Plant miR397 and its functions
Shili Huang A , Jiajie Zhou A , Lei Gao A and Yulin Tang A B
A B
A Guangdong Provincial Key Laboratory for Plant Epigenetics, Shenzhen Key Laboratory of Marine Bioresource and Eco-environmental Science, Longhua Institute of Innovative Biotechnology, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen 518060, Guangdong province, China.
B Corresponding author. Email: yltang@szu.edu.cn
Functional Plant Biology - https://doi.org/10.1071/FP20342
Submitted: 31 October 2020 Accepted: 23 November 2020 Published online: 18 December 2020
Journal Compilation © CSIRO 2020 Open Access CC BY
Abstract
MicroRNAs (miRNAs) are noncoding, small RNAs of 20–24 nucleotides (nt) and function critically at the post-transcriptional level to regulate gene expression through cleaving mRNA targets or interfering with translation of the target mRNAs. They are broadly involved in many biological processes in plants. The miR397 family in plants contains several conserved members either in 21-nt or in 22-nt that mainly target the laccase (LAC) genes functioning in lignin synthesis and are involved in the development of plants under various conditions. Recent findings showed that miR397b in Arabidopsis could also target to Casein Kinase II Subunit Beta 3 (CKB3) and mediate circadian regulation and plant flowering. This review aims to summarise recent updates on miR397 and provides the available basis for understanding the functional mechanisms of miR397 in plant growth and development regulation and in response to external adverse stimulation.
Keywords: agricultural production, CKB3, LAC, lignin, microRNA, miR397, plant development, post-transcriptional, stress responses.
Introduction
MiRNAs are a class of endogenous, noncoding, small RNA molecules with 20–24 nucleotides (nt). They function critically at the post-transcriptional level to regulate gene expression through cleaving RNA targets or interfering with the translation of the target mRNAs and have extensive regulatory roles in many biological processes. MiR397 was initially reported as a novel and abiotic stress-regulated miRNA (miR397a and miR397b) in Arabidopsis by Sunkar and Zhu (2004), and it was identified as one of the differentially expressed-miRNAs in plant post-embryogenic development at 2006 (Luo et al. 2006). Recently, several studies showed that miR397 was broadly involved in plant growth, development and stress responses. It is involved in the regulation of flowering (Feng et al. 2020) and flower, seed and fruit development (Zhang et al. 2013; Burklew et al. 2014; Wang et al. 2014; Zhang et al. 2016; Xue et al. 2019). It is responsive to different stresses such as drought (Zhou et al. 2010), low temperature (Dong and Pei 2014), nitrogen starvation (Xu et al. 2011; Liang et al. 2012), low copper (Cu) availability (Abdel-Ghany and Pilon 2008; Lu et al. 2011a), and it may mediate regulatory mechanisms of tolerance to different stresses. It functions most likely by downregulating the expression levels of its targets LAC2, LAC4, LAC17 (Zhang et al. 2013; Wang et al. 2014; Swetha et al. 2018), and CKB3 (Feng et al. 2020). The biological functions and the regulatory patterns of miR397 and its targets may provide important evidence for genetic manipulation to improve crop traits. In this paper, the recent research updates of miR397 are summarised and the functional mechanisms of miR397 are discussed.
MiR397 and its target genes in plants
Members of the miR397 family
MiR397 is usually 21-nt in length in most plants, but a 22-nt miR397 form has also been identified in Oryza sativa (Jeong et al. 2011; Swetha et al. 2018).
In Arabidopsis, two members of 21-nt miR397 with only one base variance in their mature sequences, miR397a (ucauugagugcagcguugaug) and miR397b (ucauugagugcaucguugaug), were identified. Both of them are matured from the 5′ arms of miRNA precursors (Abdel-Ghany and Pilon 2008). The 21-nt miR397 is also present in other species, such as Pinus taeda (Lu et al. 2007), Picea abies (Yakovlev et al. 2010), Oryza sativa (Zhou et al. 2010), Zea mays (Xu et al. 2011), Citrus reticulata (Zhang et al. 2016), and Carya cathayensis (Sun et al. 2017). Sixty-eight members of the miR397 family and their mature sequences are recorded in 33 species by the miRBase database (http://www.mirbase.org, ver. 22.1, accessed 25 November 2020) (Kozomara and Griffiths-Jones 2014). Specifically, there are twenty-one members in Picea abies; three members in Populus trichocarpa; two in Arabidopsis thaliana, Oryza sativa, Brassica napus, Brachypodium distachyon, Zea mays, Arabidopsis lyrate, Hordeum vulgare, Glycine max, Malus domestica, Manihot esculenta, Amborella trichopoda, Eugenia uniflora, and Linum usitatissimum; and one in Vitis vinifera, Solanum lycopersicum, Sorghum bicolor, Ricinus communis, Citrus sinensis, Cucumis melo, Lotus japonicus, Medicago truncatula, Solanum tuberosum, Prunus persica, Triticum aestivum, Vriesea carinata, Fragaria vesca, Camelina sativa, Digitalis purpurea, Theobroma cacao, Nicotiana tabacum, and Salvia sclarea.
In rice, besides the 21-nt version of miR397, a 22-nt miR397, with a shift of 3 nt and an extension by 1 nt when compared with other miR397 (uugagugcagcguugaugaacc), was identified. It was found to be expressed at high levels in two wild relatives of indica rice (O. nivara and O. rufipogon) (Swetha et al. 2018).
miR397 mostly negatively regulates LACs which participate in lignin synthesis and is involved in the related biological processes.
Target genes of miR397
Some members of the LAC family are predicted or confirmed as the target genes of miR397 in plants (Jones-Rhoades and Bartel 2004; Lu et al. 2013; Wang et al. 2014; Swetha et al. 2018). In Arabidopsis, LAC2, LAC4 and LAC17 were identified as the target genes of miR397 by bioinformatic prediction and 5′-rapid amplification of cDNA ends (5′-RACE) analysis (Jones-Rhoades and Bartel 2004). In rice, at least fifteen of the 30 LACs were predicted as the potential targets of miR397, and several of them (such as OsLAC3, OsLAC6, OsLAC7, and OsLAC19) were confirmed in the Indica line O. nivara by degradome analysis (Swetha et al. 2018). In Eucalyptus grandis, 17 LAC genes were predicted as the target genes of miR397, among which LAC1, LAC4, LAC10, LAC11, LAC12 and LAC20 were highly expressed in xylem and involved in the biosynthesis of xylem lignin and maintaining mechanical toughness of stems (Arcuri et al. 2020). In pears, 27 LAC genes identified from genomic sequences were potential target genes of PbrmiR397a (Xue et al. 2019). LACs, with four Cu atoms, belong to the blue Cu oxidase family and are functionally involved in the oxidation of substrate (Thurston 1994; Claus and Filip 1997). A large number of studies show that LACs in plants are positive regulators in lignin polymerisation, polymerising monolignols into lignin (Bao et al. 1993; Ranocha et al. 2002). They play important roles in cell wall lignin biosynthesis and in maintaining plant mechanical toughness and resistance to lodging and external adverse stimulation. It was reported that overexpressing Malus hupehensis LAC7 in tobacco improved the lignin content level and enhanced the resistance against pathogens (Yu et al. 2020). Double mutant lac4 lac17 in Arabidopsis showed a great reduction in lignin content (Berthet et al. 2011). miR397 negatively regulates the transcript levels of target LACs and may further inhibit cell wall lignification and be involved in the related biological processes. Intriguingly, it seems that not all LACs function positively in the synthesis of lignin. Khandal et al. (2020) found that the Arabidopsis lac2 mutants and the lines with miR397-overexpression had the enhanced lignin deposition in root vasculature, whereas the lines with LAC2-overexpression or the lines of miR397-STTM had the opposite phenotype (Khandal et al. 2020; Yu 2020). However, the reason for the inhibition of lignin deposition by LAC2 is unclear.
Some other genes are also suggested to be the targets of miR397. CKB3 in Arabidopsis is targeted by miR397b to regulate circadian rhythm (Feng et al. 2020). A sister chromatid mucin in longan is targeted by miR397a and regulates cell division (Xu et al. 2019). A transcription factor ICE1 in wheat is also identified as a target of miR397 and functions in cold adaption (Gupta et al. 2014). Furthermore, multiple miR397-targeted genes related to plant growth, metabolism and signal transduction were predicted online using ‘psRNA Target’ (http://plantgrn.noble.org/psRNATarget/, accessed 25 November 2020; expectation ≦ 2.5). The RRA1, RRA2, DPA, CLPP3, GNS1/SUR4, and 14-3-3 family protein genes are also probably targeted and cleaved by miR397. Genes encoding SRF4, ribosomal protein S3 family protein, O-glycosyl hydrolases family 17 protein and transcription factor jumonji (JmjC) domain-containing protein may be inhibited at the translation level by miRNA targeting. However, all these predictions still need to be verified.
Roles of miR397
Roles of miR397 in plant growth and development
Embryogenic development
Somatic embryogenesis (SE) is a common method of asexual reproduction to develop plantlets. Many experiments have proved that miRNAs played indispensable roles in the SE process. It was reported that miR397 was expressed highly in undifferentiated embryogenic callus (EC) but lowly in other differentiated tissues in rice (Luo et al. 2006). In sweet oranges, however, expression of miR397 was undetectable in EC whereas the highest expression level emerged in the period of globular-shaped embryo formation. It was then decreased but remained at a relatively high level after the formation of a cotyledon-shaped somatic embryo (Wu et al. 2011). Expression of miR397 showed a higher level in non-embryogenic callus (NEC) without SE capability than in EC, and an unexpected miR397-LAC expression pattern was exhibited: UC46_6145 (laccase), a verified target gene of sweet orange miR397, expressed at a very low level in EC and at higher level in NEC (Wu et al. 2011). Although insufficient evidences can prove that the miR397-LAC regulatory pathway is involved in the SE process, miR397 is indeed required for the transition from EC to mature embryo and the maintenance of SE capability. As for the unexpected LAC expression trend in EC and NEC, which is not antagonistic to that of miR397, one hypothesis is that there are other unknown factors regulate LAC expression. Another explanation is that miR397 may work through other potential targets. But, the regulatory mechanism of miR397 in SE still needs further study.
Flower organ and seed development
miR397 is involved in the regulation of the development of reproductive organs. Overexpression of miR397 either in Arabidopsis (Wang et al. 2014) or in O. sativa japonica (Zhang et al. 2013) can increase the number of seeds and enlarge seed size, accompanying with the downregulation of its target LACs. Transgenic plants with overexpression of the target gene LAC showed the opposite phenotypes. At the grain-filling stage in rice, miR397 showed higher expression in the superior spikelets than in the inferior spikelets (Peng et al. 2014). As an important regulator of seed development, miR397 had a strong negative correlation with the expression of its target genes LACs. The inhibition of miR397 on its targets LACs normally reduced the lignin biosynthesis, and the reduction of lignification in seeds and fruits at the development stage may promote cell elongation and be further beneficial to seed and fruit development. The above studies demonstrate a common molecular mechanism for regulating plant yield by miR397-LACs.
However, the effect of the 22-nt miR397 on grain yield in indica rice (Swetha et al. 2018) is opposite to that of the 21-nt miR397 (miR397a and miR397b) in japonica rice. Transgenic plants with miR397 overexpression in cultivated line backgrounds displayed de-domestication phenotypes (lower seed setting rate and less seeds per panicle), whereas LAC3-overexpressing plants were stronger and healthier and had increased yield. This was contrary to the observation of that the overexpression of 21-nt miR397 in japonica rice, in which the yield was increased. The 22-nt miR397 mediated indica rice domestication with negative regulation of yield-traits. Previously, Jeong et al. (2011) identified more abundant 22-nt miR397 instead of 21-nt miR397 in rice plants (O. sativa ssp. japonica cv. Nipponbare). It can target the L-ascorbate oxidase mRNA (Jeong et al. 2011). Up to now, the 22-nt form of miR397 has rarely been found in other plants expect for rice and maize (Jeong et al. 2011).
Usually, 21-nt miR397 functions in inhibiting expression by cleaving target mRNAs, whereas 22-nt miR397 can lead to the production of secondary siRNAs (small interfering RNAs) (Cuperus et al. 2010). In that pathway, the single 22-nt miRNA directs cleavage of the mRNA target and generates a long dsRNA (double-stranded RNA) product via RDR (RNA-dependent RNA polymerases), which is then cleaved by DCL (DICER-LIKE) protein to produce secondary siRNAs. These secondary siRNAs are then incorporated into the RISC by loading an AGO protein, and function in gene-silencing far beyond their original targets (Chen et al. 2007; Fei et al. 2013). In indica rice, the 22-nt miR397 could target and cleave LAC mRNAs and generate secondary siRNAs, then induced a cascade silencing effect and could result in robust silencing of LACs, including LAC3, LAC7, LAC12 and LAC13 (Swetha et al. 2018). The differential responses of species demonstrate a genotypic background-dependent regulatory mechanism related to miR397. But it is unclear whether 22-nt miR397 in japonica rice has similar effects to indica rice, though 22-nt miR397 has been found to be abundant in japonica rice (Jeong et al. 2011). Further, overexpression of miR397 in japonica rice can increase yield (Zhang et al. 2013), but showed opposite phenotype in miR397-overexpressing indica rice lines (Swetha et al. 2018).
Fruit development
miR397 also regulates fruit development. In Citrus, granulation is universally observed at the harvest and storage stage in fruit, which seriously reduces fruit quality and commercial value. Previous studies have shown that the granulation level in fruit is positively related to the lignin content (Wu et al. 2014). Gene annotation data from Citrus and real-time quantitative reverse transcription PCR (qRT-PCR) analysis showed that the expression of miR397 was decreased as the granulation level increased and was significantly correlated with the level of granulation. Also, the target of miR397, LAC gene Cs6 g06890.1, exhibited a significant increase in granulation of the juice sac and was positively correlated with the lignin content at different granulation levels (Zhang et al. 2016).
In addition, Xue et al. (2019) confirmed the function of miR397 in regulating stone cell formation at the early development stage of pear fruit. Stone cells with lignin deposition in cell walls accumulate in fruit flesh and seriously reduce fruit quality. Pear varieties with low levels of stone cells showed a higher abundance of PbrmiR397a and lower levels of PbrLACs and lignin compared with pear varieties with high levels of stone cells (Xue et al. 2019). The observations that the reduced lignin content and stone cell number in pear plants overexpressing PbmiR397a transiently or with silencing of the three LACs (LAC1, LAC2, and LAC18) simultaneously were consistent with the role of miR397 in regulating lignin and stone cell content in pear fruit (Xue et al. 2019).
Either 21-nt or 22-nt miR397 is related to sex reproductive organ development via a miR397-LAC pathway. Manipulation of the miR397 levels may be an effective pathway for the improvement in yield or product quality-related traits.
Flowering
Many studies have proved that miRNAs are involved in flowering regulation. In tobacco, miR397 is maintained at a very low expression level in flowers, but the function of low expression of miR397 in flowers is not clear (Burklew et al. 2014). Feng et al. (2020) found that overexpression of miR397b in Arabidopsis significantly delayed its flowering and revealed a regulatory role of miR397b in flowering by targeting and cleaving CKB3 mRNA instead of LACs, because the single mutant plants lac2, lac4, or lac17 exhibited no change in flowering time whereas the ckb3 knockout plants exhibited a late-flowering phenotype similar to the miR397b-overexpressing plants, and overexpression of CKB3 displayed early-flowering (Feng et al. 2020). As a component of the circadian oscillator, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) regulates the circadian clock through phosphorylation by CKB3 (Lu et al. 2011b). The transcription of miR397b can be inhibited by CCA1 through binding on its promoter (Feng et al. 2020). Thus, miR397b is involved in a feedback circuit of miR397b-CKB3-CCA1 in the circadian clock and affects flowering time.
Roles of miR397 in abiotic stress responses
Drought stress
Drought stress is a severe environmental constraint on plant growth and development. During the long course of evolution, plants have developed a series of mechanisms to adapt to the stress. Many drought stress-related miRNAs have been identified.
miR397 was shown to be downregulated by drought stress in some plants, such as the tillering stage rice (Zhou et al. 2010), 2-week-old wheat seedlings (Gupta et al. 2014) and potato plants (Shin et al. 2017). Conversely, miR397 showed a different expression pattern in some cases. When 3-month-old greenhouse-grown sugarcane cultivars with higher drought tolerance or lower drought tolerance were subjected to drought stress, miR397 was found to be induced in the unfolded young leaf of both cultivars after 2 days of drought, but it was differentially downregulated in higher drought tolerance sugarcane at 4 days of drought and could be recovered after an additional 2 days of rehydration (Ferreira et al. 2012). In Arabidopsis, miR397 was slightly induced when 2-week-old seedlings grown on MS-ager plates were exposed to dry air for 10 h (Sunkar and Zhu 2004). In tomatoes, the transcripts of miR397 increased after drought stress, and overexpression of tomato miR397 conferred drought tolerance to Arabidopsis (Xiang et al. 2016). However, the mechanisms of how the miR397 responses to drought and what the regulatory roles of miR397 in drought stressed plants have not been elucidated in the above studies.
Recently, Khandal et al. (2020) found that the LAC2, a negative regulator of lignin deposition, was downregulated by the elevated miR397b in Arabidopsis roots under water-deficit conditions (Khandal et al. 2020). It has been reported that low-lignin xylem reduced water transport efficiency (Kitin et al. 2010). An increased lignin deposition in root vascular tissue caused by the downregulation of the miR397b-targeted LAC2 might be helpful for plants to save water during drought stress.
In addition, some predicted miR397 target genes, such as Casein kinase II and L-ascorbate oxidase precursor, have been shown to be involved in drought stress response (Mehta et al. 2009; Kang and Udvardi 2012).
The diverse expression patterns of miR397 under drought stress in different cases imply that the mechanisms of plants in responding to drought stress vary with plant genetic background and growth stages due to their different water demand. Furthermore, it seems likely that plant at different growth stage has different regulatory pathway. For example, at seedling stage, Arabidopsis with overexpression of miR397b have higher lignin level than that in wild type (Wang et al. 2014), whereas an opposite trend was exhibited during the germination period (Khandal et al. 2020). The up- and downstream pathways of miR397 in the response to stress remain unclear and need to be clarified.
Low temperature stress
miR397 could be induced by cold treatment in some plants such as Arabidopsis (Sunkar and Zhu 2004) and Hemerocallis fulva (An et al. 2014). Moreover, overexpression of miR397a in Arabidopsis significantly improved plant tolerance to chilling and freezing stresses. The cold-regulated CBF2 and the downstream COR gene targets, which are attributed to plant tolerance to cold stress (Van Buskirk and Thomashow 2006), had higher transcript levels in these transgenic plants than in wild-type plants when they were subjected to chilling (Dong and Pei 2014).
Interestingly, an opposite phenomenon was observed in wheat seedlings (Gupta et al. 2014). Analysis of the potential target genes showed that wheat miR397 could target the transcription factor ICE1 in addition to LAC. ICE1 is an inducer of the cold stress- responsive factor CBF. Lower expression levels of miR397 in cold treatment led to the upregulation of ICE1 genes and further positively regulated the expression of CBF to help plant adapt to cold stress. Thus, different molecular mechanisms related to miR397 may be involved in the response to cold stress in Arabidopsis and wheat.
Heavy metal stress
Copper (Cu) is an essential micronutrient for plant growth. Either deficient or excessive Cu inhibits plant growth. Several miRNAs, including miR397, miR398, miR408 and miR857, were identified as Cu-responsive miRNAs. All these miRNAs have abundant Cu-responsive cis-elements (GTAC) in their promoter region. SPL7 can bind to these GTAC motifs and activate their expression under Cu deficiency (Abdel-Ghany and Pilon 2008; Yamasaki et al. 2009). Expression of miR397 in plants is generally induced by Cu deficiency and repressed by excessive Cu stress. Expression of miR397 was markedly induced in Arabidopsis (Abdel-Ghany and Pilon 2008) and bananas (Patel et al. 2019) in Cu deficiency condition. Correspondingly, the expression of several LACs, the target genes of miR397, were downregulated. In Populus trichocarpa (Lu et al. 2011a) and grapevines (Leng et al. 2017), the transcript levels of miR397 were decreased by excessive Cu treatment. LAC protein generally contains four Cu atoms and belongs to the Cu protein family. It is speculated that the increase in miR397 under Cu deficiency with concomitant downregulation of its target LACs is helpful for saving Cu for basal and important metabolism, such as photosynthesis which requires Cu-plastocynin (PC) (Abdel-Ghany and Pilon 2008). In response to excessive Cu stress, the repressed expression of miR397 results in the accumulation of LACs. The upregulation of LACs normally increases lignin synthesis and deposition in cell walls. It further leads to the stronger binding of Cu to cell walls and a reduction of the Cu up-taking into cells. In contrast, the large amounts of LACs should consume much Cu. This suggests a Cu homeostasis regulation by miR397-LACs during Cu stress.
Some studies based on genome-wide transcriptome analysis showed that miR397 could be upregulated by other heavy metal stresses. It was reported that miR397 was upregulated by cadmium (Cd) stress in Brassica napus (Zhou et al. 2012; Fu et al. 2019), Brassica parachinensis (Zhou et al. 2017; Liu et al. 2020), and soybeans (Fang et al. 2013), as well as by chromium (Cr) in rice (Dubey et al. 2020).
Gielen et al. (2016) found that Cd-exposure led to a decrease in Cu levels in Arabidopsis plants compared with the control (Gielen et al. 2016). Moreover, miR397 and other Cu-responsive miRNA (miR395, miR398, and miR857), as well as their upstream regulator SPL7, were upregulated, and targets of miR397 (LAC2, LAC4, and LAC17) were downregulated. A similar response was induced by Cd stress and Cu deficiency. Furthermore, supply of additional Cu to Cd-exposed plants could counteract the Cu deficiency response. It is speculated that Cd stress affects Cu homeostasis, induces Cu deficiency and further upregulates miR397.
Nutrition stress
Using miRNA transcriptome analysis, repression of miR397 expression was detected in Arabidopsis (Liang et al. 2012), maize (Xu et al. 2011; Zhao et al. 2012) and common bean (Váldes-López et al. 2010) under nitrogen deficiency conditions. In potatoes, the reduced expression of miR397 was also detected in roots concomitant with the upregulation of its target gene LAC in low nitrogen conditions (Tiwari et al. 2020). In the case of nitrogen deficiency, maintaining of the C : N balance in plants is essential for normal growth. It seems that miR397 may be involved in the regulation of N homeostasis or the adaptation to nitrogen limitation by maintaining C : N homeostasis through increasing LAC expression and incorporating excess fixed C into lignin. However, the regulatory mechanisms and the functions of miR397 during nitrogen starvation are still unclear.
Boron (B) is also an essential micronutrient for plant. High-throughput sequencing of Citrus grown in sand after excessive B treatment for 15 weeks showed that miR397a was downregulated in the B-tolerant Citrus leaves with concomitant upregulation of the verified targets of miR397a, LAC4 and LAC17. Further, an increase of secondary deposition of cell-wall polysaccharides in vessel elements was detected. Conversely, significant upregulation of miR397a and poor development of vessel elements in the vascular bundles was observed in the B-intolerant Citrus (Huang et al. 2016). In trifoliate oranges and barley leaves, reductions in the miR397 expression levels were observed after excessive B treatment (Ozhuner et al. 2013; Jin et al. 2016). The expression of LAC7 was markedly induced and higher LAC activity and lignin content was detected in trifoliate oranges.
One important role of B is to maintain structure stabilisation of cell walls and regulate polyphenoloxidase activity and cell wall lignification. B-deficiency or B-toxicity may influence the cell wall structure. The above reports reveal a conservative response mechanism in plants in adaptation to B toxicity: in general, the miR397- targeted gene LACs are upregulated by the downregulated miR397 when plants are subjected to excessive B. This leads to changes in the compositions and structures of the xylem cell walls and may alleviate the B-toxicity to the plant. The opposite expression patterns of miR397 and LACs in B-intolerant Citrus lead to its more sensitive response to B toxicity.
Other abiotic stresses
It was reported that miR397 could also function in the herbicide resistance of plants. The miR397 transcript level was constitutively higher in fenoxaprop-P-ethyl-resistant Beckmannia syzigachne populations with years of fenoxaprop-P-ethyl use history than in the susceptible B. syzigachne population, whereas the LAC expression and enzyme activity were higher in the susceptible population (Pan et al. 2017). The miR397-overexpressing rice showed improved fenoxaprop-P-ethyl tolerance and higher expression of oxidase/peroxides genes. The functional mechanism of miR397 and its targets in herbicide resistance should be divulged and which will provide theoretical basis and novel strategies for creating the new varieties resistance to the herbicide.
Roles of miR397 in plant-microbe interactions
Biological interactions occur frequently in the processes such as pathogen infection stress and plant-microbe symbiosis. Many miRNAs play vital roles in the host reaction during these processes. For example, miR168, which could be induced by virus infection, is involved in a feedback regulation to control the accumulation of AGO1 and repress host-mounted RNA-silencing (Várallyay et al. 2010). In order to better understand the function of miRNAs in pathogen infection, many studies were conducted to identify the differential expressed miRNAs induced by infection through high-throughput sequencing. MiR397 was identified as one of the delegates (Li et al. 2017; Xia et al. 2018). However, further investigation is lacking. Recently, Yu et al. (2020) found that transient overexpression of miR397b in Malus hupehensis leaves triggered the increased sensitivity to Botryosphaeria dothidea owing to the reduced LAC7 and lignin content, whereas the inhibition of miR397b in leaves transformed with STTM397 had opposite effects (Yu et al. 2020).
miR397 targets LACs and further influences the synthesis of lignin and the cell wall structure. Serving as the first physical barrier, cell walls play important roles in pathogen resistance by protecting plants from pathogen attack. Downregulation of miR397 expression may be helpful for pathogen resistance. The regulation mode of miR397-LAC-lignification in pathogen defence provides a clue for future research on plant virus disease resistance.
Many miRNAs are involved in the regulation of plant-microbe symbiosis. The symbiotic relationship can be formed between the legume Lotus japonicus and Mesorhizobium loti and enhance the nitrogen-fixing ability of L. japonicus. De Luis et al. found that miR397 was accumulated at high levels in the M. loti-infected active nodules with strong nitrogen-fixing ability but was kept at low levels in the noninfected, inactive and spontaneous nodules (De Luis et al. 2012). When a mutant with deficient nitrogen fixation capacity was inoculated with M. loti, only weak expression levels of miR397 could be detected, whereas an opposite case was occurred in the wild-type plants with normal nitrogen fixation capacity. Upregulated expression of miR397 in maize was also found after inoculated with diazotrophic bacteria (Thiebaut et al. 2014). Unlike other pathogen infection, plants being symbiotic with diazotrophic bacteria do not show the activate defence response against the diazotrophic bacteria colonisation. Instead, they reduce the synthesis of lignin regulated by miR397 and maintain a stable symbiotic relationship to improve crop production. These observations demonstrate the function of miR397 in regulating the relationship of plant-microbe symbiosis.
Symbiosis of arbuscular mycorrhiza (AM) fungi with maize roots can improve maize yield and resistance. Xu et al. (2018) found that the inoculation of AM fungi on maize roots could downregulate the expression of miR397 and might further regulate genes functioning in fatty acid metabolism and promote lipid delivery from plants to AM fungi (Xu et al. 2018). It suggests that miR397 could regulate the symbiosis of plant and microbe through different targets.
Conclusions
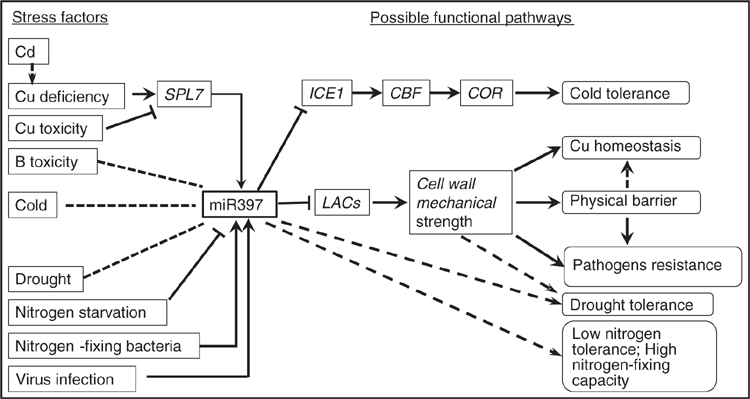
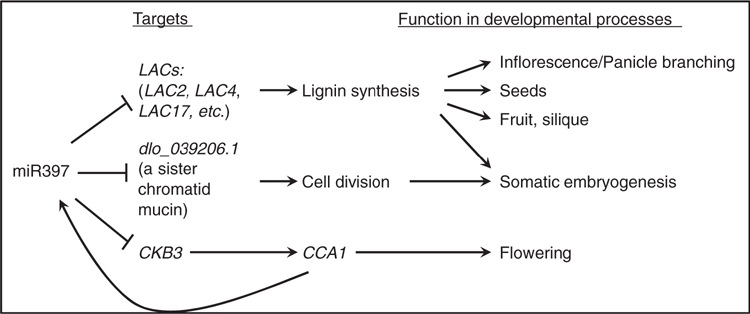
MiR397 has been identified mostly in the form of 21-nt or the exception of 22-nt in plants. 21-nt miR397 generally downregulates the transcript levels of its universal target LACs and inhibits laccase activity and cell wall lignification, further influencing cell wall mechanical toughness, and is involved in various biological processes such as cell growth, reproduction organ development and plant resistance to external adverse stresses (Table 1; Figs 1, 2). 22-nt miR397 can similarly target and cleave LAC mRNAs and even generate secondary siRNAs, further inducing a robust cascade silencing of LACs. In this way, 22-nt miR397 mediated indica rice domestication with negative regulation of yield-related traits.

|
|
The 21-nt miR397 can target and cleave CKB3 (Fig. 1), which can phosphorylate a component of the circadian oscillator CCA1. However, CCA1 can bind to the promoter of miR397 and inhibit its transcription. Thus, miR397 can be involved in a feedback circuit of miR397b-CKB3-CCA1 in the circadian clock and manipulate plant flowering.
In longan, miR397 targets not only LACs but also a sister chromatid mucin (Xu et al. 2019) (Fig. 1), which functions during chromosome replication and is involved in cell division regulation. miR397 may also regulate genes functioning in fatty acid metabolism that are involved in microbe-plant interactions. In addition, several genes related to plant growth, metabolism and signal transduction, such as RRA1, RRA2, DPA, CLPP3, GNS1/SUR4, 14-3-3s, SRF4, and JmjC, may be cleaved or inhibited at the translation level by miR397 targeting as predicted online, but this needs to be verified.
In the regulatory pathway related to miR397, some transcription regulators function in upstream of miR397 and regulate its expression (Figs. 1, 2). For example, CCA1 functions as a transcription inhibitor of miR397 during plant flowering regulation. Another transcription regulator, SPL7, can bind on the Cu-responsive cis elements (GTAC) in the promoter of miR397 and regulate its expression when plants perceive the stress of deficient or excessive Cu.
Overall, this review provides information about the possible roles of miR397 in plant development (Fig. 1) and in response to external adverse stimulation (Fig. 2). However, evidence of miR397 controlling its targets when participating in particular biological processes is still lacking. The related regulatory mechanism is still unclear and needs to be better understood. Further studies characterising the up- and downstream components of miR397 and their cross-linked genes should be performed. These studies would be helpful for understanding the function of miR397 and divulging its application prospects for increasing agricultural production as well as its ability to enhance plant adaptation to abiotic stresses.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This work was supported by the Natural Science Foundation of Guangdong province (2020A1515010309) and the Guangdong Innovation Research Team Fund (2014ZT05S078).
References
Abdel-Ghany SE, Pilon M (2008) MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. Journal of Biological Chemistry 283, 15932–15945.| MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
An F, Lu B, Liang M, Tang H, Li F (2014) Bioinformatics, expression and functional analysis of microRNAs in response to low temperature in Hemerocallis fulva (L.) L. Chih Wu Sheng Li Hsueh T’ung Hsun 50, 483–487.
Arcuri MLC, Fialho LC, Nunes-Laitz AV, Fuchs-Ferraz MCP, Wolf IR, Valente GT, Marino CL, Maia IG (2020) Genome-wide identification of multifunctional laccase gene family in Eucalyptus grandis: potential targets for lignin engineering and stress tolerance. Trees 34, 745–758.
| Genome-wide identification of multifunctional laccase gene family in Eucalyptus grandis: potential targets for lignin engineering and stress tolerance.Crossref | GoogleScholarGoogle Scholar |
Bao W, Omalley DM, Whetten R, Sederoff RR (1993) A laccase associated with lignification in loblolly-pine xylem. Science 260, 672–674.
| A laccase associated with lignification in loblolly-pine xylem.Crossref | GoogleScholarGoogle Scholar | 17812228PubMed |
Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cezard L, Le Bris P, Borrega N, Herve J, Blondet E, Balzergue S, Lapierre C, Jouanin L (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. The Plant Cell 23, 1124–1137.
| Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems.Crossref | GoogleScholarGoogle Scholar | 21447792PubMed |
Burklew CE, Xie F, Ashlock J, Zhang B (2014) Expression of microRNAs and their targets regulates floral development in tobacco (Nicotiana tabacum). Functional & Integrative Genomics 14, 299–306.
| Expression of microRNAs and their targets regulates floral development in tobacco (Nicotiana tabacum).Crossref | GoogleScholarGoogle Scholar |
Chen H-M, Li Y-H, Wu S-H (2007) Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 104, 3318–3323.
| Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 17360645PubMed |
Claus H, Filip Z (1997) The evidence of a laccase-like enzyme activity in a Bacillus sphaericus strain. Microbiological Research 152, 209–216.
| The evidence of a laccase-like enzyme activity in a Bacillus sphaericus strain.Crossref | GoogleScholarGoogle Scholar |
Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC (2010) Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nature Structural & Molecular Biology 17, 997–1003.
| Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis.Crossref | GoogleScholarGoogle Scholar |
De Luis A, Markmann K, Cognat V, Holt DB, Charpentier M, Parniske M, Stougaard J, Voinnet O (2012) Two microRNAs linked to nodule infection and nitrogen-fixing ability in the Legume Lotus japonicus. Plant Physiology 160, 2137–2154.
| Two microRNAs linked to nodule infection and nitrogen-fixing ability in the Legume Lotus japonicus.Crossref | GoogleScholarGoogle Scholar | 23071252PubMed |
Dong C-H, Pei H (2014) Over-expression of miR397 improves plant tolerance to cold stress in Arabidopsis thaliana. Journal of Plant Biology 57, 209–217.
| Over-expression of miR397 improves plant tolerance to cold stress in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar |
Dubey S, Saxena S, Chauhan AS, Mathur P, Rani V, Chakrabaroty D (2020) Identification and expression analysis of conserved microRNAs during short and prolonged chromium stress in rice (Oryza sativa). Environmental Science and Pollution Research International 27, 380–390.
| Identification and expression analysis of conserved microRNAs during short and prolonged chromium stress in rice (Oryza sativa).Crossref | GoogleScholarGoogle Scholar | 31792790PubMed |
Fan G, Zhai X, Niu S, Ren Y (2014) Dynamic expression of novel and conserved microRNAs and their targets in diploid and tetraploid of Paulownia tomentosa. Biochimie 102, 68–77.
| Dynamic expression of novel and conserved microRNAs and their targets in diploid and tetraploid of Paulownia tomentosa.Crossref | GoogleScholarGoogle Scholar | 24565810PubMed |
Fang X, Zhao Y, Ma Q, Huang Y, Wang P, Zhang J, Nian H, Yang C (2013) Identification and comparative analysis of cadmium tolerance-associated miRNAs and their targets in two soybean genotypes. PLoS One 8, e81471
| Identification and comparative analysis of cadmium tolerance-associated miRNAs and their targets in two soybean genotypes.Crossref | GoogleScholarGoogle Scholar | 24386213PubMed |
Fei Q, Xia R, Meyers BC (2013) Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. The Plant Cell 25, 2400–2415.
| Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks.Crossref | GoogleScholarGoogle Scholar | 23881411PubMed |
Feng Y-Z, Yu Y, Zhou Y-F, Yang Y-W, Lei M-Q, Lian J-P, He H, Zhang Y-C, Huang W, Chen Y-Q (2020) A natural variant of miR397 mediates a feedback loop in circadian rhythm. Plant Physiology 182, 204–214.
| A natural variant of miR397 mediates a feedback loop in circadian rhythm.Crossref | GoogleScholarGoogle Scholar | 31694901PubMed |
Ferreira TH, Gentile A, Vilela RD, Lacerda Costa GG, Dias LI, Endres L, Menossi M (2012) microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.). PLoS One 7, e46703
| microRNAs associated with drought response in the bioenergy crop sugarcane (Saccharum spp.).Crossref | GoogleScholarGoogle Scholar | 23071617PubMed |
Fu Y, Mason AS, Zhang Y, Lin B, Xiao M, Fu D, Yu H (2019) MicroRNA-mRNA expression profiles and their potential role in cadmium stress response in Brassica napus. BMC Plant Biology 19, 570
| MicroRNA-mRNA expression profiles and their potential role in cadmium stress response in Brassica napus.Crossref | GoogleScholarGoogle Scholar | 31856702PubMed |
Ghorecha V, Patel K, Ingle S, Sunkar R, Krishnayya NSR (2014) Analysis of biochemical variations and microRNA expression in wild (Ipomoea campanulata) and cultivated (Jacquemontia pentantha) species exposed to in vivo water stress. Physiology and Molecular Biology of Plants 20, 57–67.
| Analysis of biochemical variations and microRNA expression in wild (Ipomoea campanulata) and cultivated (Jacquemontia pentantha) species exposed to in vivo water stress.Crossref | GoogleScholarGoogle Scholar | 24554839PubMed |
Gielen H, Remans T, Vangronsveld J, Cuypers A (2016) Toxicity responses of Cu and Cd: the involvement of miRNAs and the transcription factor SPL7. BMC Plant Biology 16, 145
| Toxicity responses of Cu and Cd: the involvement of miRNAs and the transcription factor SPL7.Crossref | GoogleScholarGoogle Scholar | 27352843PubMed |
Gupta OP, Meena NL, Sharma I, Sharma P (2014) Differential regulation of microRNAs in response to osmotic, salt and cold stresses in wheat. Molecular Biology Reports 41, 4623–4629.
| Differential regulation of microRNAs in response to osmotic, salt and cold stresses in wheat.Crossref | GoogleScholarGoogle Scholar | 24682922PubMed |
Huang J-H, Qi Y-P, Wen S-X, Guo P, Chen X-M, Chen L-S (2016) Illumina microRNA profiles reveal the involvement of miR397a in Citrus adaptation to long-term boron toxicity via modulating secondary cell-wall biosynthesis. Scientific Reports 6, 22900
| Illumina microRNA profiles reveal the involvement of miR397a in Citrus adaptation to long-term boron toxicity via modulating secondary cell-wall biosynthesis.Crossref | GoogleScholarGoogle Scholar | 26962011PubMed |
Jeong DH, Park S, Zhai J, Gurazada SG, De Paoli E, Meyers BC, Green PJ (2011) Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. The Plant Cell 23, 4185–4207.
| Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage.Crossref | GoogleScholarGoogle Scholar | 22158467PubMed |
Jin L-F, Liu Y-Z, Yin X-X, Peng S-A (2016) Transcript analysis of citrus miRNA397 and its target LAC7 reveals a possible role in response to boron toxicity. Acta Physiologiae Plantarum 38, 18
| Transcript analysis of citrus miRNA397 and its target LAC7 reveals a possible role in response to boron toxicity.Crossref | GoogleScholarGoogle Scholar |
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Molecular Cell 14, 787–799.
| Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA.Crossref | GoogleScholarGoogle Scholar | 15200956PubMed |
Kang Y, Udvardi M (2012) Global regulation of reactive oxygen species scavenging genes in alfalfa root and shoot under gradual drought stress and recovery. Plant Signaling & Behavior 7, 539–543.
| Global regulation of reactive oxygen species scavenging genes in alfalfa root and shoot under gradual drought stress and recovery.Crossref | GoogleScholarGoogle Scholar |
Kayihan DS, Kayihan C, Ciftci YO (2019) Moderate level of toxic boron causes differential regulation of microRNAs related to jasmonate and ethylene metabolisms in Arabidopsis thaliana. Turkish Journal of Botany 43, 167–172.
| Moderate level of toxic boron causes differential regulation of microRNAs related to jasmonate and ethylene metabolisms in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar |
Khandal H, Singh AP, Chattopadhyay D (2020) The microRNA397b -LACCASE2 module regulates root lignification under water and phosphate deficiency. Plant Physiology 182, 1387–1403.
| The microRNA397b -LACCASE2 module regulates root lignification under water and phosphate deficiency.Crossref | GoogleScholarGoogle Scholar | 31949029PubMed |
Kitin P, Voelker SL, Meinzer FC, Beeckman H, Strauss SH, Lachenbruch B (2010) Tyloses and phenolic deposits in xylem vessels impede water transport in low-lignin transgenic poplars: a study by cryo-fluorescence microscopy. Plant Physiology 154, 887–898.
| Tyloses and phenolic deposits in xylem vessels impede water transport in low-lignin transgenic poplars: a study by cryo-fluorescence microscopy.Crossref | GoogleScholarGoogle Scholar | 20639405PubMed |
Koc I, Filiz E, Tombuloglu H (2015) Assessment of miRNA expression profile and differential expression pattern of target genes in cold-tolerant and cold-sensitive tomato cultivars. Biotechnology, Biotechnological Equipment 29, 851–860.
| Assessment of miRNA expression profile and differential expression pattern of target genes in cold-tolerant and cold-sensitive tomato cultivars.Crossref | GoogleScholarGoogle Scholar |
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research 42, D68–D73.
| miRBase: annotating high confidence microRNAs using deep sequencing data.Crossref | GoogleScholarGoogle Scholar | 24275495PubMed |
Leng X, Wang P, Zhao P, Wang M, Cui L, Shangguan L, Wang C (2017) Conservation of microRNA-mediated regulatory networks in response to copper stress in grapevine. Plant Growth Regulation 82, 293–304.
| Conservation of microRNA-mediated regulatory networks in response to copper stress in grapevine.Crossref | GoogleScholarGoogle Scholar |
Li L, Yang K, Wang S, Lou Y, Zhu C, Gao Z (2017) Integrated mRNA and microRNA transcriptome analysis reveals miRNA regulation in response to PVA in potato. Scientific Reports 7, 16925
| Integrated mRNA and microRNA transcriptome analysis reveals miRNA regulation in response to PVA in potato.Crossref | GoogleScholarGoogle Scholar |
Li Y, Hu X, Chen J, Wang W, Xiong X, He C (2020) Genome-wide analysis of laccase genes in moso bamboo highlights PeLAC10 involved in lignin biosynthesis and in response to abiotic stresses. Plant Cell Reports 39, 751–763.
| Genome-wide analysis of laccase genes in moso bamboo highlights PeLAC10 involved in lignin biosynthesis and in response to abiotic stresses.Crossref | GoogleScholarGoogle Scholar |
Liang G, He H, Yu D (2012) Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS One 7, e48951
| Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 23185505PubMed |
Liang G, Ai Q, Yu D (2015) Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis. Scientific Reports 5, 11813
| Uncovering miRNAs involved in crosstalk between nutrient deficiencies in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 26134148PubMed |
Lin Y, Lai Z (2013) Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.). PLoS One 8, e60337
| Comparative analysis reveals dynamic changes in miRNAs and their targets and expression during somatic embryogenesis in longan (Dimocarpus longan Lour.).Crossref | GoogleScholarGoogle Scholar | 24391812PubMed |
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14, 836–843.
| Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 18356539PubMed |
Liu Y, Liu S, Deng Y, You C, Zhang W, Zhou J, Chen X, Gao L, Tang Y (2020) Genome-wide mRNA and small RNA transcriptome profiles uncover cultivar- and tissue-specific changes induced by cadmium in Brassica parachinensis. Environmental and Experimental Botany 180, 104207
| Genome-wide mRNA and small RNA transcriptome profiles uncover cultivar- and tissue-specific changes induced by cadmium in Brassica parachinensis.Crossref | GoogleScholarGoogle Scholar |
Lu S, Sun Y-H, Amerson H, Chiang VL (2007) MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. The Plant Journal 51, 1077–1098.
| MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development.Crossref | GoogleScholarGoogle Scholar | 17635765PubMed |
Lu S, Yang C, Chiang VL (2011a) Conservation and diversity of microRNA-associated copper-regulatory networks in Populus trichocarpa. Journal of Integrative Plant Biology 53, 879–891.
| Conservation and diversity of microRNA-associated copper-regulatory networks in Populus trichocarpa.Crossref | GoogleScholarGoogle Scholar | 22013976PubMed |
Lu SX, Liu H, Knowles SM, Li J, Ma L, Tobin EM, Lin C (2011b) A role for protein kinase casein kinase2 alpha-subunits in the Arabidopsis circadian clock. Plant Physiology 157, 1537–1545.
| A role for protein kinase casein kinase2 alpha-subunits in the Arabidopsis circadian clock.Crossref | GoogleScholarGoogle Scholar | 21900482PubMed |
Lu S, Li Q, Wei H, Chang MJ, Tunlaya-Anukit S, Kim H, Liu J, Song J, Sun YH, Yuan L, Yeh TF, Peszlen I, Ralph J, Sederoff RR, Chiang VL (2013) Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa. Proceedings of the National Academy of Sciences of the United States of America 110, 10848–10853.
| Ptr-miR397a is a negative regulator of laccase genes affecting lignin content in Populus trichocarpa.Crossref | GoogleScholarGoogle Scholar | 23754401PubMed |
Luo Y-C, Zhou H, Li Y, Chen J-Y, Yang J-H, Chen Y-Q, Qu L-H (2006) Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development. FEBS Letters 580, 5111–5116.
| Rice embryogenic calli express a unique set of microRNAs, suggesting regulatory roles of microRNAs in plant post-embryogenic development.Crossref | GoogleScholarGoogle Scholar | 16959252PubMed |
Mehta PA, Rebala KC, Venkataraman G, Parida A (2009) A diurnally regulated dehydrin from Avicennia marina that shows nucleo-cytoplasmic localization and is phosphorylated by casein kinase II in vitro. Plant Physiology and Biochemistry 47, 701–709.
| A diurnally regulated dehydrin from Avicennia marina that shows nucleo-cytoplasmic localization and is phosphorylated by casein kinase II in vitro.Crossref | GoogleScholarGoogle Scholar | 19398349PubMed |
Ozhuner E, Eldem V, Ipek A, Okay S, Sakcali S, Zhang B, Boke H, Unver T (2013) Boron stress responsive microRNAs and their targets in barley. PLoS One 8, e59543
| Boron stress responsive microRNAs and their targets in barley.Crossref | GoogleScholarGoogle Scholar | 23555702PubMed |
Pan L, Zhao H, Yu Q, Bai L, Dong L (2017) miR397/laccase gene mediated network improves tolerance to fenoxaprop-p-ethyl in Beckmannia syzigachne and Oryza sativa. Frontiers in Plant Science 8, 879
| miR397/laccase gene mediated network improves tolerance to fenoxaprop-p-ethyl in Beckmannia syzigachne and Oryza sativa.Crossref | GoogleScholarGoogle Scholar | 28588605PubMed |
Patel P, Yadav K, Srivastava AK, Suprasanna P, Ganapathi TR (2019) Overexpression of native Musa-miR397 enhances plant biomass without compromising abiotic stress tolerance in banana. Scientific Reports 9, 16434
| Overexpression of native Musa-miR397 enhances plant biomass without compromising abiotic stress tolerance in banana.Crossref | GoogleScholarGoogle Scholar | 31712582PubMed |
Peng T, Sun H, Qiao M, Zhao Y, Du Y, Zhang J, Li J, Tang G, Zhao Q (2014) Differentially expressed microRNA cohorts in seed development may contribute to poor grain filling of inferior spikelets in rice. BMC Plant Biology 14, 196
| Differentially expressed microRNA cohorts in seed development may contribute to poor grain filling of inferior spikelets in rice.Crossref | GoogleScholarGoogle Scholar | 25052585PubMed |
Ranocha P, Chabannes M, Chamayou S, Danoun S, Jauneau A, Boudet AM, Goffner D (2002) Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar. Plant Physiology 129, 145–155.
| Laccase down-regulation causes alterations in phenolic metabolism and cell wall structure in poplar.Crossref | GoogleScholarGoogle Scholar | 12011346PubMed |
Shin S-J, Lee J-H, Kwon H-B (2017) Genome-wide identification and characterization of drought responsive microRNAs in Solanum tuberosum L. Genes & Genomics 39, 1193–1203.
| Genome-wide identification and characterization of drought responsive microRNAs in Solanum tuberosum L.Crossref | GoogleScholarGoogle Scholar |
Sun M-x, Lin Y, Lai Z (2017) Genome-wide analysis of miRNAs in Carya cathayensis. BMC Plant Biology 17, 228
| Genome-wide analysis of miRNAs in Carya cathayensis.Crossref | GoogleScholarGoogle Scholar |
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. The Plant Cell 16, 2001–2019.
| Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 15258262PubMed |
Swetha C, Basu D, Pachamuthu K, Tirumalai V, Nair A, Prasad M, Shivaprasad PV (2018) Major domestication-related phenotypes in Indica rice are due to loss of miRNA-mediated laccase silencing. The Plant Cell 30, 2649–2662.
| Major domestication-related phenotypes in Indica rice are due to loss of miRNA-mediated laccase silencing.Crossref | GoogleScholarGoogle Scholar | 30341147PubMed |
Thiebaut F, Rojas CA, Grativol C, Motta MR, Vieira T, Regulski M, Martienssen RA, Farinelli L, Hemerly AS, Ferreira PCG (2014) Genome-wide identification of microRNA and siRNA responsive to endophytic beneficial diazotrophic bacteria in maize. BMC Genomics 15, 766
| Genome-wide identification of microRNA and siRNA responsive to endophytic beneficial diazotrophic bacteria in maize.Crossref | GoogleScholarGoogle Scholar | 25194793PubMed |
Thurston CF (1994) The structure and function of fungal laccases. Microbiology-Sgm 140, 19–26.
| The structure and function of fungal laccases.Crossref | GoogleScholarGoogle Scholar |
Tiwari JK, Buckseth T, Zinta R, Saraswati A, Singh RK, Rawat S, Chakrabarti SK (2020) Genome-wide identification and characterization of microRNAs by small RNA sequencing for low nitrogen stress in potato. PLoS One 15, e0233076
| Genome-wide identification and characterization of microRNAs by small RNA sequencing for low nitrogen stress in potato.Crossref | GoogleScholarGoogle Scholar | 32428011PubMed |
Valdes-Lopez O, Yang SS, Aparicio-Fabre R, Graham PH, Reyes JL, Vance CP, Hernandez G (2010) MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity. New Phytologist 187, 805–818.
| MicroRNA expression profile in common bean (Phaseolus vulgaris) under nutrient deficiency stresses and manganese toxicity.Crossref | GoogleScholarGoogle Scholar |
Van Buskirk HA, Thomashow MF (2006) Arabidopsis transcription factors regulating cold acclimation. Physiologia Plantarum 126, 72–80.
| Arabidopsis transcription factors regulating cold acclimation.Crossref | GoogleScholarGoogle Scholar |
Várallyay E, Válóczi A, Ágyi A, Burgyán J, Havelda Z (2010) Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO Journal 29, 3507–3519.
| Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation.Crossref | GoogleScholarGoogle Scholar |
Wang C-Y, Zhang S, Yu Y, Luo Y-C, Liu Q, Ju C, Zhang Y-C, Qu L-H, Lucas WJ, Wang X, Chen Y-Q (2014) MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis. Plant Biotechnology Journal 12, 1132–1142.
| MiR397b regulates both lignin content and seed number in Arabidopsis via modulating a laccase involved in lignin biosynthesis.Crossref | GoogleScholarGoogle Scholar | 24975689PubMed |
Wang M, Yuan D, Tu L, Gao W, He Y, Hu H, Wang P, Liu N, Lindsey K, Zhang X (2015) Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.). New Phytologist 207, 1181–1197.
| Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.).Crossref | GoogleScholarGoogle Scholar |
Wu X-M, Liu M-Y, Ge X-X, Xu Q, Guo W-W (2011) Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange. Planta 233, 495–505.
| Stage and tissue-specific modulation of ten conserved miRNAs and their targets during somatic embryogenesis of Valencia sweet orange.Crossref | GoogleScholarGoogle Scholar | 21103993PubMed |
Wu J-L, Pan T-F, Guo Z-X, Pan D-M (2014) Specific lignin accumulation in granulated juice sacs of Citrus maxima. Journal of Agricultural and Food Chemistry 62, 12082–12089.
| Specific lignin accumulation in granulated juice sacs of Citrus maxima.Crossref | GoogleScholarGoogle Scholar | 25419620PubMed |
Xia Z, Zhao Z, Li M, Chen L, Jiao Z, Wu Y, Zhou T, Yu W, Fan Z (2018) Identification of miRNAs and their targets in maize in response to Sugarcane mosaic virus infection. Plant Physiology and Biochemistry 125, 143–152.
| Identification of miRNAs and their targets in maize in response to Sugarcane mosaic virus infection.Crossref | GoogleScholarGoogle Scholar | 29453091PubMed |
Xiang J, Lin P, Li X, Li S, Liu M, Zhang L, Guo Y (2016) Overexpression of tomato Sly-miR397 gene enhances drought tolerance in Arabidopsis thaliana. Zhongguo Nongye Daxue Xuebao 21, 51–58.
Xu Z, Zhong S, Li X, Li W, Rothstein SJ, Zhang S, Bi Y, Xie C (2011) Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots. PLoS One 6, e28009
| Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots.Crossref | GoogleScholarGoogle Scholar | 22194943PubMed |
Xu Y, Zhu S, Liu F, Wang W, Wang X, Han G, Cheng B (2018) Identification of arbuscular mycorrhiza fungi responsive microRNAs and their regulatory network in maize. International Journal of Molecular Sciences 19, 3201
| Identification of arbuscular mycorrhiza fungi responsive microRNAs and their regulatory network in maize.Crossref | GoogleScholarGoogle Scholar |
Xu X, Liao Q, Chen X, Chen X, Lin Y, Lai Z (2019) Molecular characteristics and expression analysis of miR397 family members during the early somatic embryogenesis in Dimocarpus longan Lour. Guoshu Xuebao 36, 567–577.
Xue C, Yao JL, Qin MF, Zhang MY, Allan AC, Wang DF, Wu J (2019) PbrmiR397a regulates lignification during stone cell development in pear fruit. Plant Biotechnology Journal 17, 103–117.
| PbrmiR397a regulates lignification during stone cell development in pear fruit.Crossref | GoogleScholarGoogle Scholar | 29754465PubMed |
Yakovlev IA, Fossdal CG, Johnsen O (2010) MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce. New Phytologist 187, 1154–1169.
| MicroRNAs, the epigenetic memory and climatic adaptation in Norway spruce.Crossref | GoogleScholarGoogle Scholar |
Yamasaki H, Hayashi M, Fukazawa M, Kobayashi Y, Shikanai T (2009) SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. The Plant Cell 21, 347–361.
| SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 19122104PubMed |
Yang X, Wang L, Yuan D, Lindsey K, Zhang X (2013) Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. Journal of Experimental Botany 64, 1521–1536.
| Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis.Crossref | GoogleScholarGoogle Scholar | 23382553PubMed |
Yu Y (2020) LACCASE2 negatively regulates lignin deposition of Arabidopsis roots. Plant Physiology 182, 1190–1191.
| LACCASE2 negatively regulates lignin deposition of Arabidopsis roots.Crossref | GoogleScholarGoogle Scholar | 32127430PubMed |
Yu X, Gong H, Cao L, Hou Y, Qu S (2020) MicroRNA397b negatively regulates resistance of Malus hupehensis to Botryosphaeria dothidea by modulating MhLAC7 involved in lignin biosynthesis. Plant Science 292, 110390
| MicroRNA397b negatively regulates resistance of Malus hupehensis to Botryosphaeria dothidea by modulating MhLAC7 involved in lignin biosynthesis.Crossref | GoogleScholarGoogle Scholar | 32005395PubMed |
Zandkarimi H, Bedre R, Solis J, Mangu V, Baisakh (2015) Sequencing and expression analysis of salt-responsive miRNAs and target genes in the halophyte smooth cordgrass (Spartina alternifolia Loisel). Molecular Biology Reports 42, 1341–1350.
| Sequencing and expression analysis of salt-responsive miRNAs and target genes in the halophyte smooth cordgrass (Spartina alternifolia Loisel).Crossref | GoogleScholarGoogle Scholar | 25976974PubMed |
Zhang J, Zhang S, Han S, Wu T, Li X, Li W, Qi L (2012) Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis. Planta 236, 647–657.
| Genome-wide identification of microRNAs in larch and stage-specific modulation of 11 conserved microRNAs and their targets during somatic embryogenesis.Crossref | GoogleScholarGoogle Scholar | 22526500PubMed |
Zhang Y-C, Yu Y, Wang C-Y, Li Z-Y, Liu Q, Xu J, Liao J-Y, Wang X-J, Qu L-H, Chen F, Xin P, Yan C, Chu J, Li H-Q, Chen Y-Q (2013) Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nature Biotechnology 31, 848
| Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching.Crossref | GoogleScholarGoogle Scholar | 23873084PubMed |
Zhang J, Wang M, Cheng F, Dai C, Sun Y, Lu J, Huang Y, Li M, He Y, Wang F, Fan B (2016) Identification of microRNAs correlated with citrus granulation based on bioinformatics and molecular biology analysis. Postharvest Biology and Technology 118, 59–67.
| Identification of microRNAs correlated with citrus granulation based on bioinformatics and molecular biology analysis.Crossref | GoogleScholarGoogle Scholar |
Zhao M, Tai H, Sun S, Zhang F, Xu Y, Li WX (2012) Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency. PLoS One 7, e29669
| Cloning and characterization of maize miRNAs involved in responses to nitrogen deficiency.Crossref | GoogleScholarGoogle Scholar | 23209616PubMed |
Zheng Y, Chen C, Liang Y, Sun R, Gao L, Liu T, Li D (2019) Genome-wide association analysis of the lipid and fatty acid metabolism regulatory network in the mesocarp of oil palm (Elaeis guineensis Jacq.) based on small noncoding RNA sequencing. Tree Physiology 39, 356–371.
| Genome-wide association analysis of the lipid and fatty acid metabolism regulatory network in the mesocarp of oil palm (Elaeis guineensis Jacq.) based on small noncoding RNA sequencing.Crossref | GoogleScholarGoogle Scholar | 30137626PubMed |
Zhong J, He W, Peng Z, Zhang H, Li F, Yao J (2020) A putative AGO protein, OsAGO17, positively regulates grain size and grain weight through OsmiR397b in rice. Plant Biotechnology Journal 18, 916–928.
| A putative AGO protein, OsAGO17, positively regulates grain size and grain weight through OsmiR397b in rice.Crossref | GoogleScholarGoogle Scholar | 31529568PubMed |
Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. Journal of Experimental Botany 61, 4157–4168.
| Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa.Crossref | GoogleScholarGoogle Scholar | 20729483PubMed |
Zhou ZS, Song JB, Yang ZM (2012) Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium. Journal of Experimental Botany 63, 4597–4613.
| Genome-wide identification of Brassica napus microRNAs and their targets in response to cadmium.Crossref | GoogleScholarGoogle Scholar | 22760473PubMed |
Zhou Q, Yang Y-C, Shen C, He C-T, Yuan J-G, Yang Z-Y (2017) Comparative analysis between low- and high-cadmium-accumulating cultivars of Brassica parachinensis to identify difference of cadmium-induced microRNA and their targets. Plant and Soil 420, 223–237.
| Comparative analysis between low- and high-cadmium-accumulating cultivars of Brassica parachinensis to identify difference of cadmium-induced microRNA and their targets.Crossref | GoogleScholarGoogle Scholar |