Proteomic analysis of young sugarcane plants with contrasting salt tolerance
Denise A. Chiconato A C , Marília G. de Santana Costa B , Tiago S. Balbuena B , Rana Munns
B , Tiago S. Balbuena B , Rana Munns  C D E and Durvalina M. M. dos Santos A
C D E and Durvalina M. M. dos Santos A
A Department of Biologia Aplicada à Agropecuária, Universidade Estadual Paulista ‘Julio de Mesquita Filho’, 14884-900 Jaboticabal, SP, Brasil.
B Department of Tecnologia, Universidade Estadual Paulista ‘Julio de Mesquita Filho’, 14884-900 Jaboticabal, SP, Brasil.
C CSIRO Agriculture and Food, GPO Box 1700, Canberra, ACT 2601, Australia.
D School of Agriculture and Environment, and ARC Centre of Excellence in Plant Energy Biology, University of Western Australia, Crawley, WA 6009, Australia.
E Corresponding author. Email: rana.munns@csiro.au
Functional Plant Biology 48(6) 588-596 https://doi.org/10.1071/FP20314
Submitted: 8 October 2020 Accepted: 19 January 2021 Published: 15 February 2021
Journal Compilation © CSIRO 2021 Open Access CC BY-NC-ND
Abstract
Soil salinity affects sugarcane (Saccharum officinale L.) production in arid and semiarid climates, severely reducing productivity. This study aimed to identify differentially regulated proteins in two cultivars that differ markedly in tolerance of saline soil. Plants were grown for 30 days and then subjected to treatments of 0 and 160 mM NaCl for 15 days. The tolerant cultivar showed a 3-fold upregulation of lipid metabolising enzymes, GDSL-motif lipases, which are associated with defence to abiotic stress, and which were not upregulated in the sensitive cultivar. Lipoxygenase was 2-fold upregulated in the tolerant cultivar but not in the sensitive cultivar, as were Type III chlorophyll a/b binding proteins. Other differences were that in the sensitive cultivar, the key enzyme of C4 photosynthesis, phosphoenolpyruvate carboxylase was downregulated, along with other chloroplast enzymes. Na+ concentrations had not reached toxic concentrations in either cultivar by this time of exposure to salt, so these changes would be in response to the osmotic effect of the soil salinity, and likely be in common with plants undergoing drought stress.
Keywords: Saccharum, acylhydrolase, peroxiredoxin, sodium exclusion, sugarcane, salinity.
Introduction
Soil salinity affects 7% of the world’s arable land, leading to significant losses of productivity in agricultural crops (Munns and Tester 2008; FAO 2011). Sugarcane (Saccharum officinale L., Poaceae) is an important crop for sugar and ethanol production, being cultivated in several tropical and subtropical areas of the world, with Brazil the largest producer of these products. Sugarcane is moderately sensitive to salinity, and it is estimated that one million hectares of land planted with this crop are affected by salt (Patade et al. 2011). Sugarcane cultivation in salinised soils may lead to a productivity decrease of 50% or more when compared with non-saline soils (Suprasanna et al. 2011; Sengar et al. 2013; Kumar et al. 2014).
Salinity affects plants in two ways, through its osmotic and its ionic effects (Munns and Tester 2008). The high concentration of salt in the soil solution lowers the soil water potential and reduces water uptake and growth rate. This osmotic effect results in decreased stomatal conductance and hence photosynthesis. Plants also regulate the uptake of Na+ and Cl– to avoid these ions accumulating to toxic levels. Plants exclude ~95% of the salt in the soil solution while they take up water (Munns et al. 2020a). Exclusion by roots, along with the regulation of transport and cellular compartmentation of the 5% of salt taken up, is needed to sequester the salt within vacuoles and prevent it building up in the cytoplasm. This is energy demanding and has a high energy cost, possibly consuming 10% of the ATP produced by respiration (Munns et al. 2020b). In addition, the reduction in CO2 assimilation may increase the production of reactive oxygen species (ROS), leading to lipid peroxidation and electrolyte leakage, which causes increased leaf senescence and eventually death of the whole plant (Gupta and Huang 2014; Negrão et al. 2017). Knowledge of key enzymes and protective mechanisms involved in resistance to soil salinity can be gained by comparing closely related genotypes differing in salt tolerance. In this study, we have compared the response of two sugarcane cultivars with contrasting tolerance: cv. SP 81–3250, which showed no reduction in growth or photosynthesis after a month in 160 mM NaCl and cv. IAC 87–3396, which was markedly affected (Chiconato et al. 2019).
Plant tolerance to salinity is a complex set of mechanisms and involves many genes and proteins directly or indirectly related to plant protection. For this reason, proteomics is an appropriate and efficient technology for studies of plant tolerance to salinity stress as it allows the identification of many types of stress defence proteins. These proteins perform a large range of functions, such as protection, ion transport, energy capture and transfer, and signalling (Rodziewicz et al. 2014).
In the ‘omics’ context in relation to crop improvement, studies on metabolomics and proteomics are superseding transcriptomics in usefulness. Gene expression alone does not provide complete information on protein synthesis or activity, as post-transcriptional and post-translational modifications regulate the function and activity of many proteins (Chen and Harmon 2006). The status of sugarcane proteomics has been reviewed by Barnabas et al. (2015) and Ali et al. (2019). Proteomics can provide an analysis of protein functions in complex and dynamic applications, such as those related to cell signalling and interactions between proteins (Cox and Mann 2007). Quantification by spectral counting is a label-free quantification method, which is efficient and low cost (Liu et al. 2004; Old et al. 2005). Proteomic studies have been used to study developmental changes in sugarcane (Maranho et al. 2019), and salt stress was shown to induce changes in the proteomic profile of micro-propagated sugarcane shoots (Passamani et al. 2017). However, there are no published proteomic studies on sugarcane plants growing in saline soil.
The aim of this work was to identify differentially regulated proteins in young plants of two sugarcane cultivars that differ in salt tolerance. Results from previous experiments (Chiconato et al. 2019) had shown that salinity up to 160 mM NaCl for 30 days did not inhibit growth or photosynthesis of the tolerant cultivar SP 81–3250 but that growth and photosynthesis of the sensitive cultivar IAC 87–3396 was reduced by half (Fig. S1). After 15 days of salt treatment, growth of the sensitive cultivar was affected as shown by decreases in both shoot and root biomass. However, at this time the younger leaves showed no sign of injury: chlorophyll content and chlorophyll fluorescence were normal, and the Na+ concentrations were still low (Chiconato et al. 2019). Even at 30 days, the reduction in photosynthesis in the sensitive cultivar was due to stomatal limitations, not damage to the photosystem. Consequently, in this study the youngest fully expanded leaf of plants of both cultivars were sampled at 15 days for proteomic analysis, to identify proteins that were adaptive rather than those merely responding to injury.
Materials and methods
Plant material and growing conditions
The experiments were conducted at the State University of São Paulo, Campus Jaboticabal, São Paulo, Brazil, in a greenhouse with natural light and average temperature of 30°C and relative humidity of 40%. Two sugarcane cultivars, SP 81–3250 and IAC 87–3396, were provided as small stem cuttings (setts or mini-totes) by São Paulo Agribusiness Technology Agency. The cv. SP 81–3250 has much higher tolerance to salinity than cv. IAC 87–3396 (Chiconato et al. 2019) and are referred to below as the tolerant and the sensitive cultivar respectively. The plants from the stem cuttings were transplanted into 2-L pots filled with soil (Dystrophic Red Latosol) with a chemical and textural characterisation as described previously (Chiconato et al. 2019). Soil fertilisation was carried out in three stages every 10 days using 0.3 g of NH4H2PO4 and 0.35 g of KH2PO4 per pot (incorporated into soil before transplanting), then 0.45 g of urea and 0.35 g of K2SO4 (added after 10 and 20 days) with daily irrigation to restore the water loss through evapotranspiration.
Experimental design was completely randomised, in a 2 × 2 factorial scheme, with two sugarcane cultivars (SP 81–3250 and IAC 87–3396) × two NaCl concentrations (0 and 160 mM), with four replicates per treatment. After 30 days, the salt treatment started and plants were harvested after 15 days in 160 mM NaCl.
Extraction and digestion of proteins
Analyses were performed on the first leaf from the top with clearly visible dewlap. Foliar proteins were extracted using the phenolic method (Hurkman and Tanaka 1986). Samples containing 1 g of pulverised plant material macerated in liquid nitrogen were transferred to fresh tubes and 4 mL of extraction buffer (500 mM Tris, pH 8.0, 50 mM EDTA, 700 mM sucrose, 100 mM KCl and 1% β-mercaptoethanol) were added. After 1 h of incubation, an equal volume of equilibrated phenol solution (Sigma-Aldrich) was added to the extract and homogenised for 5 min at 4°C. After 15 min of centrifugation at 3200g the organic and aqueous phases were separated. Proteins were precipitated from the phenol phase by the addition of 10 mL of 0.1 M ammonium acetate in methanol for 16 h at –20°C followed by centrifugation at 3200g for 15 min at 4°C. Subsequently, the protein pellets were washed twice in 2 mL of ice-cold acetone, followed by centrifugation at 3200g for 15 min. The proteins were then resuspended in 100 μL of sample buffer (6 M urea in 25 mM Ambic). Protein extract concentration was determined by the method described by Bradford (1976) using bovine serum albumin concentrations (0.0625, 0.125, 0.25, 0.5 mg L–1) for the standard curve.
For all samples, 100-μg aliquots of the digested protein solution were used, which was carried out for 12 h at 37°C with the addition of trypsin at 1:50 (enzyme:protein) ratio. Then the samples were vacuum dried, desalted using Millipore Ziptip C18 columns (Sigma-Aldrich) as recommended by the manufacturer, and resuspended in 0.1% formic acid at the time of the liquid chromatographic separation coupled to the mass spectrometer.
Mass spectrometric analysis
Peptides were separated through a C18 column (15 cm, 3 μm, 120Å) by reverse phase liquid chromatography, with a 120-min gradient of 5–70% of formic acid in acetonitrile at a flow of 500 nL min–1. Mass spectrometer analysis was performed using a Q-Exactive (Thermo Fisher Scientific), operated in the data dependent acquisition (DDA) positive ion mode with cycles consisting of a full scan at 70 000 FWHM (full width at half maximum) (400–2000 m/z) followed by 10 cycles of ‘data dependent scans’ at 35 000 FWHM. Peptide fragmentation was obtained by HCD (higher-energy collisional dissociation) fragmentation using a collision energy of 27eV, with dynamic exclusion of 30 s for the selected ions.
Protein identification and data analysis
Protein identification was performed using the spectral counting method with sorghum (Sorghum bicolor L.) protein sequences, available from the Phytozome site (Goodstein et al. 2012). The sorghum genome database was chosen as being more complete and searchable than the sugarcane genome and was more comparable than maize (Zea mays L.) or rice (Oryza sativa L.). Identifications were made using the integrated PatternLab for Proteomics platform (Carvalho et al. 2016). For the spectral correlation, the Comet tool (Eng et al. 2015), available in the platform was used with the following modifications: cysteine carbamidomethylation and methionine oxidation, as static and variable modification, respectively. Every spectral alignments were filtered with SePro (Carvalho et al. 2012) and adjusted to 1% false– positive FDR (false discovery rate). Proteins that shared common peptides were grouped according to the principle of maximum parsimony and protein abundance were evaluated using the normalised spectral abundance factor (NSAF) approach (Paoletti et al. 2006).
In order to obtain a general analysis of the proteome response of young sugarcane plants submitted to salinity (160 mM NaCl), differentially regulated proteins were determined using the fold-change criterion of at least 1.5 times and P < 0.05, according to the Kruskal–Wallis non-parametric method.
Statistical analysis
Statistical analysis was performed by the statistical software AgroStat (Barbosa and Maldonado 2011). The data were submitted to variance analysis by the F test and the significant differences between the treatments were compared by the Tukey test at 5% probability.
Results
This study compared the proteome response of young leaves of the salt-tolerant cv. SP 81–3250 with the salt-sensitive cv. IAC 87–3396 grown for 15 days in 160 mM NaCl. Previous studies had found that this treatment reduced growth of the sensitive cultivar in terms of shoot and root biomass, but no injury had occurred in the younger leaves (Chiconato et al. 2019). Consequently, the youngest fully expanded leaves of both cultivars were sampled at 15 days for proteomic analysis to catch protein changes that would adapt plants to salt stress rather than those merely responding to injury. Proteins that were up or downregulated in response to NaCl were identified, then their differential expression was compared between the two cultivars.
Salt-tolerant cv. SP 81–3250
After 15 days at 160 mM NaCl, eight proteins were identified with a significant increase in their abundance, and 10 proteins with a significant reduction, in relation to controls grown over the same period of time without the addition of NaCl (Fig. 1).
Among the differentially regulated proteins, two proteoforms of GDSL-motif Lipase/Acylhydrolase showed greatest abundance (Fig. 1) with ‘fold-change’ of 2.96 and 2.30 respectively (Table S1) when under salt stress conditions. In parallel, another protein involved in lipid metabolism, lipoxygenase enzyme 9-LOX, showed an increase of 1.71-fold when grown in saline conditions (Fig. 1; Table S1).
Other upregulated proteins were Histone H2A, 50S ribosomal protein L12, Type III chlorophyll a/b-binding protein, H+-transporting ATP synthase chain 9, and the chloroplast precursor peroxiredoxin Q (Fig. 1).
Proteins downregulated were mainly concerned with photosynthetic carbon assimilation and metabolism (Fig. 1; Table S1). These included chloroplastic pyruvate, phosphate dikinase, ATP synthase subunit gamma, Photosystem II P680 chlorophyll a apoprotein, and ferredoxin. The most downregulated protein was the chloroplast translational elongation factor Tu (Fig. 1).
Salt-sensitive cv. IAC 87–3396
For young plants of cv. IAC 87–3396, 10 proteins were identified with a significant increase in their abundance and 23 proteins that were downregulated under salinity (Fig. 2).
Among the proteins showing increased expression, triosephosphate isomerase (TPI) showed the greatest (2.99-fold) increase (Fig. 2;Table S2). Other chloroplastic enzymes were also upregulated: ATP synthase β subunit, ATP synthase delta chain, and 2-Cys peroxiredoxin BAS1. Malate dehydrogenase [NADP] showed upregulation for two proteoforms. The stress-related protein, Heatshock 70 increased in this cultivar (Fig. 2; Table S2).
Among downregulated proteins were Type III chlorophyll a/b-binding, which was upregulated in the tolerant cultivar. The other downregulated proteins were specific to the sensitive cultivar and were largely involved in carbohydrate metabolism and chloroplast function. Notably, PEP carboxylase, the primary CO2 acceptor in C4 photosynthesis, was downregulated. The greatest degree of downregulation was for proteins concerned with protein synthesis during cell division and elongation (Fig. 2; Table S2).
Comparison of tolerant versus sensitive cultivars
A quantitative analysis was made of the eight proteins that were upregulated in the tolerant cultivar when exposed to salt (Fig. 1) compared with their expression in the sensitive cultivar. Fig. 3 shows that the major differences in the proteomic salt response between the two sugarcane genotypes were in GDSL-motif lipases, a lipoxygenase and a chlorophyll a/b-binding protein.
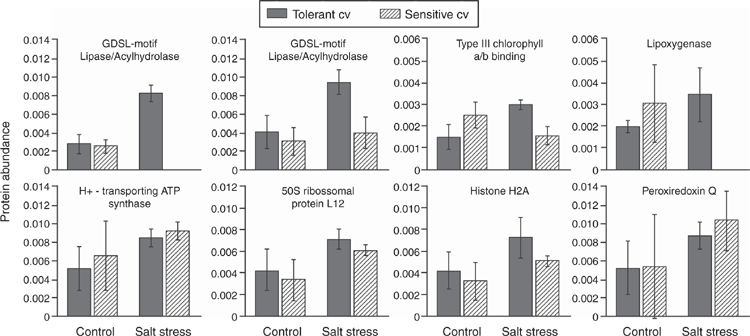
|
For the two proteoforms of GDSL-motif lipases (Sobic number 008G007800.1 and 008G007800.2), both of which were upregulated in the tolerant cultivar under salt stress, one was not expressed in the salt-treated sensitive cultivar, and the other showed no change in expression in response to salt stress. Type III chlorophyll a/b-binding protein (Sobic 010G189300.2) was downregulated in response to stress in the sensitive cultivar, and lipoxygenase (Sobic 003G305900.1) was not expressed in the salt-treated sensitive cultivar (Fig. 3). The other four proteins that were upregulated in the tolerant plants, the H+-transporting ATPase (Sobic 001G417200.1), 50S ribosomal protein (Sobic 003G246400.1), Histone 2A (Sobic 002G276000.1) and peroxyredoxin Q (Sobic 010G072900.1) were similarly upregulated in the sensitive plants.
Discussion
The differentially regulated proteins identified in response to salt stress were all related to photosynthesis and energy generation, antioxidant activity, and lipid metabolism. Additional energy is needed by plants in saline soil to regulate the uptake and transport of Na+ and Cl– and to provide osmotic adjustment by compartmentation of Na+ and Cl– within vacuoles of cells, along with the synthesis of organic solutes to avoid toxic accumulation of Na+ or Cl– (Munns et al. 2020b). Antioxidant activity is needed to control the excessive production of reactive oxygen species (ROS) produced by plants under stress (Demidchik 2015).
Proteins upregulated in the tolerant cv. SP 81–3250
The proteins with the greatest increase for the tolerant cultivar were two proteoforms of GDSL-motif lipase/acylhydrolase. The protein abundance (Fig. 3) shows that the two proteoforms of this enzyme, although they were present in the control plants of both cultivars, only showed increased expression under salt stress in tolerant plants. These GDSL-lipases are a large gene family that hydrolyse lipids and with multiple functions in plants widely related to pest attack and abiotic stress (Hong et al. 2008; Tan et al. 2014), as well as plant morphogenesis, development and seed germination (Ling et al. 2006). Enzymes of the lipase group are possibly anchored in the plasma membrane, releasing fatty acids from the lipids, and have an important function in cell signalling, contributing to defence mechanisms of salinity tolerance (Naranjo et al. 2006). In Arabidopsis thaliana L., a gene from the GDSL-motif lipase group, AtLTL1, was induced by salinity and associated with salt tolerance as shown by overexpression studies (Naranjo et al. 2006). In rice, GDSL-like Lipase/Acylhydrolase gene expression was among the drought and salt-responsive cell wall-related genes induced in both leaves and roots (Landi et al. 2017).
Also upregulated were proteins that protect plants from lipid peroxidation. Lipoxygenases (LOX) catalyse the oxygenation of long chain polyunsaturated fatty acids into short chain and volatile compounds from which are derived jasmonic acid and other signalling agents. Lipoxygenase enzyme is involved in many physiological processes during plant development and under stress, acting to modulate ROS accumulation, lipid peroxidation and gene expression, and conferring tolerance to biotic and abiotic stresses such as pathogen attack, drought, salinity and oxidative stress (Lim et al. 2015; Shaban et al. 2018). Oxidative stress results from every abiotic and biotic stress that affects plant growth (Demidchik 2015), with accumulation of ROS caused by the increased electron transport activity in the photosystems and the reduced carbon metabolism under salt stress (Silveira and Carvalho 2016). In Capsicum annuum L., CaLOX1-overexpressing plants exhibited tolerance phenotypes to drought and high salt stresses and showed more rapid scavenging of ROS and higher expression of ABA- and stress-responsive marker genes (Lim et al. 2015).
Peroxiredoxins (Prx) are a widespread family of one of the major ROS-scavenging enzymes that act as antioxidants in ROS elimination, specifically on hydroperoxidases, protecting the plant from lipid peroxidation. Peroxiredoxin Q is a member of this family and overexpression of this gene from the halophyte Suaeda salsa L. conferred salt tolerance to a non-halophytic species Eustoma grandiflorum Shinn (Guan et al. 2014). These enzymes are responsive to salinity and may be an essential response to the increase in ROS. Our results for Peroxiredoxin Q corroborate those found by Xu et al. (2015), who observed the same increase of PrxQ in plants exposed to salinity for 1 h, promoting stronger defence against salt-induced oxidative stress. Nevertheless, these authors reported the action of this enzyme in the regulation of other enzymes of the oxidising system and cell redox balance.
In addition to enzymes related to oxidative stress tolerance, other upregulated proteins in the tolerant cultivar are involved in energy production. Type III chlorophyll a/b-binding proteins, although they were present in the sensitive cultivar under salinity, only showed increased activity in the tolerant one (Fig. 3). These proteins are involved in the photosynthetic light reactions of plants. Their upregulation suggests that photosynthetic mechanism is not impaired and able to respond to increased demand for energy production under salt stress. Sui et al. (2015) studied two sorghum genotypes submitted to salinity. These authors found that gene expression for chlorophyll a/b-binding proteins and other chloroplast proteins were downregulated in the sensitive cultivar, whereas some transcripts were either not changed or upregulated in the tolerant cultivar. In addition, plants of this cultivar showed increase of ATP synthase chain 9, a protein needed to supply additional energy for plant under salt stress, such as for the regulation of Na+ transport into cell compartments, avoiding the excessive accumulation of Na+ in the cytosol (Xu et al. 2015).
Histones organise DNA into nucleosomes and so influence the transcription of the nuclear genome. Histone H2A can affect ABA signalling (Liu et al. 2009; Zhu et al. 2012; Luo et al. 2017) and consequently stomatal closure, which would reduce the loss of water by leaves and maintain water status. Histone interaction with DNA confers a greater reinforcement in the structure of the nucleosomes before the ionic strength in concentrated solutions, and so can regulate transcription in response to ABA (Liu et al. 2009; Zhu et al. 2012).
The chloroplastic 50S ribosomal protein L12 is a component of the chloroplast translation machinery for the synthesis of chloroplast-encoded proteins for the photosynthetic apparatus. Fatehi et al. (2012) found upregulation of this protein for two genotypes of barley (Hordeum vulgare L.) under salt stress and concluded that these plants showed tolerance to the NaCl inhibitory effect in protein synthesis. The downregulation of this protein in soybean leaves was associated with a reduction in plant growth (Ahmad et al. 2016). Upregulation of 50S ribosomal protein L12-in both sugarcane cultivars (Fig. 3) indicates that protein synthesis had increased in response to the stress.
Proteins upregulated in the sensitive cv. IAC 87–3396
Plants of the sensitive cultivar, having reduced growth in saline soil, showed upregulation of proteins involved in energy production and protection. Triosephosphate isomerase (TPI) is an important enzyme in primary metabolism, which catalyses the reaction between dihydroxyacetone phosphate and 3-P glyceraldehyde which feeds into the glycolysis pathway. Gao et al. (2011) found 3.3-fold increase in the level of that same enzyme in wheat (Triticum aestivum L.) plants under salinity. These authors concluded that the increase in expression of this enzyme, related to the regulation of glucose metabolism, was due to the additional energetic needs of the plant under stress. Furthermore, upregulation of the triosephosphate isomerase enzyme was found in barley plants under salt stress (Alikhani et al. 2013; Shen et al. 2017), in wheat under salt stress (Gao et al. 2011) and in maize under water deficiency (Riccardi et al. 1998).
Other proteins concerned with energy production were upregulated under salinity. Among them, ATP synthase β subunit and ATP synthase γ chain, both related to glucose metabolism in the production of ATP and consequently to the TPI mentioned above. Gao et al. (2011) found an increase in ATPase subunits proteins in wheat plants under salinity and discussed their importance in biological processes under salt stress, during which there is an increased demand for energy for ion transport mechanisms and osmotic adjustment with organic solutes (Munns and Tester 2008), and a continued supply of ATP from chloroplast or mitochondrion for the activity of the H+-ATPases at the plasma membrane and other membranes.
Another important upregulated stress protein in the sensitive cultivar was 2-Cys peroxiredoxin BAS1. It plays an important role in the elimination of hydroperoxides, both alkyl hydroperoxides formed from lipids by lipoxygenase and of hydrogen peroxide (Kitajima 2008). These are reactive oxygen species and show increased production when the plant is undergoing salinity stress. The upregulation of this protein was observed by Lv et al. (2016) in wheat plants during salinity and recovery. Heatshock 70 kDa, also upregulated, is commonly expressed in response to abiotic stresses which can cause oxidative stress. Heat shock proteins are molecular chaperones which protect protein structure in plants under environmental stress. This may be through a mechanism involving ROS scavenging: their expression during oxidative stress may be induced by H2O2 functioning as a signalling molecule (Sun et al. 2002).
In cv. IAC 87–3396, we also observed downregulation over 6-fold of proteins directly involved with translation and cell growth, and to a lesser extent those involved with photosynthetic and carbon metabolism. Among the listed downregulated proteins is Phosphoenolpyruvate carboxylase 1 (PEPCase), an important enzyme of C4 plants and the primary acceptor of CO2. Eprintsev et al. (2011) found inhibition of malate dehydrogenase [NADP] (MDH) in maize subjected to 1 h of NaCl stress. These authors commented on the damage caused in the C4 photosynthetic cycle of these plants, but even so, there was a high rate of CO2 absorption, confirmed by the reduction of phosphoenolpyruvate. Also in maize, Omoto et al. (2012) found upregulation of MDH, together with PEPCase, and attributed this response to an adaptation to salinity. Our results with the sensitive cultivar were different: although there was upregulation of MDH, there was downregulation of PEPCase, demonstrating that there were severe losses in the photosynthetic activity of these plants. Malate is a metabolite that coordinates the functioning of tissues such as mesophyll and vascular bundle sheath and, therefore enzymes such as malate dehydrogenase are important in plant responses to environmental changes. MDH is also involved in the reduced dependence on glucose for energetic synthesis, with rapid mobilisation and storage of organic acids (Eprintsev et al. 2011).
Concluding remarks
Here we sampled the most recently expanded leaves at a time (15 days in 160 mM NaCl) that was long enough for differences in salt tolerance between cultivars to be seen in terms of growth reductions yet early enough that the younger leaves were healthy and without injury (Chiconato et al. 2019). Changes in the proteome in the youngest fully expanded leaf would therefore be expected to involve signalling and energy production, as well as dealing with increased ROS. Although the tolerant cultivar showed a greater ability to exclude Na+ from leaves, at 15 days Na+ had not reached a concentration in leaves or roots of either cultivar that could be considered toxic (Chiconato et al. 2019). Changes in the proteome therefore would be in response to the osmotic stress not the salt-specific stress, and so likely to be in common with drought or water stress. Both the control of ROS and the energy demands for synthesis of compatible solutes are in common with drought and salinity (Munns 2011).
In conclusion, sugarcane plants of the tolerant cultivar showed an increase in the production of proteins associated with defence to abiotic stress. These enzymes are involved in signalling pathways and antioxidant systems, their effectiveness evidenced by little reduction in photosynthesis and shoot biomass in that cultivar. The sensitive cultivar showed upregulation of important stress proteins, however, those directly related to photosynthesis and development were downregulated in these plants, which is consistent with the reduction in growth and biomass of the plants. We suggest that the upregulated proteins in the tolerant cultivar provided adaptation that allowed continued growth and energy production, whereas those that were upregulated in the more sensitive cultivar protected the plant from stress as it grew more slowly and are in factor markers of salt-stress. These markers have the potential for selecting sugarcane genotypes that differ in salt tolerance, and for breeding more salt-tolerant sugarcane cultivars.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
The authors acknowledge the CAPES program (Coordination for the Improvement of Higher Education Personnel) for providing a doctorate and research scholarship for D.C., and support from São Paulo Research Foundation (FAPESP), process n. 2011/11650–0.
References
Ahmad P, Latef AAHA, Rasool S, Akram NA, Ashraf M, Gucel S (2016) Role of proteomics in crop stress tolerance. Frontiers in Plant Science 7, 1336| Role of proteomics in crop stress tolerance.Crossref | GoogleScholarGoogle Scholar | 27660631PubMed |
Ali A, Khan M, Sharif R, Mujtaba M, Gao SJ (2019) Sugarcane omics: an update on the current status of research and crop improvement. Plants 8, 344
| Sugarcane omics: an update on the current status of research and crop improvement.Crossref | GoogleScholarGoogle Scholar |
Alikhani M, Khatabi B, Sepehri M, Nekouei MK, Mardi M, Salekdeh G (2013) A proteomics approach to study the molecular basis of enhanced salt tolerance in barley (Hordeum vulgare L.) conferred by the root mutualistic fungus Piriformospora indica. Molecular BioSystems 9, 1498–1510.
| A proteomics approach to study the molecular basis of enhanced salt tolerance in barley (Hordeum vulgare L.) conferred by the root mutualistic fungus Piriformospora indica.Crossref | GoogleScholarGoogle Scholar | 23545942PubMed |
Barbosa JC, Maldonado W, Junior (2011) AgroEstat - Sistema para análises estatísticas de ensaios agronômicos versão 1.1.0.694. Jaboticabal: FCAV/UNESP.
Barnabas L, Ramadass A, Amalraj RS, Palaniyandi M, Rasappa V (2015) Sugarcane proteomics: an update on current status, challenges, and future prospects. Proteomics 15, 1658–1670.
| Sugarcane proteomics: an update on current status, challenges, and future prospects.Crossref | GoogleScholarGoogle Scholar | 25641866PubMed |
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254.
| A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding.Crossref | GoogleScholarGoogle Scholar | 942051PubMed |
Carvalho PC, Fischer JS, Xu T, Yates JR, Barbosa VC (2012) PatternLab: from mass spectra to label‐free differential shotgun proteomics. Current Protocols in Bioinformatics 40, 13.19.1–13.19.18.
| PatternLab: from mass spectra to label‐free differential shotgun proteomics.Crossref | GoogleScholarGoogle Scholar |
Carvalho PC, Lima DB, Leprevost FV, Santos MD, Fischer JS, Aquino PF, Moresco JJ, Yates JR, Barbosa VC (2016) Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0. Nature Protocols 11, 102–117.
| Integrated analysis of shotgun proteomic data with PatternLab for proteomics 4.0.Crossref | GoogleScholarGoogle Scholar | 26658470PubMed |
Chen SX, Harmon AC (2006) Advances in plant proteomics. Proteomics 6, 5504–5516.
| Advances in plant proteomics.Crossref | GoogleScholarGoogle Scholar |
Chiconato DA, da Silveira Sousa G, dos Santos DMM, Munns R (2019) Adaptation of sugarcane plants to saline soil. Environmental and Experimental Botany 162, 201–211.
| Adaptation of sugarcane plants to saline soil.Crossref | GoogleScholarGoogle Scholar |
Cox J, Mann M (2007) Is proteomics the new genomics? Cell 130, 395–398.
| Is proteomics the new genomics?Crossref | GoogleScholarGoogle Scholar | 17693247PubMed |
Demidchik V (2015) Mechanisms of oxidative stress in plants: from classical chemistry to cell biology. Environmental and Experimental Botany 109, 212–228.
| Mechanisms of oxidative stress in plants: from classical chemistry to cell biology.Crossref | GoogleScholarGoogle Scholar |
Eng JK, Hoopmann MR, Jahan TA, Egertson JD, Noble WS, MacCoss MJ (2015) A deeper look into Comet – implementation and features. Journal of the American Society for Mass Spectrometry 26, 1865–1874.
| A deeper look into Comet – implementation and features.Crossref | GoogleScholarGoogle Scholar | 26115965PubMed |
Eprintsev AT, Fedorina OS, Bessmeltseva YS (2011) Response of the malate dehydrogenase system of maize mesophyll and bundle sheath to salt stress. Russian Journal of Plant Physiology: a Comprehensive Russian Journal on Modern Phytophysiology 58, 448–453.
| Response of the malate dehydrogenase system of maize mesophyll and bundle sheath to salt stress.Crossref | GoogleScholarGoogle Scholar |
FAO (2011) The state of the world’s land and water resources for food and agriculture – Managing systems at risk. Food and Agriculture Organization of the United Nations, Rome and Earthscan, London. http://www.fao.org/ag/agl/agll/spushisk
Fatehi F, Hosseinzadeh A, Alizadeh H, Brimavandi T, Struik PC (2012) The proteome response of salt-resistant and salt-sensitive barley genotypes to long-term salinity stress. Molecular Biology Reports 39, 6387–6397.
| The proteome response of salt-resistant and salt-sensitive barley genotypes to long-term salinity stress.Crossref | GoogleScholarGoogle Scholar | 22297690PubMed |
Gao L, Yan X, Li X, Guo G, Hu Y, Ma W, Yan Y (2011) Proteome analysis of wheat leaf under salt stress by two-dimensional difference gel electrophoresis (2D-DIGE). Phytochemistry 72, 1180–1191.
| Proteome analysis of wheat leaf under salt stress by two-dimensional difference gel electrophoresis (2D-DIGE).Crossref | GoogleScholarGoogle Scholar | 21257186PubMed |
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–D1186.
| Phytozome: a comparative platform for green plant genomics.Crossref | GoogleScholarGoogle Scholar | 22110026PubMed |
Guan C, Liu X, Song X, Wang G, Ji J, Jin C (2014) Overexpression of a peroxiredoxin Q gene, SsPrxQ, in Eustoma grandiflorum Shinn enhances its tolerance to salt and high light intensity. Molecular Breeding 33, 657–667.
| Overexpression of a peroxiredoxin Q gene, SsPrxQ, in Eustoma grandiflorum Shinn enhances its tolerance to salt and high light intensity.Crossref | GoogleScholarGoogle Scholar |
Gupta B, Huang B (2014) Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. International Journal of Genomics 2014, 701596
| Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization.Crossref | GoogleScholarGoogle Scholar | 24804192PubMed |
Hong JK, Choi HW, Hwang IS, Kim DS, Kim NH, Choi DS, Kim YJ, Hwang BK (2008) Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 227, 539–558.
| Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance.Crossref | GoogleScholarGoogle Scholar | 17929052PubMed |
Hurkman WJ, Tanaka CK (1986) Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiology 81, 802–806.
| Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis.Crossref | GoogleScholarGoogle Scholar | 16664906PubMed |
Kitajima S (2008) Hydrogen peroxide‐mediated inactivation of two chloroplastic peroxidases, ascorbate peroxidase and 2‐cys peroxiredoxin. Photochemistry and Photobiology 84, 1404–1409.
| Hydrogen peroxide‐mediated inactivation of two chloroplastic peroxidases, ascorbate peroxidase and 2‐cys peroxiredoxin.Crossref | GoogleScholarGoogle Scholar | 19067962PubMed |
Kumar T, Khan MR, Jan SA, Ahmad N, Niaz Ali N, Zia MA, Roomi S, Iqbal A, Ali GM (2014) Efficient regeneration and genetic transformation of sugarcane withAVP1 gene. American-Eurasian Journal of Agricultural & Environmental Sciences 14, 165–171.
Landi S, Hausman JF, Guerriero G, Esposito S (2017) Poaceae vs. abiotic stress: focus on drought and salt stress, recent insights and perspectives. Frontiers in Plant Science 8, 1214
| Poaceae vs. abiotic stress: focus on drought and salt stress, recent insights and perspectives.Crossref | GoogleScholarGoogle Scholar | 28744298PubMed |
Lim CW, Han SW, Hwang IS, Kim DS, Hwang BK, Lee SC (2015) The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant & Cell Physiology 56, 930–942.
| The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response.Crossref | GoogleScholarGoogle Scholar |
Ling H, Zhao J, Zuo K, Qiu C, Yao H, Qin J, Sun X, Tang K (2006) Isolation and expression analysis of a GDSL-like lipase gene from Brassica napus L. Journal of Biochemistry and Molecular Biology 39, 297–303.
Liu H, Liu H, Sadygov RG, Sadygov RG, Yates JR (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Analytical Chemistry 76, 4193–4201.
| A model for random sampling and estimation of relative protein abundance in shotgun proteomics.Crossref | GoogleScholarGoogle Scholar | 15253663PubMed |
Liu ZQ, Gao J, Dong AW, Shen WH (2009) A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3–2 mutant. Molecular Plant 2, 688–699.
| A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3–2 mutant.Crossref | GoogleScholarGoogle Scholar | 19825649PubMed |
Luo M, Cheng K, Xu Y, Yang S, Wu K (2017) Plant responses to abiotic stress regulated by histone deacetylases. Frontiers in Plant Science 8, 2147
| Plant responses to abiotic stress regulated by histone deacetylases.Crossref | GoogleScholarGoogle Scholar | 29326743PubMed |
Lv DW, Zhu GR, Zhu D, et al (2016) Proteomic and phosphoproteomic analysis reveals the response and defense mechanism in leaves of diploid wheat T. monococcum under salt stress and recovery. Journal of Proteomics 143, 93–105.
| Proteomic and phosphoproteomic analysis reveals the response and defense mechanism in leaves of diploid wheat T. monococcum under salt stress and recovery.Crossref | GoogleScholarGoogle Scholar | 27095598PubMed |
Maranho RC, Benez MM, Maranho GB, et al (2019) Proteomic analysis of axillary buds of sugarcane at different cutting stages: evidence for alterations in axillary bud gene expression. Crop and Pasture Science 70, 622–633.
| Proteomic analysis of axillary buds of sugarcane at different cutting stages: evidence for alterations in axillary bud gene expression.Crossref | GoogleScholarGoogle Scholar |
Munns R (2011) Plant Adaptations to Salt and Water Stress: Differences and Commonalities. Advances in Botanical Research 57, 1–32.
| Plant Adaptations to Salt and Water Stress: Differences and Commonalities.Crossref | GoogleScholarGoogle Scholar |
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annual Review of Plant Biology 59, 651–681.
| Mechanisms of salinity tolerance.Crossref | GoogleScholarGoogle Scholar | 18444910PubMed |
Munns R, Passioura JB, Colmer TD, Byrt CS (2020a) Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytologist 225, 1091–1096.
| Osmotic adjustment and energy limitations to plant growth in saline soil.Crossref | GoogleScholarGoogle Scholar |
Munns R, Day DA, Fricke W, et al (2020b) Energy costs of salt tolerance in crop plants. New Phytologist 225, 1072–1090.
| Energy costs of salt tolerance in crop plants.Crossref | GoogleScholarGoogle Scholar |
Naranjo MA, Forment J, Roldan M, Serrano R, Vicente O (2006) Overexpression of Arabidopsis thaliana LTL1, a salt‐induced gene encoding a GDSL‐motif lipase, increases salt tolerance in yeast and transgenic plants. Plant, Cell & Environment 29, 1890–1900.
| Overexpression of Arabidopsis thaliana LTL1, a salt‐induced gene encoding a GDSL‐motif lipase, increases salt tolerance in yeast and transgenic plants.Crossref | GoogleScholarGoogle Scholar |
Negrão S, Schmöckel SM, Tester M (2017) Evaluating physiological responses of plants to salinity stress. Annals of Botany 119, 1–11.
| Evaluating physiological responses of plants to salinity stress.Crossref | GoogleScholarGoogle Scholar | 27707746PubMed |
Old WM, Meyer-Arend K, Aveline-Wolf L, Pierce KG, Mendonza A, Sevinsky JR, Resing KA, Ahn NG (2005) Comparison of 14 label-free methods for quantifying human proteins by shotgun proteomics. Molecular & Cellular Proteomics 4, 1487–1502.
| Comparison of 14 label-free methods for quantifying human proteins by shotgun proteomics.Crossref | GoogleScholarGoogle Scholar |
Omoto E, Mitsutaka T, Hiroshi M (2012) Adaptation responses in C4 photosynthesis of maize under salinity. Journal of Plant Physiology 169, 469–477.
| Adaptation responses in C4 photosynthesis of maize under salinity.Crossref | GoogleScholarGoogle Scholar | 22209164PubMed |
Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proceedings of the National Academy of Sciences of the United States of America 103, 18928–18933.
| Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors.Crossref | GoogleScholarGoogle Scholar | 17138671PubMed |
Passamani LZ, Barbosa RR, Reis RS, et al (2017) Salt stress induces changes in the proteomic profile of micropropagated sugarcane shoots. PLoS One 12, e0176076
| Salt stress induces changes in the proteomic profile of micropropagated sugarcane shoots.Crossref | GoogleScholarGoogle Scholar | 28419154PubMed |
Patade VY, Bhargava S, Suprasanna P (2011) Salt and drought tolerance of sugarcane under iso-osmotic salt and water stress: growth, osmolytes accumulation, and antioxidant defense. Journal of Plant Interactions 6, 275–282.
| Salt and drought tolerance of sugarcane under iso-osmotic salt and water stress: growth, osmolytes accumulation, and antioxidant defense.Crossref | GoogleScholarGoogle Scholar |
Riccardi F, Gazeau P, Vienne D, Zivy M (1998) Protein changes in response to progressive water deficit in maize quantitative variation and polypeptide identification. Plant Physiology 117, 1253–1263.
| Protein changes in response to progressive water deficit in maize quantitative variation and polypeptide identification.Crossref | GoogleScholarGoogle Scholar | 9701581PubMed |
Rodziewicz P, Swarcewicz B, Chmielewska K, Wojakowska A, Stobiecki M (2014) Influence of abiotic stresses on plant proteome and metabolome changes. Acta Physiologiae Plantarum 36, 1–19.
| Influence of abiotic stresses on plant proteome and metabolome changes.Crossref | GoogleScholarGoogle Scholar |
Sengar K, Sengar RS, Singh A (2013) Biotechnological and genomic analysis for salinity tolerance in sugarcane. International Journal of Biotechnology and Bioengineering Research 4, 407–414.
Shaban M, Ahmed MM, Sun H, Ullah A, Zhu L (2018) Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses. BMC Genomics 19, 599
| Genome-wide identification of lipoxygenase gene family in cotton and functional characterization in response to abiotic stresses.Crossref | GoogleScholarGoogle Scholar | 30092779PubMed |
Shen QF, Fu LB, Qiu L, Xue F, Zhang GP (2017) Time course of ionomic and proteomic analysis of a Tibetan wild barley at early stage under salt stress. Plant Growth Regulation 81, 11–21.
| Time course of ionomic and proteomic analysis of a Tibetan wild barley at early stage under salt stress.Crossref | GoogleScholarGoogle Scholar |
Silveira JAG, Carvalho FEL (2016) Proteomics, photosynthesis and salt resistance in crops: An integrative view. Journal of Proteomics 143, 24–35.
| Proteomics, photosynthesis and salt resistance in crops: An integrative view.Crossref | GoogleScholarGoogle Scholar | 26957143PubMed |
Sui N, Yang Z, Liu M, Wang B (2015) Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genomics 16, 534
| Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves.Crossref | GoogleScholarGoogle Scholar | 26186930PubMed |
Sun W, Montagu MV, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochimica et Biophysica Acta (BBA) – Gene Structure and Expression 1577, 1–9.
| Small heat shock proteins and stress tolerance in plants.Crossref | GoogleScholarGoogle Scholar |
Suprasanna P, Patade VY, Desai NS, Devarumath RM, Kawar PG, Pagariya MC, Babu KH (2011) Biotechnological developments in sugarcane improvement: an overview. Sugar Tech 13, 322–335.
| Biotechnological developments in sugarcane improvement: an overview.Crossref | GoogleScholarGoogle Scholar |
Tan X, Yan S, Tan R, Zhang Z, Wang Z, Chen J (2014) Characterization and expression of a GDSL-like lipase gene from Brassica napus in Nicotiana benthamiana. The Protein Journal 33, 18–23.
| Characterization and expression of a GDSL-like lipase gene from Brassica napus in Nicotiana benthamiana.Crossref | GoogleScholarGoogle Scholar | 24363150PubMed |
Xu J, Lan H, Fang H, Huang X, Zhang H, Huang J (2015) Quantitative proteomic analysis of the rice (Oryza sativa L.) salt response. PLoS One 10, e0120978
| Quantitative proteomic analysis of the rice (Oryza sativa L.) salt response.Crossref | GoogleScholarGoogle Scholar | 26720634PubMed |
Zhu Y, Dong A, Shen W (2012) Histone variants and chromatin assembly in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms 1819, 343–348.
| Histone variants and chromatin assembly in plant abiotic stress responses.Crossref | GoogleScholarGoogle Scholar |




