Identification of salt tolerance QTL in a wheat RIL mapping population using destructive and non-destructive phenotyping
Muhammad A. Asif A B , Melissa Garcia A B C , Joanne Tilbrook A B , Chris Brien A D E , Kate Dowling A D , Bettina Berger B D , Rhiannon K. Schilling A B , Laura Short A B , Christine Trittermann A B , Matthew Gilliham B F , Delphine Fleury A B C G , Stuart J. Roy
A D E , Kate Dowling A D , Bettina Berger B D , Rhiannon K. Schilling A B , Laura Short A B , Christine Trittermann A B , Matthew Gilliham B F , Delphine Fleury A B C G , Stuart J. Roy  A B C H and Allison S. Pearson A B F
A B C H and Allison S. Pearson A B F
A Australian Centre for Plant Functional Genomics, PMB 1, Glen Osmond, SA 5064, Australia.
B School of Agriculture, Food and Wine & Waite Research Institute, The University of Adelaide, PMB 1, Glen Osmond, SA 5064, Australia.
C ARC Industrial Transformation Research Hub for Wheat in a Hot and Dry Climate, The University of Adelaide, PMB1, Glen Osmond, SA 5064, Australia.
D Australian Plant Phenomics Facility, The Plant Accelerator, The University of Adelaide, SA 5064, Australia.
E School of Information Technology and Mathematical Sciences, The University of South Australia, GPO Box 2471, Adelaide, SA 5001, Australia.
F ARC Centre of Excellence in Plant Energy Biology, Waite Research Institute, The University of Adelaide, PMB 1, Glen Osmond, SA 5064, Australia.
G Innolea, 6 chemin de Panedautes, 31700, Mondonville, France.
H Corresponding author. Email: stuart.roy@adelaide.edu.au
Functional Plant Biology 48(2) 131-140 https://doi.org/10.1071/FP20167
Submitted: 12 June 2020 Accepted: 31 July 2020 Published: 24 August 2020
Journal Compilation © CSIRO 2021 Open Access CC BY-NC-ND
Abstract
Bread wheat (Triticum aestivum L.) is one of the most important food crops, however it is only moderately tolerant to salinity stress. To improve wheat yield under saline conditions, breeding for improved salinity tolerance of wheat is needed. We have identified nine quantitative trail loci (QTL) for different salt tolerance sub-traits in a recombinant inbred line (RIL) population, derived from the bi-parental cross of Excalibur × Kukri. This population was screened for salinity tolerance subtraits using a combination of both destructive and non-destructive phenotyping. Genotyping by sequencing (GBS) was used to construct a high-density genetic linkage map, consisting of 3236 markers, and utilised for mapping QTL. Of the nine mapped QTL, six were detected under salt stress, including QTL for maintenance of shoot growth under salinity (QG(1-5).asl-5A, QG(1-5).asl-7B) sodium accumulation (QNa.asl-2A), chloride accumulation (QCl.asl-2A, QCl.asl-3A) and potassium : sodium ratio (QK:Na.asl-2DS2). Potential candidate genes within these QTL intervals were shortlisted using bioinformatics tools. These findings are expected to facilitate the breeding of new salt tolerant wheat cultivars.
Additional keywords: chloride, non-destructive phenotyping, potassium, salinity, shoot ion-independent tolerance, sodium, quantitative trait locus, wheat.
Introduction
Wheat plays a major role in food security across the globe and the demand of wheat is expected to increase by 60% by 2050 (Nelson et al. 2010; Shiferaw et al. 2013). However, one of the major constraints for the productivity of wheat is soil salinity, which greatly reduces yield (Rengasamy 2002, 2006; Roy et al. 2014; Munns and Gilliham 2015). In Australia, it is estimated that 69% of the Australian wheatbelt is affected by salinity to some degree (Rengasamy 2002). Hence, to meet the growing demands of wheat consumption, there is an urgent need to identify novel salt tolerance subtraits and develop salinity tolerant wheat cultivars.
To survive saline conditions, plants rely on a range of different tolerance mechanisms, including, but not limited to: excluding Na+ and Cl- from the shoot (Rashid et al. 1999; Poustini and Siosemardeh 2004; Roy et al. 2014; Ismail and Horie 2017); maintaining a high K+ : Na+ ratio in the leaves (Shabala and Cuin 2008; Shabala and Pottosin 2014; Hanin et al. 2016; Ali and Yun 2017); vacuolar sequestration of toxic ions (Hasegawa et al. 2000; Flowers and Colmer 2008; Munns et al. 2016; Ismail and Horie 2017); compatible solutes synthesis (Munns et al. 2016, 2020; van Zelm et al. 2020); and maintenance of plant growth (Roy et al. 2014; Al-Tamimi et al. 2016; Tilbrook et al. 2017; Asif et al. 2018). Reductions in plant growth have been shown to occur during the first few minutes of salt stress, before ions can accumulate to high levels in the shoot tissue, and the ability of plants to maintain shoot growth is called shoot ion-independent tolerance or osmotic tolerance (Roy et al. 2014; Asif et al. 2018). Of the described tolerance mechanisms, little is known about the genes that control the mechanisms of shoot ion-independent tolerance and/or Cl- accumulation in bread wheat, although several candidate genes have been proposed in recent studies (Genc et al. 2014; Asif et al. 2018). Shoot ion-independent tolerance is considered a complex mechanism and long-distance signalling (reactive oxygen species and Ca2+ signalling or long distance electrical signalling) is believed to be involved in this tolerance mechanism (Kudla et al. 2010; Roy et al. 2014; Al-Tamimi et al. 2016; van Zelm et al. 2020). Chloride, in contrast, is an essential micronutrient with metabolic functions in enzyme activation, photosynthesis, osmoregulation and movement of stomata, but it is toxic at high concentrations and can result in leaf chlorosis, growth reduction and yield loss (White and Broadley 2001; Tavakkoli et al. 2010; Li et al. 2017; Wege et al. 2017; Geilfus 2018). Identification and exploitation of genetic variation for these tolerance mechanisms have great potential to breed new salt tolerant wheat cultivars. Therefore, a forward genetics approach to identify quantitative trait loci (QTL) followed by fine mapping could lead to the detection of novel genes for these tolerance mechanisms.
QTL mapping is a valuable tool for studying complex polygenic traits like salinity tolerance in various crops (Ortiz 1998; Ruttan 1999; Roy et al. 2011). QTL for salinity tolerance have been identified in a large number of plant species including bread wheat (Asif et al. 2019); however, limited success has been made so far regarding the identification of candidate genes and development of new salt tolerant wheat cultivars (Gilliham et al. 2017; Asif et al. 2019). Hence, more studies are needed to identify and understand salt tolerance mechanisms and the underlying genes responsible for such traits. Here, we describe the genetic characterisation of a new recombinant inbred line (RIL) population and QTL associated with different salt tolerance subtraits. We speculate on the potential candidate genes within the QTL intervals which could be tested to develop new salt tolerant cultivars.
Materials and methods
Plant materials
An F2:F6 population of RILs, derived from a cross between single plants of two Australian wheat cultivars Excalibur-198 (RAC177/Uniculm492//RAC311S) and Kukri-199 (76ECN44/76ECN36//RAC549/Madden/6*RAC177) were utilised for this study. Excalibur is a salt- and drought-tolerant cultivar with a high yield potential under South Australian conditions, but it has low grain quality and is susceptible to rust. Kukri produces excellent quality grain and is resistant to rust (Izanloo et al. 2008; Asif et al. 2018). Seeds of RILs and parents (Excalibur and Kukri) were provided by Australian Centre for Plant Functional Genomics (ACPFG), Australia.
Non-destructive glasshouse-based phenotyping
A phenotyping experiment was conducted on 128 RILs using the automated high-throughput imaging facility at The Plant Accelerator, Adelaide, South Australia (longitude 138.64, latitude –34.97). The 128 RILs and parents (Excalibur and Kukri) were phenotyped during late winter to early spring, 14 August to 16 September 2013 using a partially replicated (20%) split-plot design, using the same method described by Asif et al. (2018).
Plants were germinated and planted as described by Asif et al. (2018) with some minor changes. Salt treatment was applied by adding 212 mL of 170 mM NaCl at 27% (w/w) soil water content (573 ml) to the saucer of each salt stress pot to a final concentration of 100 mM NaCl in the soil after drying down to 17% (w/w) soil water (361 mL). Control pots received 212 mL of water on the same day to reach a soil water content of 27% (w/w) and allow drying down to 17% (w/w), the same as salt treated pots. After salt application, each pot was weighed and watered automatically, on a daily basis on the electronic conveyor system to maintain the soil water content at 17% (w/w) and 100 mM NaCl concentration in soil. Plant imaging, shoot ion-independent tolerance calculations and statistical analysis were performed as described previously by Asif et al. (2018).
Measurement of leaf Na+, K+ and Cl- concentration
The fourth leaf, which was the fully developed leaf blade under salt stress treatment, was harvested 13 days after salt treatment and used to measure Na+, K+ and Cl- contents. Fresh weight of leaf samples was recorded before samples were dried in an oven for two days at 65°C and the dry weight recorded. Samples were digested in 10 mL of 1% (v/v) HNO3 at 85°C for 4 h in a 54-well Hotblock (Environmental Express). The concentration of Na+ and K+ were measured using a flame photometer (Model 420 Sherwood), while Cl- was measured using a chloride analyser (Model 926 Sherwood).
Genotyping of the RIL population
Genomic DNA was extracted from leaf material using a phenol/chloroform method described previously by Rogowsky et al. (1991) and Pallotta et al. (2000). The DNA concentration was quantified by PicoGreen (Ahn et al. 1996). 272 RILs were genotyped using genotyping by sequencing (GBS) to identify single nucleotide polymorphism (SNP) markers for high density genetic map construction. GBS libraries were prepared using protocols described in Elshire et al. (2011) and Poland et al. (2012). DNA samples of all the RILs and parents were digested with two restriction enzymes (PstI – CTGCAG and MspI – CCGG) for complexity reduction and barcoded with DNA adapters, designed following the criteria described by Poland et al. (2012) (Table S1, available as Supplementary Material to this paper). Three multiplex GBS libraries, each having 96 samples (93 RILs, two parents and one negative control), were sequenced using the Illumina NextSeq500 platform at the Australian Genome Research Facility (AGRF, Adelaide, South Australia). Sequencing data was processed using the Universal Network Enabled Analysis Kit (UNEAK), which is the non-reference GBS SNP calling pipeline and an extension of the Java program of TASSEL (Lu et al. 2012). The heterozygous SNP calls were assigned as missing data and only the SNP markers which contained less than 20% missing data were used for map construction.
RILs were further genotyped for the phenology genes Ppd-2A, Ppd-2B and Vrn-A1 and 16 Kompetitive Allele Specific PCR (KASP) polymorphic markers by a KASP assay (https://www.biosearchtech.com/products/pcr-kits-and-reagents/genotyping-assays/kasp-genotyping-chemistry, accessed 4 August 2020) (Table S2).
Genetic map construction
Map construction was performed using R/ASMap following the instructions outlined by Taylor (2015). Genotypic data was checked for lines with missing data (>25%), segregation distortion (an allele frequency of either parent at <0.4 and >0.6, at P = 0.05) and clonal individuals (similarity of >90%) using the appropriate functions in R/ASMap (Taylor 2015). The data outside the threshold ranges were removed and remaining markers and lines were used for map construction. Recombination fractions were converted to cM distances using the Kosambi mapping function and the final map was constructed using 3236 markers for the 128 RILs that were phenotyped. The total length of the genetic map was 3084 cM, with a marker density of 0.95 cM per marker (Table 1). Chromosome numbers were assigned to each linkage group (LG) using the sequence reads that were outputted from the UNEAK pipeline for each of the SNPs that were identified. These sequences were assigned to each of the chromosomes using an in-house BLAST portal with a BLASTn performed against the IWGSC RefSeq v1.0 (IWGSC, https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies, accessed 2 August 2020). The limit of acceptance of assignment was based on the percentage of similarity (>96%) and the final percentage of matches (80–100%) between the query (the SNP markers) and the hit from the sequence database.

|
The A and B genome were well represented among all of the LGs. A total of 1478 markers were assigned to the A genome, which consisted of 1415 cM of the genetic map (0.96 cM per marker) (Table 1). The B genome accounted for 1007 cM with 1459 markers (0.69 cM per marker) (Table 1). The D genome was the least represented group with 299 markers spanning 662 cM (2.22 cM per marker) (Table 1).
QTL analysis
Composite interval mapping (CIM) was performed on 128 RILs using WinQTLCart-vers. 2.5 (Model 6 standard analysis with five control markers and a window size of 10 cM) (Wang et al. 2012) (Table S3). Log of odds (LOD) value thresholds were determined with 1000 times permutations (Churchill and Doerge 1994) at a 1 cM walk speed (P = 0.05). Significant QTL were summarised with their position on a linkage group, LOD score, magnitude and directions of their estimated additive effects and their contribution to the genetic variance. Map graphics and QTL positions were drawn using MapChart 2.1 (Voorrips 2002). The notation for individual QTL followed the format previously described by Asif et al. (2018). A QTL region was defined as unique if it was further than 15 cM from a neighbouring QTL (Sewell et al. 2000; Sewell et al. 2002).
Physical mapping of the QTL
To determine the potential candidate genes within the QTL intervals, all the markers (Tables S4, S5) that were up to two LOD drops from the maximum likelihood value of selected QTL were used for BLASTn against the IWGSC RefSeq v1.0 (IWGSC, as above) using an in-house BLAST portal as described previously by Asif et al. (2018). Only the query sequence having a cumulative identity percentage of similarity (>96%) and a cumulative alignment length percentage of matches (90–100%) to the hit from the sequence database were shortlisted. All the scaffolds that were within the QTL intervals were retrieved from the BLAST results and used to find expressed genes on the scaffolds, using DAWN (diversity among wheat genomes) (Watson-Haigh et al. 2018) and POTAGE (PopSeq Ordered Triticum aestivum Gene Expression) (Suchecki et al. 2017). DAWN integrates data from the Triticum aestivum Chinese Spring IWGSC RefSeq v1.0 genome with public whole genome sequencing and exome data from 17 and 62 bread wheat accessions, respectively (Watson-Haigh et al. 2018) and RNA-Seq expression data from five wheat tissues (root, leaf, stem, spike and grain), taken at three developmental stages (seedling, vegetative and flowering). POTAGE integrates map location with gene expression and inferred functional annotation and visualises these data through a web browser interface (Suchecki et al. 2017).
Results
Glasshouse phenotyping of RILs
To assess the responses of plants to salinity, we exposed a total of 128 RILs and parents (Excalibur and Kukri) to 100 mM NaCl salt stress for 13 days; their growth during this period was monitored using high-throughput, non-destructive imaging. Images were used to extract the projected shoot area (PSA), which in turn was used to estimate the relative growth rate (RGR) of plants in the interval of 1-5 days after salt treatment. The ratio of RGR (RGR salt/RGR control) during this period was used to calculate the shoot ion-independent tolerance. The range of shoot ion-independent tolerance within the RIL population varied from 0.51 to 1.16 (Table 2).

|
The ionic contents (Na+, K+, and Cl–) in the fourth leaf of the RILs growing in 100 mM saline soil for 13 days revealed considerable variation between the lines (Table 2). The mean and standard error of shoot Na+ accumulation for this population was 215 ± 21 µmol g–1 DW. The majority of lines had a concentration between 50 and 600 µmol g–1 DW, with seven lines having a leaf Na+ concentration greater than 600 µmol g–1 DW and the highest concentration observed at 1877 µmol g–1 DW (Table 2). By contrast, K+ accumulation in the leaf followed a closer to normal distribution and had a population mean of 1013 ± 14 µmol g–1 DW and ranged from 364 to 1377 µmol g–1 DW (Table 2). The fourth leaf Cl- accumulation exhibited a distribution similar to that of Na+ with a population mean of 458 ± 21 µmol g–1 DW (Table 2).
QTL mapping
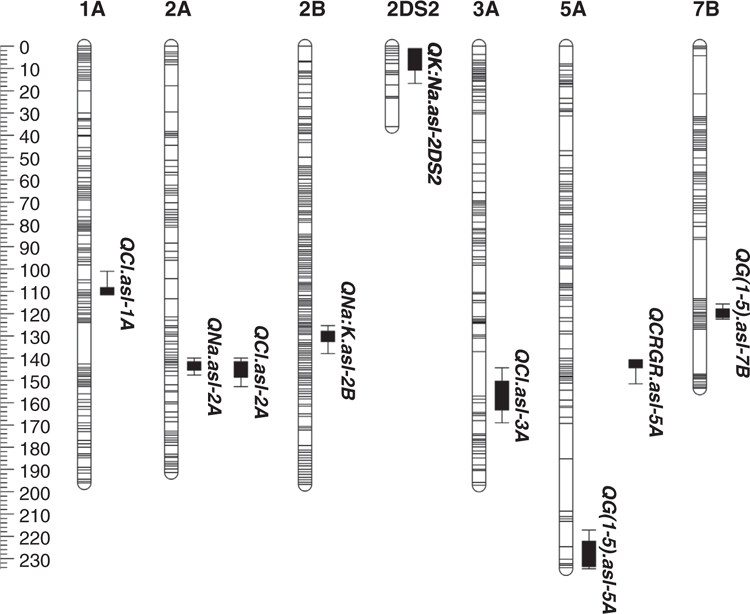
A total of nine QTL at eight unique locations on seven linkage groups were detected under salt stress (100 mM NaCl; six QTL) and control (0 mM NaCl; three QTL) treatments (Table 3; Fig. 1). The phenotypic variation explained by a single QTL ranged between 8.7 and 14.6% (Table 3).
The ability to maintain growth under salinity (shoot ion-independent tolerance) was mapped to two QTL on chromosomes 5A (QG(1-5).asl-5A) and 7B (QG(1-5).asl-7B) (Table 3; Fig. 1). The LOD score of the 5A QTL was 3.8 and explained 10.9% of the phenotypic variation. The second QTL on chromosomes 7B had a LOD score of 3.4 and accounted for 8.7% of the phenotypic variation. The 5A QTL had a positive effect from the Excalibur parent, while the favourable allele of the 7B QTL was from Kukri (Table 3). Under control treatment a significant QTL for relative growth rate (RGR) was also identified in another region on chromosome 5A (QCRGR.asl-5A) with a LOD score of 4.1 and explaining 11.2% of the phenotypic variation. The additive effect of this QTL was very small (0.003) with the positive allele inherited from the Excalibur parent (Table 3).
A single QTL for fourth leaf Na+ accumulation under salt treatment was mapped on chromosome 2A (QNa.asl-2A) with a LOD score of 3.5. It accounted for 10.3% of the phenotypic variation with the allele for Na+ exclusion linked to Excalibur (Table 3).
A total of three QTL were associated with fourth leaf Cl– exclusion including two QTL in salt treated plants and one under control (Table 3; Fig. 1). The QTL detected under the salt treatment were mapped on chromosome 2A (QCl.asl-2A) and 3A (QCl.asl-3A) with LOD scores of 3.4 and 3.6, respectively. These two QTL accounted for 9.7 and 10.6% of the phenotypic variation and the Cl– exclusion allele for both was inherited from the Excalibur parent (Table 3). A single QTL detected on chromosome 1A under control treatment had a LOD score of 3.2 which accounted for 9.2% of the phenotypic variation with the favourable allele for Cl– exclusion derived from Kukri (Table 3).
One QTL was identified for Na+ : K+ DW under control treatment on chromosome 2B (QNa:K.asl-2B). This QTL had a LOD score of 4.9 and explained 14.6% of the phenotypic variation with the allele for Na+ exclusion derived from the Kukri parent (Table 3). One QTL for K+ : Na+ DW was detected under salt stress on chromosome 2DS2 (QK:Na.asl-2DS2) with a LOD score of 3.6 and phenotypic variance of 10.2%. The beneficial allele for this QTL was inherited from the Excalibur parent (Table 3).
Predicted genes within the QTL intervals
All the scaffolds from the bread wheat IWGSC RefSeq v1.0 (IWGSC, as above) that were within two LOD drops from the maximum likelihood value of the QTL observed under salt treatment (QG(1-5).asl-5A, QG(1-5).asl-7B, QNa.asl-2A, QCl.asl-2A, QCl.asl-3A, QK:Na.asl-2DS2) were retrieved from BLAST results and used to investigate the presence of potential candidate genes using DAWN (Watson-Haigh et al. 2018) and POTAGE (Suchecki et al. 2017). Potential candidate genes were selected based on their role in salinity tolerance as published in literature.
The first QTL for shoot ion-independent tolerance or maintenance of growth under salinity on chromosome 5A (QG(1-5).asl-5A) spanned six scaffolds (scaffold45192, scaffold15805, scaffold4453, scaffold43014, scaffold21530, scaffold34444–3) and contained 184 expressed genes (Table S6) which contained several potential candidates for the phenotype including the purple acid phosphatases (PAPs) and a nitrate transporter (NRT1.1) (Table 4). The second QTL for this tolerance sub-trait identified on chromosome 7B (QG(1-5).asl-7B) was located on four scaffolds (scaffold101790, scaffold101708, scaffold138378, scaffold85256) containing a total of 88 expressed genes with a calmodulin like (CML) protein shortlisted as a potential candidate (Table S6).
Two scaffolds (scaffold52744, scaffold78889) were found within the QTL region for QNa.asl-2A, which contained 280 expressed genes (Table S6). Candidate genes selected within this interval include those coding for a vacuolar type H+-ATPase (V-ATPase) and calcium/calmodulin-dependent protein kinases (CaMKs) based on their role in salt tolerance (Kirsch et al. 1996; Dietz et al. 2001; Pandey et al. 2002; Beyenbach and Wieczorek 2006; Yang et al. 2011; Lv et al. 2017) (Table 4).
The Cl- accumulation QTL (QCl.asl-2A) identified under salinity stress was located across four scaffolds (scaffold52744, scaffold78889, scaffold13951, scaffold21915) containing 317 expressed genes (Table S6). There are an additional 37 genes located within this interval compared with QNa.asl-2A, however none of these genes appear as additional candidates to those listed above. For QCl.asl-3A, a total of four scaffolds (scaffold87465, scaffold60640, scaffold10162, scaffold29358) were identified having 108 expressed genes (Table S6). A Multidrug and Toxic Compound Extrusion (MATE) protein was shortlisted as a candidate gene based on its role in Cl- transport (Zhang et al. 2017) (Table 4).
For the K+ : Na+ DW QTL (QK:Na.asl-2DS2), a total of five scaffolds (scaffold65451, scaffold 42730, scaffold87109, scaffold32556, scaffold38944) with 170 expressed genes were retrieved (Table S6). Within this QTL region a sodium/hydrogen exchanger 7 (NHX7) or a salt overly sensitive 1 (SOS1) was selected as a potential candidate due to its role in Na+ homeostasis (Zhu 2003; Olías et al. 2009; Ullah et al. 2016) (Table 4).
Discussion
QTL detection using the newly constructed genetic map revealed a total of nine QTL at eight unique locations on seven different chromosomes for several salt tolerance sub-traits. These include novel QTL for shoot ion-independent tolerance or maintenance of shoot growth under salinity (QG(1-5).asl-7B), Cl- accumulation (QCl.asl-3A) and K+ : Na+ DW (QK:Na.asl-2DS2) (Table 3; Fig. 1) with mining of the bread wheat reference sequence allowing the identification of candidate genes within these regions.
To date, a limited number of studies have been conducted to identify QTL linked with shoot ion-independent tolerance and/or Cl- accumulation in bread wheat, with the majority of studies focusing on the identification of QTL for shoot ion accumulation (mostly Na+ exclusion and K+ accumulation) (Dubcovsky et al. 1996; Ma et al. 2007; Genc et al. 2010, 2013; Díaz De León et al. 2011; Oyiga et al. 2018; Asif et al. 2019). Shoot ion-independent tolerance is an important tolerance mechanism and helps plants in maintaining tissue expansion and tillering during the initial phase of salt stress before salt accumulates to toxic levels in the shoot (Roy et al. 2014). Recent advancements in non-destructive imaging technology have helped to study this tolerance mechanism in more detail and identifying a QTL in bread wheat on chromosome 7A (Asif et al. 2018).
In this study, two QTL were detected for shoot ion-independent tolerance on chromosomes 5A (QG(1-5).asl-5A) and 7B (QG(1-5).asl-7B) (Table 3; Fig. 1). The physical position of the QTL QG(1-5).asl-5A is in the same QTL region identified by Oyiga et al. (2018) for shoot DW under salt stress signifying the importance of this locus in maintenance of plant biomass under salinity and is some distance away from the known developmental gene, vernalisation gene Vrn-A1. However, another QTL on Chromosome 5A controlling plant biomass (QCRGR.asl-5A) was detected in the a region of Vrn-A1. This shows that Vrn-A1 gene could have an effect on plant biomass.
To the best of our knowledge QG(1-5).asl-7B is novel, and no other QTL in this region have been reported before under salt treatment. Analysis of shoot ion-independent QTL, QG(1-5).asl-5A, in DAWN and POTAGE showed several salt tolerance genes within the region under the QTL, such as, PAPs, and NRT1.1 (Table 4). PAPs belong to a diverse group of acid phosphatases and are found in plants, animals and microorganisms (Schenk et al. 2000; Olczak et al. 2003). In plants, the majority of PAPs catalyse the hydrolysis of phosphate esters and anhydrides (Zhang et al. 2011; Schenk et al. 2013); however, recent studies also showed their role in improving plant growth and alleviating oxidative damage during salt stress (Li et al. 2008; Deng et al. 2014) which makes them attractive candidates from this study. The second candidate, NRT1.1 modulates the nitrate-dependent Na+ transport in Arabidopsis under saline conditions and helps in osmotic adjustment, which prevents water loss and wilting during salt stress (Álvarez‐Aragón and Rodríguez‐Navarro 2017). Among 88 genes within the interval of QG(1-5).asl-7B a CML protein was shortlisted as a potential candidate (Table 4). CML protein belongs to calcium-binding EF-hand family proteins, which play an important role in cellular calcium signalling cascades through the regulation of numerous target proteins (Ranty et al. 2006; Shi and Du 2020). This protein has a role in salinity tolerance and genes related to CML are shown to be upregulated under salt stress (Zeng et al. 2015; Dubrovina et al. 2019; Shi and Du 2020) and has improved the salt tolerance of Arabidopsis by affecting abscisic acid mediated pathways (Magnan et al. 2008).
A novel QTL for Cl- concentration was detected on chromosome 3A QCl.asl-3A under 100 mM salt stress in the glasshouse (Table 3; Fig. 1). Previously a QTL for Cl- accumulation has been mapped on chromosome 3A under hydroponics and field conditions (Genc et al. 2014) but its physical location is different to the QCl.asl-3A reported in this study. For QCl.asl-3A, the MATE transporters were identified as a potential candidate (Table 4). MATE are widely accepted as transporters of organic compounds (Li et al. 2002; Marinova et al. 2007; Dobritzsch et al. 2016; Zhang et al. 2017); however, a recent study has also shown the role of two tonoplast MATE-type proteins in sequestration of Cl- into the vacuole which can be helpful in controlling the toxic Cl- concentration in the cytoplasm under saline conditions (Zhang et al. 2017).
Using the new IWGSC reference sequence, (as above), it appears that the QTL for leaf Na+ accumulation (QNa.asl-2A) identified in this study is not in the same position as the Nax1 locus (35 Mbp away) (Lindsay et al. 2004), Q.Na2A locus (88 Mbp away) (Genc et al. 2010, 2013) and significant Na+ concentration SNP locus on chromosome 2A (46 Mbp away) (Genc et al. 2019). However, QNa.asl-2A may be the same locus as Na+ content and K+ : Na+ QTL described by (Oyiga et al. 2018), as their markers sit on the same IWGSC scaffold (52744). The QNa.asl-2A is also away (74.7 cM) from the photoperiodinsensitive gene Ppd-A1, which means this genes has no effect on leaf Na+ accumulation. The QTL (QNa.asl-2A) was associated with fourth leaf Na+ exclusion under salinity stress in the glasshouse and is co-located with a Cl- exclusion QTL (QCl.asl-2A) (Table 3; Fig. 1). The physical position of both QNa.asl-2A and QCl.asl-2A based on the IWGSC reference sequence 1.0, (as above) indicates that these are on the long arm of chromosome 2A where other QTL have previously been detected for leaf Na+ accumulation (Genc et al. 2010, 2013; Oyiga et al. 2018), K+ : Na+ (Oyiga et al. 2018), Cl- accumulation (Genc et al. 2014), maturity (Díaz De León et al. 2011), tiller number (Díaz De León et al. 2011) and seedling biomass (Genc et al. 2010, 2014) under salt stress in bread wheat. Co-location of ion accumulation (Na+, Cl-) and biomass related QTL indicates that more than one gene for the salt tolerance sub-traits may be present within this region. These may include genes such as V-ATPase and CaMKs which are shortlisted as a candidate for QNa.asl-2A (Table 4). V-ATPases are involved in pumping protons into the vacuole and establishing an electro chemical gradient used by the Na+/H+ antiporters to sequester Na+ into the vacuole which in return improves plant performance under saline conditions (Kirsch et al. 1996; Dietz et al. 2001; Beyenbach and Wieczorek 2006; Lv et al. 2017). CaMKs are Ca2+-regulated protein kinases and play a key role in stress signalling (Zhang and Lu 2003; Wang et al. 2004). These kinases do not directly bind Ca2+ by themselves, but instead interact with a specific Ca2+ sensor, such as calmodulin (CaM) or calcineurin B-like protein (CBL) (Zhang and Lu 2003; Wang et al. 2004) known for their role in salt tolerance (Pandey et al. 2002; Yang et al. 2011).
A novel QTL for K+ : Na+ DW was detected on the short arm of chromosome 2D (QK:Na.asl-2DS2) (Table 3; Fig. 1). Previous studies (Ma et al. 2007; Genc et al. 2010, 2019; Oyiga et al. 2018), found QTL for other salt tolerance subtraits on chromosome 2D including plant biomass, chlorophyll content, leaf chlorosis and leaf Na+ accumulation, however, at a different region of the chromosome based on the physical position. Of the 170 genes within the region of QK:Na.asl-2DS2, a NHX7 was shortlisted as potential candidate based on its role in salt tolerance by limiting the Na+ accumulation in plants (Shi et al. 2003; Ullah et al. 2016) (Table 4).
It should be noted that DAWN (Watson-Haigh et al. 2018) and POTAGE (Suchecki et al. 2017) bioinformatics tools use databases that have genes expressed only under non-saline conditions, hence, it is possible that there could be other salt responsive genes present in these intervals that have not been detected due to a lack of gene expression data of wheat under salinity stress.
Phenotyping experiments conducted under controlled environments do not always mimic field conditions. Detection of salt stress related traits in the greenhouse under a limited stress period and at a specific growth stage will not necessarily translate to improved yields under field conditions. A single salt tolerance sub-trait cannot always guarantee yield improvement. Traits such as early vigour, at the seedling stage, may be detrimental to final grain yield under field conditions, as the plant may accumulate greater biomass, therefore having reduced water use efficiency. Hence, multiple salinity tolerance subtraits are required over the lifespan of the plant to contribute towards improved yield. Future studies are needed to develop NILs around the key salt tolerance QTL identified in the present study, to evaluate them under a range of salinity levels in the field, and to identify the best alleles for future breeding programs, as was done by Asif et al. (2018).
In summary, novel QTL have been identified for shoot ion-independent tolerance (QG(1-5).asl-7B), Cl- accumulation (QCl.asl-3A) and K+ : Na+ DW (QK:Na.asl-2DS2) in bread wheat. The detection of shoot ion-independent tolerance and Cl- accumulation QTL in this study will help to better understand the genetic control of these mechanisms in bread wheat and has the potential to speed up breeding for these sub-traits. Future work should focus on studying the effect of these loci on yield under saline field conditions followed by fine mapping and differential expression of candidate gene(s) between the two parents.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgements
This project was funded by the Grains Research and Development Corporation (GRDC): Project UA00118 to SJR; UA00145 to SJR, BB, MG; ARC Centre of Excellence (CE1400008) and International wheat yield partnership (IWYP)/GRDC Project IWYP60/ANU00027 to MG. MAA thanks the University of Adelaide (UofA) for Adelaide Scholarships International (ASI) and the Australian Centre for Plant Functional Genomics (ACPFG) for his PhD stipend, also acknowledged the financial support from the Australian Society of Plant Scientists (ASPS), the Crop Science Society of South Australia Inc. and the Plant Nutrition Trust to attend conferences. The Plant Accelerator, Australian Plant Phenomics Facility, is funded under the National Collaborative Research Infrastructure Strategy (NCRIS). Authors declare that none of the funding bodies have any role in the design of the study and collection, analysis, and interpretation of data as well as in writing the manuscript. We acknowledged the help of Dr Christa Niemietz (UofA), and The Plant Accelerator staff who assisted with the phenotyping experiments. The bread wheat genome assembly was accessed through the International Wheat Genome Sequencing Consortium at https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies.
References
Ahn SJ, Costa J, Rettig Emanuel J (1996) PicoGreen quantitation of DNA: effective evaluation of samples pre-or post-PCR. Nucleic Acids Research 24, 2623–2625.| PicoGreen quantitation of DNA: effective evaluation of samples pre-or post-PCR.Crossref | GoogleScholarGoogle Scholar | 8692708PubMed |
Al-Tamimi N, Brien C, Oakey H, Berger B, Saade S, Ho YS, Schmöckel SM, Tester M, Negrão S (2016) Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping. Nature Communications 7, 13342
| Salinity tolerance loci revealed in rice using high-throughput non-invasive phenotyping.Crossref | GoogleScholarGoogle Scholar | 27853175PubMed |
Ali A, Yun D-J (2017) Salt stress tolerance; what do we learn from halophytes? Journal of Plant Biology 60, 431–439.
| Salt stress tolerance; what do we learn from halophytes?Crossref | GoogleScholarGoogle Scholar |
Álvarez‐Aragón R, Rodríguez‐Navarro A (2017) Nitrate‐dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions. The Plant Journal 91, 208–219.
| Nitrate‐dependent shoot sodium accumulation and osmotic functions of sodium in Arabidopsis under saline conditions.Crossref | GoogleScholarGoogle Scholar | 28370621PubMed |
Asif MA, Schilling RK, Tilbrook J, Brien C, Dowling K, Rabie H, Short L, Trittermann C, Garcia A, Barrett-Lennard EG, Berger B, Mather DE, Gilliham M, Fleury D, Tester M, Roy SJ, Pearson AS (2018) Mapping of novel salt tolerance QTL in an Excalibur × Kukri doubled haploid wheat population. Theoretical and Applied Genetics 131, 2179–2196.
| Mapping of novel salt tolerance QTL in an Excalibur × Kukri doubled haploid wheat population.Crossref | GoogleScholarGoogle Scholar | 30062653PubMed |
Asif MA, Pearson AS, Schilling RK, Roy SJ (2019) Opportunities for developing salt-tolerant wheat and barley varieties. Annual Plant Reviews 2, 1–61.
| Opportunities for developing salt-tolerant wheat and barley varieties.Crossref | GoogleScholarGoogle Scholar |
Beyenbach KW, Wieczorek H (2006) The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. Journal of Experimental Biology 209, 577–589.
| The V-type H+ ATPase: molecular structure and function, physiological roles and regulation.Crossref | GoogleScholarGoogle Scholar | 16449553PubMed |
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971.
Deng L, Chen F, Jiang L, Lam HM, Xiao G (2014) Ectopic expression of GmPAP3 enhances salt tolerance in rice by alleviating oxidative damage. Plant Breeding 133, 348–355.
| Ectopic expression of GmPAP3 enhances salt tolerance in rice by alleviating oxidative damage.Crossref | GoogleScholarGoogle Scholar |
Díaz De León JL, Escoppinichi R, Geraldo N, Castellanos T, Mujeeb-Kazi A, Röder MS (2011) Quantitative trait loci associated with salinity tolerance in field grown bread wheat. Euphytica 181, 371–383.
| Quantitative trait loci associated with salinity tolerance in field grown bread wheat.Crossref | GoogleScholarGoogle Scholar |
Dietz K-J, Tavakoli N, Kluge C, Mimura T, Sharma S, Harris G, Chardonnens A, Golldack D (2001) Significance of the V‐type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level. Journal of Experimental Botany 52, 1969–1980.
| Significance of the V‐type ATPase for the adaptation to stressful growth conditions and its regulation on the molecular and biochemical level.Crossref | GoogleScholarGoogle Scholar | 11559732PubMed |
Dobritzsch M, Lübken T, Eschen-Lippold L, Gorzolka K, Blum E, Matern A, Marillonnet S, Böttcher C, Dräger B, Rosahl S (2016) MATE transporter-dependent export of hydroxycinnamic acid amides. The Plant Cell 28, 583–596.
| MATE transporter-dependent export of hydroxycinnamic acid amides.Crossref | GoogleScholarGoogle Scholar | 26744218PubMed |
Dubcovsky J, Santa Maria G, Epstein E, Luo M-C, Dvořák J (1996) Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theoretical and Applied Genetics 92, 448–454.
| Mapping of the K+/Na+ discrimination locus Kna1 in wheat.Crossref | GoogleScholarGoogle Scholar | 24166270PubMed |
Dubrovina AS, Aleynova OA, Ogneva ZV, Suprun AR, Ananev AA, Kiselev KV (2019) The effect of abiotic stress conditions on expression of calmodulin (CaM) and calmodulin-like (CML) genes in wild-growing grapevine Vitis amurensis. Plants 8, 602
| The effect of abiotic stress conditions on expression of calmodulin (CaM) and calmodulin-like (CML) genes in wild-growing grapevine Vitis amurensis.Crossref | GoogleScholarGoogle Scholar |
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6, e19379
| A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species.Crossref | GoogleScholarGoogle Scholar | 21573248PubMed |
Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytologist 179, 945–963.
| Salinity tolerance in halophytes.Crossref | GoogleScholarGoogle Scholar | 18565144PubMed |
Geilfus C-M (2018) Chloride: from nutrient to toxicant. Plant & Cell Physiology 59, 877–886.
| Chloride: from nutrient to toxicant.Crossref | GoogleScholarGoogle Scholar |
Genc Y, Oldach K, Verbyla A, Lott G, Hassan M, Tester M, Wallwork H, McDonald G (2010) Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theoretical and Applied Genetics 121, 877–894.
| Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress.Crossref | GoogleScholarGoogle Scholar | 20490443PubMed |
Genc Y, Oldach K, Gogel B, Wallwork H, McDonald GK, Smith AB (2013) Quantitative trait loci for agronomic and physiological traits for a bread wheat population grown in environments with a range of salinity levels. Molecular Breeding 32, 39–59.
| Quantitative trait loci for agronomic and physiological traits for a bread wheat population grown in environments with a range of salinity levels.Crossref | GoogleScholarGoogle Scholar |
Genc Y, Taylor J, Rongala J, Oldach K (2014) A major locus for chloride accumulation on chromosome 5A in bread wheat. PLoS One 9, e98845
| A major locus for chloride accumulation on chromosome 5A in bread wheat.Crossref | GoogleScholarGoogle Scholar | 24893005PubMed |
Genc Y, Taylor J, Lyons GH, Li Y, Cheong J, Appelbee M, Oldach K, Sutton T (2019) Bread wheat with high salinity and sodicity tolerance. Frontiers in Plant Science 10, 1280
| Bread wheat with high salinity and sodicity tolerance.Crossref | GoogleScholarGoogle Scholar | 31695711PubMed |
Gilliham M, Able JA, Roy SJ (2017) Translating knowledge in abiotic stress tolerance to breeding programs. The Plant Journal 90, 898–917.
| Translating knowledge in abiotic stress tolerance to breeding programs.Crossref | GoogleScholarGoogle Scholar | 27987327PubMed |
Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K (2016) New insights on plant salt tolerance mechanisms and their potential use for breeding. Frontiers in Plant Science 7, 1787
| New insights on plant salt tolerance mechanisms and their potential use for breeding.Crossref | GoogleScholarGoogle Scholar | 27965692PubMed |
Hasegawa PM, Bressan RA, Zhu J-K, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annual Review of Plant Physiology and Plant Molecular Biology 51, 463–499.
| Plant cellular and molecular responses to high salinity.Crossref | GoogleScholarGoogle Scholar | 15012199PubMed |
Ismail AM, Horie T (2017) Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annual Review of Plant Biology 68, 405–434.
| Genomics, physiology, and molecular breeding approaches for improving salt tolerance.Crossref | GoogleScholarGoogle Scholar | 28226230PubMed |
Izanloo A, Condon AG, Langridge P, Tester M, Schnurbusch T (2008) Different mechanisms of adaptation to cyclic water stress in two South Australian bread wheat cultivars. Journal of Experimental Botany 59, 3327–3346.
| Different mechanisms of adaptation to cyclic water stress in two South Australian bread wheat cultivars.Crossref | GoogleScholarGoogle Scholar | 18703496PubMed |
Kirsch M, Zhigang A, Viereck R, Löw R, Rausch T (1996) Salt stress induces an increased expression of V-type H+-ATPase in mature sugar beet leaves. Plant Molecular Biology 32, 543–547.
| Salt stress induces an increased expression of V-type H+-ATPase in mature sugar beet leaves.Crossref | GoogleScholarGoogle Scholar | 8980504PubMed |
Kudla J, Batistič O, Hashimoto K (2010) Calcium signals: the lead currency of plant information processing. The Plant Cell 22, 541-5–63.
| Calcium signals: the lead currency of plant information processing.Crossref | GoogleScholarGoogle Scholar |
Li L, He Z, Pandey GK, Tsuchiya T, Luan S (2002) Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. Journal of Biological Chemistry 277, 5360–5368.
| Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification.Crossref | GoogleScholarGoogle Scholar | 11739388PubMed |
Li WYF, Shao G, Lam HM (2008) Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses. New Phytologist 178, 80–91.
| Ectopic expression of GmPAP3 alleviates oxidative damage caused by salinity and osmotic stresses.Crossref | GoogleScholarGoogle Scholar |
Li B, Tester M, Gilliham M (2017) Chloride on the move. Trends in Plant Science 22, 236–248.
| Chloride on the move.Crossref | GoogleScholarGoogle Scholar | 28081935PubMed |
Lindsay MP, Lagudah ES, Hare RA, Munns R (2004) A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Functional Plant Biology 31, 1105–1114.
| A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat.Crossref | GoogleScholarGoogle Scholar | 32688978PubMed |
Lu F, Glaubitz J, Harriman J, Casstevens T, Elshire R (2012) TASSEL 3.0 Universal Network Enabled Analysis Kit (UNEAK) pipeline documentation. Available at https://bytebucket.org/tasseladmin/tassel-5-source/wiki/docs/TasselPipelineUNEAK.pdf?rev=2741cb808962db1cec82bbfb80aa3b0bbaf039ca [Verified 13 March 13 2017].
Lv S, Jiang P, Tai F, Wang D, Feng J, Fan P, Bao H, Li Y (2017) The V-ATPase subunit A is essential for salt tolerance through participating in vacuolar Na+ compartmentalization in Salicornia europaea. Planta 246, 1177–1187.
| The V-ATPase subunit A is essential for salt tolerance through participating in vacuolar Na+ compartmentalization in Salicornia europaea.Crossref | GoogleScholarGoogle Scholar | 28825133PubMed |
Ma L, Zhou E, Huo N, Zhou R, Wang G, Jia J (2007) Genetic analysis of salt tolerance in a recombinant inbred population of wheat (Triticum aestivum L.). Euphytica 153, 109–117.
| Genetic analysis of salt tolerance in a recombinant inbred population of wheat (Triticum aestivum L.).Crossref | GoogleScholarGoogle Scholar |
Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud JP, Aldon D (2008) Mutations in AtCML9, a calmodulin‐like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. The Plant Journal 56, 575–589.
| Mutations in AtCML9, a calmodulin‐like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid.Crossref | GoogleScholarGoogle Scholar | 18643966PubMed |
Marinova K, Pourcel L, Weder B, Schwarz M, Barron D, Routaboul J-M, Debeaujon I, Klein M (2007) The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. The Plant Cell 19, 2023–2038.
| The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat.Crossref | GoogleScholarGoogle Scholar | 17601828PubMed |
Munns R, Gilliham M (2015) Salinity tolerance of crops – what is the cost? New Phytologist 208, 668–673.
| Salinity tolerance of crops – what is the cost?Crossref | GoogleScholarGoogle Scholar | 26108441PubMed |
Munns R, James RA, Gilliham M, Flowers TJ, Colmer TD (2016) Tissue tolerance: an essential but elusive trait for salt-tolerant crops. Functional Plant Biology 43, 1103–1113.
| Tissue tolerance: an essential but elusive trait for salt-tolerant crops.Crossref | GoogleScholarGoogle Scholar | 32480530PubMed |
Munns R, Day DA, Fricke W, Watt M, Arsova B, Barkla BJ, Bose J, Byrt CS, Chen ZH, Foster KJ (2020) Energy costs of salt tolerance in crop plants. New Phytologist 225, 1072–1090.
| Energy costs of salt tolerance in crop plants.Crossref | GoogleScholarGoogle Scholar | 31004496PubMed |
Nelson GC, Rosegrant MW, Palazzo A, Gray I, Ingersoll C, Robertson R, Tokgoz S, Zhu T, Sulser TB, Ringler C (2010) ‘Food security, farming, and climate change to 2050: scenarios, results, policy options.’ (International Food Policy Research Institute: Washington, DC, USA)
Olczak M, Morawiecka B, Watorek W (2003) Plant purple acid phosphatases-genes, structures and biological function. Acta Biochimica Polonica 50, 1245–1256.
| Plant purple acid phosphatases-genes, structures and biological function.Crossref | GoogleScholarGoogle Scholar | 14740011PubMed |
Olías R, Eljakaoui Z, Li J, De Morales PA, Marin‐Manzano MC, Pardo JM, Belver A (2009) The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant, Cell & Environment 32, 904–916.
| The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs.Crossref | GoogleScholarGoogle Scholar |
Ortiz R (1998) Critical role of plant biotechnology for the genetic improvement of food crops: perspectives for the next millennium. Electronic Journal of Biotechnology 1, 152–159.
| Critical role of plant biotechnology for the genetic improvement of food crops: perspectives for the next millennium.Crossref | GoogleScholarGoogle Scholar |
Oyiga BC, Sharma RC, Baum M, Ogbonnaya FC, Léon J, Ballvora A (2018) Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat. Plant, Cell & Environment 41, 919–935.
| Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat.Crossref | GoogleScholarGoogle Scholar |
Pallotta M, Graham R, Langridge P, Sparrow D, Barker S (2000) RFLP mapping of manganese efficiency in barley. Theoretical and Applied Genetics 101, 1100–1108.
| RFLP mapping of manganese efficiency in barley.Crossref | GoogleScholarGoogle Scholar |
Pandey S, Tiwari SB, Tyagi W, Reddy MK, Upadhyaya KC, Sopory SK (2002) A Ca2+/CaM‐dependent kinase from pea is stress regulated and in vitro phosphorylates a protein that binds to AtCaM5 promoter. European Journal of Biochemistry 269, 3193–3204.
| A Ca2+/CaM‐dependent kinase from pea is stress regulated and in vitro phosphorylates a protein that binds to AtCaM5 promoter.Crossref | GoogleScholarGoogle Scholar | 12084059PubMed |
Poland JA, Brown PJ, Sorrells ME, Jannink J-L (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7, e32253
| Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach.Crossref | GoogleScholarGoogle Scholar | 22389690PubMed |
Poustini K, Siosemardeh A (2004) Ion distribution in wheat cultivars in response to salinity stress. Field Crops Research 85, 125–133.
| Ion distribution in wheat cultivars in response to salinity stress.Crossref | GoogleScholarGoogle Scholar |
Ranty B, Aldon D, Galaud J-P (2006) Plant calmodulins and calmodulin-related proteins: multifaceted relays to decode calcium signals. Plant Signaling & Behavior 1, 96–104.
| Plant calmodulins and calmodulin-related proteins: multifaceted relays to decode calcium signals.Crossref | GoogleScholarGoogle Scholar |
Rashid A, Qureshi R, Hollington P, Wyn Jones R (1999) Comparative responses of wheat (Triticum aestivum L.) cultivars to salinity at the seedling stage. Journal Agronomy & Crop Science 182, 199–208.
| Comparative responses of wheat (Triticum aestivum L.) cultivars to salinity at the seedling stage.Crossref | GoogleScholarGoogle Scholar |
Rengasamy P (2002) Transient salinity and subsoil constraints to dryland farming in Australian sodic soils: an overview. Australian Journal of Experimental Agriculture 42, 351–361.
| Transient salinity and subsoil constraints to dryland farming in Australian sodic soils: an overview.Crossref | GoogleScholarGoogle Scholar |
Rengasamy P (2006) World salinization with emphasis on Australia. Journal of Experimental Botany 57, 1017–1023.
| World salinization with emphasis on Australia.Crossref | GoogleScholarGoogle Scholar | 16510516PubMed |
Rogowsky PW, Guidet FLY, Langridge P, Shepherd KW, Koebner RMD (1991) Isolation and characterization of wheat-rye recombinants involving chromosome arm 1DS of wheat. Theoretical and Applied Genetics 82, 537–544.
| Isolation and characterization of wheat-rye recombinants involving chromosome arm 1DS of wheat.Crossref | GoogleScholarGoogle Scholar |
Roy SJ, Tucker EJ, Tester M (2011) Genetic analysis of abiotic stress tolerance in crops. Current Opinion in Plant Biology 14, 232–239.
| Genetic analysis of abiotic stress tolerance in crops.Crossref | GoogleScholarGoogle Scholar | 21478049PubMed |
Roy SJ, Negrão S, Tester M (2014) Salt resistant crop plants. Current Opinion in Biotechnology 26, 115–124.
| Salt resistant crop plants.Crossref | GoogleScholarGoogle Scholar | 24679267PubMed |
Ruttan VW (1999) The transition to agricultural sustainability. Proceedings of the National Academy of Sciences of the United States of America 96, 5960–5967.
| The transition to agricultural sustainability.Crossref | GoogleScholarGoogle Scholar | 10339524PubMed |
Schenk G, Korsinczky MLJ, Hume DA, Hamilton S, DeJersey J (2000) Purple acid phosphatases from bacteria: similarities to mammalian and plant enzymes. Gene 255, 419–424.
| Purple acid phosphatases from bacteria: similarities to mammalian and plant enzymes.Crossref | GoogleScholarGoogle Scholar | 11024303PubMed |
Schenk G, Mitić N, Hanson GR, Comba P (2013) Purple acid phosphatase: a journey into the function and mechanism of a colorful enzyme. Coordination Chemistry Reviews 257, 473–482.
| Purple acid phosphatase: a journey into the function and mechanism of a colorful enzyme.Crossref | GoogleScholarGoogle Scholar |
Sewell M, Bassoni D, Megraw R, Wheeler N, Neale D (2000) Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). I. Physical wood properties. Theoretical and Applied Genetics 101, 1273–1281.
| Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). I. Physical wood properties.Crossref | GoogleScholarGoogle Scholar |
Sewell MM, Davis MF, Tuskan GA, Wheeler NC, Elam CC, Bassoni DL, Neale DB (2002) Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). II. Chemical wood properties. Theoretical and Applied Genetics 104, 214–222.
| Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). II. Chemical wood properties.Crossref | GoogleScholarGoogle Scholar | 12582689PubMed |
Shabala S, Cuin TA (2008) Potassium transport and plant salt tolerance. Physiologia Plantarum 133, 651–669.
| Potassium transport and plant salt tolerance.Crossref | GoogleScholarGoogle Scholar | 18724408PubMed |
Shabala S, Pottosin I (2014) Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiologia Plantarum 151, 257–279.
| Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance.Crossref | GoogleScholarGoogle Scholar | 24506225PubMed |
Shi J, Du X (2020) Identification, characterization and expression analysis of calmodulin and calmodulin-like proteins in Solanum pennellii. Scientific Reports 10, 7474
| Identification, characterization and expression analysis of calmodulin and calmodulin-like proteins in Solanum pennellii.Crossref | GoogleScholarGoogle Scholar | 32366918PubMed |
Shi H, Lee B-h, Wu S-J, Zhu J-K (2003) Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nature Biotechnology 21, 81–85.
| Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana.Crossref | GoogleScholarGoogle Scholar | 12469134PubMed |
Shiferaw B, Smale M, Braun H-J, Duveiller E, Reynolds M, Muricho G (2013) Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Security 5, 291–317.
| Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security.Crossref | GoogleScholarGoogle Scholar |
Suchecki R, Watson-Haigh NS, Baumann U (2017) POTAGE: a visualisation tool for speeding up gene discovery in wheat. Scientific Reports 7, 14315
| POTAGE: a visualisation tool for speeding up gene discovery in wheat.Crossref | GoogleScholarGoogle Scholar | 29085014PubMed |
Tavakkoli E, Rengasamy P, McDonald GK (2010) High concentrations of Na+ and Cl– ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. Journal of Experimental Botany 61, 4449–4459.
| High concentrations of Na+ and Cl– ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress.Crossref | GoogleScholarGoogle Scholar | 20713463PubMed |
Taylor J (2015) Efficient linkage map construction using R/ASMap. Available at https://cran.r-project.org/web/packages/ASMap/vignettes/asmapvignette.pdf [Verified 13 March 2017].
Tilbrook J, Schilling RK, Berger B, Garcia AF, Trittermann C, Coventry S, Rabie H, Brien C, Nguyen M, Tester M, Roy SJ (2017) Variation in shoot tolerance mechanisms not related to ion toxicity in barley. Functional Plant Biology 44, 1194–1206.
| Variation in shoot tolerance mechanisms not related to ion toxicity in barley.Crossref | GoogleScholarGoogle Scholar | 32480644PubMed |
Ullah A, Dutta D, Fliegel L (2016) Expression and characterization of the SOS1 Arabidopsis salt tolerance protein. Molecular and Cellular Biochemistry 415, 133–143.
| Expression and characterization of the SOS1 Arabidopsis salt tolerance protein.Crossref | GoogleScholarGoogle Scholar | 26992907PubMed |
van Zelm E, Zhang Y, Testerink C (2020) Salt tolerance mechanisms of plants. Annual Review of Plant Biology 71, 403–433.
| Salt tolerance mechanisms of plants.Crossref | GoogleScholarGoogle Scholar | 32167791PubMed |
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93, 77–78.
| MapChart: software for the graphical presentation of linkage maps and QTLs.Crossref | GoogleScholarGoogle Scholar | 12011185PubMed |
Wang Y, Liang S, Xie Q-G, Lu Y-T (2004) Characterization of a calmodulin-regulated Ca2+-dependent-protein-kinase-related protein kinase, AtCRK1, from Arabidopsis. The Biochemical Journal 383, 73–81.
| Characterization of a calmodulin-regulated Ca2+-dependent-protein-kinase-related protein kinase, AtCRK1, from Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 15196054PubMed |
Wang S, Basten C, Zeng Z (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State Univ., Raleigh, NC. Available at http://statgen.ncsu.edu/qtlcart/WQTLCart.htm [Verified 19 March 2017].
Watson-Haigh NS, Suchecki R, Kalashyan E, Garcia M, Baumann U (2018) DAWN: a resource for yielding insights into the diversity among wheat genomes. BMC Genomics 19, 941
| DAWN: a resource for yielding insights into the diversity among wheat genomes.Crossref | GoogleScholarGoogle Scholar | 30558550PubMed |
Wege S, Gilliham M, Henderson SW (2017) Chloride: not simply a ‘cheap osmoticum’, but a beneficial plant macronutrient. Journal of Experimental Botany 68, 3057–3069.
| Chloride: not simply a ‘cheap osmoticum’, but a beneficial plant macronutrient.Crossref | GoogleScholarGoogle Scholar | 28379459PubMed |
White PJ, Broadley MR (2001) Chloride in soils and its uptake and movement within the plant: a review. Annals of Botany 88, 967–988.
| Chloride in soils and its uptake and movement within the plant: a review.Crossref | GoogleScholarGoogle Scholar |
Yang C, Li A, Zhao Y, Zhang Z, Zhu Y, Tan X, Geng S, Guo H, Zhang X, Kang Z (2011) Overexpression of a wheat CCaMK gene reduces ABA sensitivity of Arabidopsis thaliana during seed germination and seedling growth. Plant Molecular Biology Reporter 29, 681–692.
| Overexpression of a wheat CCaMK gene reduces ABA sensitivity of Arabidopsis thaliana during seed germination and seedling growth.Crossref | GoogleScholarGoogle Scholar |
Zeng H, Xu L, Singh A, Wang H, Du L, Poovaiah B (2015) Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Frontiers in Plant Science 6, 600
| Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses.Crossref | GoogleScholarGoogle Scholar | 26322054PubMed |
Zhang L, Lu Y-T (2003) Calmodulin-binding protein kinases in plants. Trends in Plant Science 8, 123–127.
| Calmodulin-binding protein kinases in plants.Crossref | GoogleScholarGoogle Scholar | 12663222PubMed |
Zhang Q, Wang C, Tian J, Li K, Shou H (2011) Identification of rice purple acid phosphatases related to phosphate starvation signalling. Plant Biology 13, 7–15.
| Identification of rice purple acid phosphatases related to phosphate starvation signalling.Crossref | GoogleScholarGoogle Scholar | 21143719PubMed |
Zhang H, Zhao F-G, Tang R-J, Yu Y, Song J, Wang Y, Li L, Luan S (2017) Two tonoplast MATE proteins function as turgor-regulating chloride channels in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 114, E2036–E2045.
| Two tonoplast MATE proteins function as turgor-regulating chloride channels in Arabidopsis.Crossref | GoogleScholarGoogle Scholar | 28202726PubMed |
Zhu J-K (2003) Regulation of ion homeostasis under salt stress. Current Opinion in Plant Biology 6, 441–445.
| Regulation of ion homeostasis under salt stress.Crossref | GoogleScholarGoogle Scholar | 12972044PubMed |