.&journal=Methods&volume=25&pages=402-408&publication_year=2001&author=K%20Livak&hl=en&doi=10.1006/meth.2001.1262" target="_blank" rel="nofollow noopener noreferrer" class="reftools">GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XhtFelt7s%3D&md5=cda22da73d6034ad2e4c74827f633baaCAS | 11846609PubMed | Marone M, Mozzetti S, De Ritis D, Pierelli L, Scambia G (2001) Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biological Procedures Online 3, 19–25.
| Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XjsFWrtQ%3D%3D&md5=93637584cbaf884926cfacd7601e919cCAS | 12734582PubMed |
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Plant Physiology 115, 493–497.
| A revised medium for rapid growth and bioassays with tobacco tissue cultures.Crossref | GoogleScholarGoogle Scholar |
Ohlrogge JB, Kuhn DN, Stumpf PK (1979) Subcelluar localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proceedings of the National Academy of Sciences of the United States of America 76, 1194–1198.
| Subcelluar localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE1MXhvVeitbo%3D&md5=7c27e1a35b7e056642aabf6d12143728CAS | 286305PubMed |
Oo KC, Stumpf PK (1979) Fatty acid biosynthesis in the developing endosperm of Cocos nucifera. Lipids 14, 132–143.
| Fatty acid biosynthesis in the developing endosperm of Cocos nucifera.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaE1MXhsVymsb8%3D&md5=eb4f088bbde96245a746ded67ee217a7CAS |
Othman A, Lazarus C, Fraser T, Stobart K (2000) Cloning of a palmitoyl–acyl carrier protein thioesterase from oil palm. Biochemical Society Transactions 28, 619–622.
| Cloning of a palmitoyl–acyl carrier protein thioesterase from oil palm.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXhsVGrtbo%3D&md5=70df1e1d1308f8e29565160bffff6ecdCAS | 11171146PubMed |
Pollard MR, Anderson L, Fan C, Hawkins DJ, Davies HM (1991) A specific acyl-ACP thioesterase implicated in medium-chain fatty acid production in immature cotyledons of Umbellularia californica. Archives of Biochemistry and Biophysics 284, 306–312.
| A specific acyl-ACP thioesterase implicated in medium-chain fatty acid production in immature cotyledons of Umbellularia californica.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3MXnvVOhuw%3D%3D&md5=ce4bd110fd586a5df161067dcca6c344CAS | 1989513PubMed |
Ramírez M, Amate L, Gil A (2001) Absorption and distribution of dietary fatty acids from different sources. Early Human Development 65, S95–S101.
| Absorption and distribution of dietary fatty acids from different sources.Crossref | GoogleScholarGoogle Scholar | 11755040PubMed |
Rivera R, Edvards KJ, Barker JHA, Aranold GM, Ayad G, Hodgkin T (1999) Isolation and characterization of polymorphic microsatellites in Cocos nucifera L. Genome 42, 668–675.
Samsudeen K, Jacob PM, Niral V, Kumaran PM, Salooja R, Moosa H (2006) Exploration and collection of coconut germplasm in Kadmat and Amini Islands of Lakshadweep, India. Genetic Resources and Crop Evolution 53, 1721–1728.
| Exploration and collection of coconut germplasm in Kadmat and Amini Islands of Lakshadweep, India.Crossref | GoogleScholarGoogle Scholar |
Sánchez-García A, Moreno-Pérez AJ, Muro-Pastor AM, Salas JJ, Garcés R, Martínez-Force E (2010) Acyl-ACP thioesterases from castor (Ricinus communis L.): an enzymatic system appropriate for high rates of oil synthesis and accumulation. Phytochemistry 71, 860–869.
| Acyl-ACP thioesterases from castor (Ricinus communis L.): an enzymatic system appropriate for high rates of oil synthesis and accumulation.Crossref | GoogleScholarGoogle Scholar | 20382402PubMed |
Shimakata T, Stumpf PK (1983) Purification and characterization of beta-ketoacyl-ACP synthetase I from Spinacia oleracea leaves. Archives of Biochemistry and Biophysics 220, 39–45.
| Purification and characterization of beta-ketoacyl-ACP synthetase I from Spinacia oleracea leaves.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaL3sXosFamtg%3D%3D&md5=a8040c027be8eb306178127a8a35e361CAS | 6830245PubMed |
Stumpf PK (1987) The biosynthesis of saturated fatty acids. The Biochemistry of Plants 9, 121–136.
Voelker T (1996) Plant acyl-ACP thioesterases: chain-length determining enzymes in plant fatty acid biosynthesis. Genetic Engineering (NY) 18, 111–133.
Voelker TA, Davies HM (1994) Alteration of the specificity and regulation of fatty acid synthesis of Escherichia coli by expression of a plant medium-chain acyl-acyl carrier protein thioesterase. Journal of Bacteriology 176, 7320–7327.
Voelker TA, Worrell AC, Anderson L, Bleibaum J, Fan C, Hawkins DJ, Radke SE, Davies HM (1992) Fatty acid biosynthesis redirected to medium chain in transgenic oilseed plants. Science 257, 72–74.
| Fatty acid biosynthesis redirected to medium chain in transgenic oilseed plants.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK3sXksVGhtbs%3D&md5=7d0a4a17a770f684a67c2d7fcc93ef80CAS | 1621095PubMed |
Voelker TA, Jones A, Cranmer AM, Davies HM, Knutzon DS (1997) Broad-range and binary-range acyl-acyl-carrier-protein thioesterases suggest an alternative mechanism for medium-chain production in seeds. Plant Physiology 114, 669–677.
| Broad-range and binary-range acyl-acyl-carrier-protein thioesterases suggest an alternative mechanism for medium-chain production in seeds.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXktVWjsrY%3D&md5=35fcb64ba9ea428dd4de1ce3383f9a8dCAS | 9193098PubMed |
Weaire PJ, Kekwick RG (1975) The synthesis of fatty acids in avocado mesocarp and cauliflower bud tissue. Biochemical Journal 146, 425–437.
Wiberg E, Banas A, Stymne S (1997) Fatty acid distribution and lipid metabolism in developing seeds of laurate-producing rape (Brassica napus L.). Planta 203, 341–348.
| Fatty acid distribution and lipid metabolism in developing seeds of laurate-producing rape (Brassica napus L.).Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DyaK2sXnt1OitLs%3D&md5=4fbd3747a38bc9051b367197040ce2acCAS | 9431681PubMed |
Wiberg E, Edwards P, Byrne J, Stymne S, Dehesh K (2000) The distribution of caprylate, caprate and laurate in lipids from developing and mature seeds of transgenic Brassica napus L. Planta 212, 33–40.
| The distribution of caprylate, caprate and laurate in lipids from developing and mature seeds of transgenic Brassica napus L.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXosVers70%3D&md5=6160d0dae7cc47064570e44bdf24359aCAS | 11219581PubMed |
Wu PZ, Lin J, Wei Q, Zeng L, Chen YP, Li MR, Jiang HW, Wu GJ (2009) Cloning and functional characterization of an acyl-acyl carrier protein thioesterase (JcFATB1) from Jatropha curcas. Tree Physiology 29, 1299–1305.
| Cloning and functional characterization of an acyl-acyl carrier protein thioesterase (JcFATB1) from Jatropha curcas.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXhtlKqtLjK&md5=ee5715cb49a53e1b7b962e1b50d39498CAS | 19671567PubMed |
Zheng X, Deng W, Luo K, Duan H, Chen Y, McAvoy R, Song S, Pei Y, Li Y (2007) The cauliflower mosaic virus (CaMV) 35S promoter sequence alters the level and patterns of activity of adjacent tissue- and organ-specific gene promoters. Plant Cell Reports 26, 1195–1203.
| The cauliflower mosaic virus (CaMV) 35S promoter sequence alters the level and patterns of activity of adjacent tissue- and organ-specific gene promoters.Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2sXotFymtLg%3D&md5=a1971e74fd0a36e13265c15b15abcf47CAS | 17340093PubMed |