Preliminary development of a genetic strategy to prevent transgene escape by blocking effective pollen flow from transgenic plants
Davinder Pal Singh A , Angelica M. Jermakow A and Stephen M. Swain A BA CSIRO Plant Industry, PMB, Merbein, Vic. 3505, Australia.
B Corresponding author. Email: steve.swain@csiro.au
Functional Plant Biology 34(12) 1055-1060 https://doi.org/10.1071/FP06323
Submitted: 7 December 2006 Accepted: 15 October 2007 Published: 27 November 2007
Abstract
Genetic modification (GM) of plants has great potential in the production of food and industrial compounds, and in molecular pharming. One of the greatest public concerns regarding this technology is effective pollen flow, in which wind- or insect-borne transgenic pollen is able to fertilise either non-GM crops of the same species, or closely related weed species, and lead to viable seed formation. In this paper we describe a novel concept, based on epigenetic inheritance (imprinting) and post-transcriptional gene silencing (PTGS)/RNA interference (RNAi), designed to prevent transgene escape via pollen flow from transgenic plants. A key advantage of this strategy is that it would allow all seeds from self-pollinated transgenic plants to be harvested and re-sown, without the need for specific treatments, while retaining all of the transgenes present in the parent. Thus, this strategy is not a Genetic Use Restriction Technology (GURT) and if implemented would not prevent seed saving by end-users.
Additional keywords: Arabidopsis, gene flow, imprinting, pollen, transgenic crops, RNAi.
Introduction
When transgenic crops first appeared on the market in the mid-1990s, many biotechnology companies and research organisations anticipated rapid and straightforward adoption of new genetically modified (GM) varieties. While advances in biotechnology have continued at a rapid pace, field trials for more than 4000 GM plants have so far resulted in the successful release of only 40 transgenic crops for commercial purposes (Daniell 1999; Brookes and Barfoot 2005). Benefits derived from these GM crops include reduced environment impact due to a decrease in the use of toxic herbicides and insecticides (Bennett et al. 2004). Other potential and demonstrated benefits include an increase in productivity, salt/drought tolerance, and disease resistance. In addition to food production, plant biotechnology has enormous potential in other areas, particularly molecular pharming for the production of valuable recombinant proteins such as pharmaceuticals, food and feed additives, biopolymers, industrial enzymes, and oils for industrial uses (Hood 2002). Despite these advantages, several highly publicised and often hypothetical environmental and safety concerns (Trewavas 1999; Dale et al. 2002; Bodulovic 2005) have limited public acceptance of GM crops in many countries. One of the most commonly raised concerns is that antibiotic selection marker(s), present in most transgenic food crops, could either inactivate oral doses of the antibiotic (Daniell 1999; Daniell et al. 2001a), or be transferred to human pathogens rendering antibiotic treatments ineffective against these microorganisms. While there is no scientific evidence that this could occur and be of medical significance, several strategies are currently being pursued to eliminate antibiotic resistant genes from future GM crops, particularly in Europe (Russell et al. 1992; Odell et al. 1994; Imantham and Day 2000; Daniell et al. 2001a, b; Hare and Chua 2002).
Another prominent environmental concern is the escape of genes via GM pollen that has potentially travelled great distances and cross-pollinated related crops or weeds, thereby creating ‘superweeds’ or causing seed contamination of ‘organic’ crops (Scott and Wilkinson 1999; Hall et al. 2000; Jemison and Vayda 2001; Estham and Sweet 2002).However in reality, this issue is often overstated (e.g. Rong et al. 2007). Ellstrand (2003) reported that 90% of the world’s 25 most important domesticated crops are able to hybridise naturally with wild relatives. While these ‘wild romances’ can act potentially as a catalyst for evolution of more difficult weeds and the extinction of wild relatives, only limited introgression of genes usually occurs. As a consequence, various approaches to minimise effective gene flow have been developed. Strategies that have been suggested to prevent transgenic crops pollinating other plants include containment in specialised pollen-proof facilities, isolation zones for GM crops (GM crops surrounded by devegetated zones or surrounded by non-insect pollinated non-GM crops to discourage the pollinators/insects from leaving the GM fields), or buffer zone/crop barriers (a non-GM crop planted around the borders of a GM crop to dilute GM pollen and increase the distance GM pollen must travel to out-cross), and the use of Genetic Use Restriction Technologies (GURTs), such as ‘terminator technology’ to prevent production of viable transgenic progeny from self-pollination or outcrossing (Kuvshinov et al. 2001; Daniell 2002). Because the potential for pollen-based out-crossing must be determined specifically for different GM crops grown in a particular environment, considerable time and money must be used to investigate these issues before a GM variety can be released. In an attempt to avoid some of these difficulties and to increase acceptance by the public and regulatory bodies, several molecular solutions have been suggested (Kuvshinov et al. 2001; Daniell 2002; Schernthaner et al. 2003). Two solutions that aim to ensure that transgenes can only be inherited through the maternally-derived genome are engineered male sterility and introduction of transgenes into the chloroplast rather than the nuclear genome (Mariani et al. 1990; Daniell 2002; Daniell et al. 2002; Huang et al. 2003a, b). Unfortunately, both approaches will not prevent wild species from pollinating the transgenic crop, potentially allowing transgenes to eventually move into the wild species by repeated pollination events. Although male sterility will prevent effective pollen flow, it is likely to cause significant problems for farmers wishing to retain seeds for next season’s crop, although in perennial horticultural crops such as some seedless citrus varieties and banana (Musa spp.), pollen in not required since vegetative rather than sexual propagation is used. Alternatively, the development of molecular techniques such as apomixis (formation of seeds without fertilisation) may allow seed propagation without pollen in field crops. Unfortunately, although considerable progress has been made (Koltunow et al. 1995), sufficient understanding of how to convert a sexually-propagated crop into an apomictic one is still several years away (Bicknell and Koltunow 2004).
An alternative and attractive approach to contain transgenes is chloroplast genetic engineering because most crops exhibit only very limited pollen transmission of chloroplast DNA (Daniell et al. 1998). However, paternal inheritance of chloroplast DNA has been reported in several higher plant species (Tilney-Bassett and Abdel-Wahab 1979; Corriveau and Coleman 1988), including tobacco (Nicotiana tabacum L.) in which 0.1–0.5% of chloroplast traits are transferred via pollen (Avni and Edelman 1991; Wang et al. 2004). Significantly, Huang et al. (2003a) have also demonstrated that transgenes can migrate from the chloroplast to the nuclear genome and subsequently be transmitted by sexual propagation, although it is not clear whether transgene function would be retained.
In this paper, we describe a novel strategy to prevent transgene escape via pollen flow from transgenic plants that could be employed either independently or as a backup for other strategies such as chloroplast transformation. A combination of epigenetic inheritance (Köhler and Grossniklaus 2002) and post-transcriptional gene silencing (PTGS)/RNA interference (RNAi) using ‘hairpin’ (hp) technology (Smith et al. 2000) is used to provide proof-of-concept for this strategy in Arabidopsis thaliana L. (Heynh.), a model plant system.
Materials and methods
FIS2 : hp-GUS construct: pSKhp-GUS was generated by cloning the hp-GUS containing 2.0 kb SmaI-XbaI DNA fragment from pMBW305 (a gift from Dr Ming Bo Wang, CSIRO Plant Industry, Canberra, Australia) at the SmaI and XbaI sites of pBluesciptSK (Stratagene, TX, USA). A 3.0-kb EcoRI-BamHI DNA fragment containing the FIS2 promoter from FIS2 : GUS (a gift from Ming Luo, CSIRO Plant Industry, Canberra) was cloned into EcoRI and BamHI digested pSKhp-GUS to obtain pSKFIS2 : hp-GUS. FIS2 : hp-GUS was constructed by cloning the 7.0-kb SalI-SacI DNA fragment from pSKFIS2 : hp-GUS at the SalI and SacI sites of the pGPTV-HPT binary vector (a gift from Dr John P. Carr, University of Cambridge, UK).
Arabidopsis thaliana transformation
Plant transformation and selection of transgenic progeny was as previously described (Singh et al. 2002). The MEA : GUS line is in the Ws ecotype and the selectable marker for super-transformation of MEA : GUS with FIS2 : hp-GUS was hygromycin (22 μg mL–1). Putative transgenic seedlings containing a single FIS2 : hp-GUS transgene locus, based on the segregation of the selectable marker on the T-DNA, were transferred to soil to obtain T2 seeds. The segregating population of T2 plants were allowed to self-pollinate and seeds obtained from T3 plants were used for crosses and other experiments. β-glucuronidase (GUS) activity was detected as previously described (Swain et al. 2002).
Results
The strategy
A novel strategy to prevent transgene escape via pollen flow is described here, involving the generation of transgenic plants containing two transgenes: the MEA (MEDEA) promoter driving expression of a seed-lethal gene and the FIS2 (Fertilisation Independent Seed 2) promoter driving expression of a ‘hairpin’ (hp) RNAi construct (Smith et al. 2000) designed to specifically silence expression of the same seed-lethal gene. MEA (also known as FIS1, Fertilisation Independent Seed 1) and FIS2 are polycomb genes expressed mainly in the endosperm where they play an essential role in seed development (Luo et al. 1999). The MEA promoter, in a MEA : GUS construct, is expressed in developing seeds 48 h after pollination when carried on the paternally-derived chromosome (from the male/pollen parent), and is also expressed both before and after fertilisation when carried on the maternally-derived chromosome (from the female parent). By contrast, the FIS2 promoter, in a FIS2 : GUS construct, is expressed from the maternally-derived chromosome throughout seed development, but is not expressed from the paternally-derived genome due to imprinting (Luo et al. 1999). On self-pollinated plants or transgenic plants fertilised by non-transgenic pollen, the seeds can survive because the FIS2 : hp-lethal-gene transgene expressed from the maternally-derived chromosome silences expression of the seed-lethal gene (Fig. 1). By contrast, seeds formed from the fertilisation by transgenic pollen on non-transgenic plants, such as non-GM crops or weedy relatives, will not survive. In this situation, the seeds will abort due to expression of the MEA : seed-lethal gene, since in this case, the FIS2 : hp-lethal-gene transgene, only present on the paternally-derived chromosome, will not be expressed (Fig. 1).

|
Proof-of-concept experiments
The strategy described above was tested using a reporter gene, β-glucuronidase (GUS), that is non-toxic and can be easily detected in plants based on the production of a blue reaction product after incubating in appropriate conditions (Jefferson et al. 1987). A MEA : GUS construct, for sense expression of GUS, and a FIS2 : hp-GUS construct (Fig. 2A) were generated with GUS taking the place of the seed-lethal gene. This hp-GUS sequence, when driven by the near-constitutive 35S promoter, has previously been shown to effectively silence expression of a 35S : GUS transgene (Smith et al. 2000). As expected (Luo et al. 1999), developing fruits (siliques) from plants homozygous for MEA : GUS possessed GUS activity in developing seeds (Fig. 2B).

|
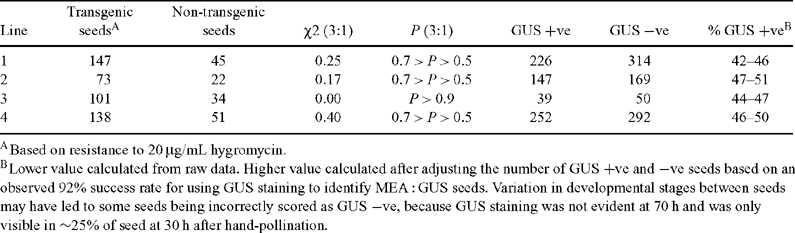
In order to determine if the FIS2 : hp-GUS transgene can silence GUS expression driven by the MEA promoter, plants homozygous for MEA : GUS were super-transformed with FIS2 : hp-GUS. Seeds from self-pollinated plants homozygous for a single locus of both the MEA : GUS and FIS2 : hp-GUS transgenes did not have detectable GUS staining, confirming that the FIS2 : hp-GUS construct can effectively silence MEA : GUS expression (Fig. 2C). This silencing persisted through at least four generations obtained by self-pollination (data not shown). Since a crucial component of the strategy described above is that MEA : GUS expression is only silenced by the FIS2 : hp-GUS transgene present on the maternally-derived chromosome, two experiments were conducted. In the first experiment, four lines homozygous for a single MEA : GUS locus, but hemizygous for an independent FIS2 : hp-GUS insert, were allowed to self pollinate. As expected and based on the presence of the selectable marker, ~75% of the progeny (i.e. the developing seeds) carried the FIS2 : hp-GUS transgene (Table 1), consistent with the presence of single FIS2 : hp-GUS locus in the homozygous MEA : GUS background. Despite the ability of the FIS2 : hp-GUS transgene to completely silence MEA : GUS expression in plants homozygous for both transgenes, GUS was only silenced in ~50% of the seeds while the remainder possessed GUS expression (Fig. 2D; Table 1). This result is consistent with maternally-derived genome-specific expression of the FIS2 : hp-GUS construct since only 50% of the progeny seeds will possess a copy of this construct on the maternally-derived chromosome, while an additional 25% of seeds will contain an inactive FIS2 : hp-GUS construct on the paternally-derived chromosome, and 25% of seeds will not contain FIS2 : hp-GUS on either chromosome. By contrast, if both copies of the FIS2 : hp-GUS construct were active, only 25% of the MEA : GUS seeds would be expected to exhibit GUS activity.
In the second experiment, pollen carrying MEA : GUS was used to hand-pollinate wild-type (WT) flowers and the silique was stained for GUS activity 48 h later. As previously reported (Luo et al. 1999), GUS activity was observed in all seeds (Fig. 2E). Furthermore, in seeds resulting from a cross between WT♀ and MEA : GUS/FIS2 : hp-GUS♂, GUS expression was also observed in all seeds confirming that MEA : GUS is active from the paternally-derived chromosome whereas FIS2 : hp-GUS is not (Fig. 2F). Since the pollen parent in this cross was grown from a seed in which MEA : GUS expression was silenced (i.e. Fig. 2C), this result also demonstrates that expression of the MEA : GUS transgene is reactivated by crossing. Importantly, we confirmed that the MEA : GUS transgene was still reactivated in progeny on a WT♀ after silencing through four generations (Table 2), demonstrating that the silencing remained dependent on the presence of the FIS2 : hp-GUS construct expressed from the maternally-derived chromosome.

|
Discussion
The possibility of pollen from transgenic crops fertilising nearby non-GM crops or weedy relatives has generated extensive public debate and has been one of the most frequently used arguments against the adoption of GM crops. In this paper, we describe and present initial conceptual development of a gene-based strategy to prevent transgene escape via pollen flow from transgenic plants. Successful development of this system in a crop plant would mean that while seed development on transgenic plants would be unaffected, transgenic seeds developing on non-transgenic female plants would abort. Constructs based on those described here, with GUS replaced by a seed-lethal gene, would be incorporated into transgenic crops in addition to transgenes designed to improve crop performance or consumer-orientated traits. An important aspect of any gene-based containment strategy is its robustness. As such, a potential issue is the possibility of transgene mutation or silencing that could subsequently enable transgene escape. In this case two techniques could be used to maximise effectiveness. First, transgenes designed for crop improvement could be tightly linked to the seed-lethal gene, ideally by being part of the same construct. Second, multiple copies of the seed-lethal construct could also be included, with different seed-specific promoters and/or seed-lethal genes, to reduce the possibility of unwanted transcriptional gene silencing inactivating the seed-lethal gene(s) (e.g. Vaucheret 2005). Complete silencing of the seed-lethal construct in self-pollinated seeds is also important to prevent unwanted yield losses in the transgenic crop. Consequently, improved forms of gene silencing constructs, such as artificial microRNAs (Alvarez et al. 2006; Schwab et al. 2006), may be required.
Although we have not tested a reciprocal system, this approach could in theory be adapted to prevent gene introgression via pollen from weedy relatives or non-transgenic crops into the transgenic crop. In this case, a promoter that was only expressed from paternally-derived chromosomes, i.e. the opposite of the FIS2 promoter, would be required to drive expression of another RNAi construct targeting a second seed-lethal gene. Pollen lacking this RNAi gene would be unable to silence the second seed-lethal gene, leading to seed abortion. Ideally, all four genes would form part of the same construct that could be added to transgenic crops. The successful implementation of both systems would mean that the transgenic crop would effectively be genetically isolated from other plants lacking this transgene system.
The approach described here, particularly the more sophisticated form that prevents outcrossing via either the male or female gametophyte, can be considered a form of transgene mitigation (Gressel 1999) since it aims to reduce the fitness of hybrid seeds developing on non-transgenic crops to zero (Lee and Natesan 2006). However, unlike existing mitigation strategies (Al-Ahmad et al. 2004), this strategy does not require any physiological change to the transgenic crop, which although often beneficial (Al-Ahmad et al. 2006), may not be desirable in all systems. The system described here also has features in common with an automatic form of another gene-containment strategy, Recoverable Block of Function (RBF; Lee and Natesan 2006). RBF involves two components, one which prevents seed germination due to activity of the Barnase RNase and the other that encodes an inducible form of Barstar, a inhibitor of Barnase (Mariani et al. 1990). To date, heat treatment has been used as the inducer of Barstar by placing expression of this gene under the control of a heatshock-inducible promoter. Thus, seed germination can only occur after heat shock of the maternal parent. While this would prevent transgene escape via unwanted hybridisation, obtaining sufficient seeds for broadacre field crops in this manner may be difficult.
Importantly, our approach is not a form of GURT and would not restrict the harvest and propagation of self-pollinated seeds from transgenic crops for subsequent planting. The inability of farmers to self-propagate seeds has been a major criticism of GURTs, including the ‘terminator’ technology that could also be used to prevent outcrossing (Kuvshinov et al. 2001). Conventional breeding of crops using this transgene containment system would also be possible, so long as the transgenic plants were used as the female parent.
The initial proof-of-concept experiments with MEA : GUS/FIS2 : hp-GUS described here suggest that it should be possible to create transgenic plants that are self-fertile and have normal or near-normal yield, but are unable to outcross via pollen. As described above, a more sophisticated system would also prevent outcrossing via the female gametophyte. Future development will require the identification and use of suitable seed-lethal gene(s) and the demonstration that silencing is sufficiently robust to allow survival of self-pollinated seeds. The required effectiveness of silencing, and hence the survival rate for seeds on self-pollinated transgenic plants, will depend on the purpose for which the transgenic crops is used. For example, broadacre production crops such as canola (Brassica napus L.) would require near-100% seed survival while some loss of yield may be tolerated in other high-value crops in which preventing transgene escape is more important. While many genes could potentially be used to prevent embryo development, including genes that when overexpressed disrupt essential functions (e.g. Singh et al. 2002), one candidate gene is Barnase (Mariani et al. 1990). In this case, Barnase expression would be driven by the MEA promoter while a Barnase RNAi construct would be driven by FIS2. However, as Barstar prevents Barnase function, an alternative strategy would be to drive Barstar expression by FIS2 instead.
Acknowledgements
We thank Dr John P. Carr for providing the binary vector pGPTV-HPT, Ming Luo for the FIS2 : GUS plasmid, and Dr Ming Bo Wang for pMBW305. We also thank Carol Sigston for technical assistance.
Al-Ahmad H,
Galili SB, Gressel J
(2004) Tandem constructs to mitigate transgene persistence: tobacco as a model. Molecular Ecology 13, 697–710.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Al-Ahmad H,
Dwyer J,
Moloney M, Gressel J
(2006) Mitigation of establishment of Brassica napus transgenes in volunteers using a tandem construct containing a selectively unfit gene. Plant Biotechnology Journal 4, 7–21.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Alvarez JP,
Pekker I,
Goldshmidt A,
Blum E,
Amsellem Z, Eshed Y
(2006) Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. The Plant Cell 18, 1134–1151.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Avni A, Edelman M
(1991) Direct selection for parental inheritance of chloroplasts in sexual progeny of Nicotiana. Molecular & General Genetics 225, 273–277.
| Crossref | GoogleScholarGoogle Scholar |

Bennett RA,
Ismael YA,
Morse SB, Shankar B
(2004) Reductions in insecticide use from adoption of Bt cotton in South Africa: impacts on economic performance and toxic load to the environment. The Journal of Agricultural Science 142, 665–674.
| Crossref | GoogleScholarGoogle Scholar |

Bicknell RA, Koltunow AM
(2004) Understanding apomixis: recent advances and remaining conundrums. The Plant Cell 16, S228–S245.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Bodulovic G
(2005) Viewpoint: is the European attitude to GM products suffocating African development? Functional Plant Biology 32, 1069–1075.
| Crossref | GoogleScholarGoogle Scholar |

Brookes G, Barfoot P
(2005) GM crops: the global economic and environmental impact – the first nine years 1996–2004. AgBio Forum 8, 187–196.

Corriveau JP, Coleman AW
(1988) Rapid screening method to detect potential biparental inheritence of plastid DNA and results for over 200 angiosperm species. American Journal of Botany 75, 1443–1458.
| Crossref | GoogleScholarGoogle Scholar |

Dale PJ,
Clarke B, Fontes EMG
(2002) Potential for the environmental impact of transgenic crops. Nature Biotechnology 20, 567–574.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Daniell H
(1999) GM crops: public perception and scientific solutions. Trends in Plant Science 4, 467–469.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Daniell H
(2002) Molecular strategies for gene containment in transgenic crops. Nature Biotechnology 20, 581–586.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Daniell H,
Datta R,
Varma S,
Gray S, Lee SB
(1998) Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nature Biotechnology 16, 345–348.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Daniell H,
Muthukumar B, Lee SB
(2001a) Engineering the chloroplast genome without the use of antibiotic selection. Current Genetics 39, 109–116.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Daniell H,
Wiebe PO, San Millan AF
(2001b) Antibiotic-free genetic engineering – an environmentally friendly approach. Trends in Plant Science 6, 237–239.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Daniell H,
Khan MS, Allison L
(2002) Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends in Plant Science 7, 84–91.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Gressel J
(1999) Tandem constructs: preventing the rise of superweeds. Trends in Biotechnology 17, 361–366.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hall LM,
Topinka K,
Huffman J,
Davis L, Good A
(2000) Pollen flow between herbicide-resistant Brassica napus is the cause of multiple resistant B. napus volunteers. Weed Science 48, 688–694.
| Crossref | GoogleScholarGoogle Scholar |

Hare PD, Chua NH
(2002) Excision of selectable marker genes from transgenic plants. Nature Biotechnology 20, 575–580.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Hood EE
(2002) From green plants to industrial enzymes. Enzyme and Microbial Technology 30, 279–283.
| Crossref | GoogleScholarGoogle Scholar |

Huang CX,
Ayliffe MA, Timmis JN
(2003a) Direct measurement of the transfer rate of chloroplast DNA into the nucleus. Nature 408, 796–815.

Huang S,
Cerny RE,
Qi Y,
Bhat D,
Aydt CM,
Hanson DD,
Malloy KP, Ness LA
(2003b) Transgenic studies on the involvement of cytokinin and gibberellin in male development. Plant Physiology 131, 1270–1282.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Imantham S, Day A
(2000) Removal of antibiotic resistance genes from transgenic tobacco plastids. Nature Biotechnology 18, 1171–1176.

Jefferson RA,
Kavanagh TA, Bevan MW
(1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907.
| PubMed |

Jemison JM, Vayda ME
(2001) Cross pollination from genetically engineered corn: wind transport and seed source. The Plant Cell 13, 1–6.
| PubMed |

Köhler C, Grossniklaus U
(2002) Epigenetic inheritance of expression states in plant development: the role of Polycomb group proteins. Current Opinion in Cell Biology 14, 773–779.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Koltunow AM,
Bicknell RA, Chaudhury AM
(1995) Apomixis: molecular strategies for the generation of genetically identical seeds without fertilization. Plant Physiology 108, 1345–1352.
| PubMed |

Kuvshinov V,
Koivu K,
Kanerva A, Pehu E
(2001) Molecular control of transgene escape from genetically modified plants. Plant Science 160, 517–522.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Lee D, Natesan E
(2006) Evaluating genetic containment strategies for transgenic plants. Trends in Biotechnology 24, 109–114.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Luo M,
Bilodeau P,
Koltunow A,
Dennis ES,
Peacock WJ, Chaudhury AM
(1999) Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 96, 296–301.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Mariani C,
DeBeuckeleer M,
Trueltner J,
Leemans J, Goldberg RB
(1990) Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature 347, 737–741.
| Crossref | GoogleScholarGoogle Scholar |

Odell JT,
Hoopes JL, Vermerris W
(1994) Seed-specific gene activation mediated by the Cre/lox site-specific recombination system. Plant Physiology 106, 447–458.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Rong J,
Lu B-R,
Song Z,
Su J,
Snow AA,
Zhang X,
Sun S,
Chen R, Wang F
(2007) Dramatic reduction of crop-to-crop gene flow within a short distance from transgenic rice fields. The New Phytologist 173, 346–353.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Russell SH,
Hoopes JL, Odell JT
(1992) Directed excision of a transgene from the plant genome. Molecular & General Genetics 234, 49–59.

Schernthaner JP,
Fabijanski SF,
Arnison PG,
Racicot M, Robert LS
(2003) Control of seed germination in transgenic plants based on the segregation of a two-component genetic system. Proceedings of the National Academy of Sciences of the United States of America 100, 6855–6859.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Schwab R,
Ossowski S,
Riester M,
Warthmann N, Weigel D
(2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell 18, 1121–1133.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Scott SE, Wilkinson MJ
(1999) Low probability of chloroplast movement from oilseed rape (Brassica napus) into wild Brassica rapa. Nature Biotechnology 17, 390–392.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Singh DP,
Jermakow AM, Swain SM
(2002) Gibberellins are required for seed development and pollen tube growth in Arabidopsis. The Plant Cell 14, 3133–3147.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Smith NA,
Singh SP,
Wang MB,
Stoutjesdijk PA,
Green AG, Waterhouse PM
(2000) Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Swain SM,
Tseng T-S,
Thornton T,
Gopalraj M, Olszewski NE
(2002) SPINDLY is a nuclear-localized repressor of gibberellin signal transduction expressed throughout the plant. Plant Physiology 129, 605–615.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Trewavas A
(1999) Gene flow and GM questions. Trends in Plant Science 4, 339.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Vaucheret H
(2005) RNA polymerase IV and transcriptional silencing. Nature Genetics 37, 659–660.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Wang T,
Li Y,
Shi Y,
Reboud X,
Darmency H, Gressel J
(2004) Low frequency transmission of a plastid-encoded trait in Setaria italica. Theoretical and Applied Genetics 108, 315–320.
| Crossref | GoogleScholarGoogle Scholar | PubMed |