Rhizoctonia solani AG groups in northeast India
P. Baiswar A C , D. D. Rosa B , S. Chandra A and S. V. Ngachan AA ICAR Research Complex for NEH Region, Umiam 793103, Meghalaya, India.
B São Paulo State University, College of Agronomic Science, Department of Plant Production – Plant Health Protection Sector, PO Box 237, Botucatu, SP 18610-307, Brazil.
C Corresponding author. Email: pbaiswar@yahoo.com
Australasian Plant Disease Notes 5(1) 85-86 https://doi.org/10.1071/DN10030
Submitted: 21 July 2009 Accepted: 16 July 2010 Published: 28 July 2010
Abstract
Rhizoctonia solani isolates collected from different crops in northeast India belonged to anastomosis group AG 2–2 IIIB (Canavalia ensiformis, Sechium edule, Glycine max and Dolichos lablab). AG 1–1A was detected on Zea mays, Rhizoctonia solani on Sechium edule and AG 4HG-II on a weed, Galinsoga parviflora, which are new records from India.
Many plant species are infected by Rhizoctonia solani (teleomorph: Thanatephorus cucumeris) in northeast India. It has also been observed that due to rising temperatures (climate change), the number of crops infected by this fungus, and severity is increasing over the years (P. Baiswar, pers. obs., 2003–08). R. solani has been classified into different anastamosis groups (AGs) on the basis of hyphal anastomosis reactions. These groups are considered to be genetically isolated (Carling 1996). At present, R. solani contains 13 AG groups (Carling et al. 2002). In addition to AG grouping, other methods like pectic zymograms (Cruickshank 1990), polymerase chain reaction – restriction fragment length polymorphism (Guillemaut et al. 2003) and fatty acid profiles (Priyatmojo et al. 2002) have also been used for classifying this fungus group.
Considering the importance of the diseases caused by this fungus, a survey was conducted for collection of different Rhizoctonia isolates from different host plants. These isolates were characterised based on anastamosis reactions.
Isolation and nuclear staining
Water agar was used for initial isolation. Plant tissues from the margins of infected and healthy portions were cut and surface sterilised using 4% NaOCl for 30 s, rinsed with sterilised water and plated on water agar for 24 h at 25°C. After incubation Rhizoctonia-like colonies were multiplied on potato dextrose agar (PDA). Actively growing hyphal cells were stained with DAPI (1,4,6-diamino-2-phenyl indole) as described by Kulik and Dery (1995). Numbers of nuclei per cell were counted in 20 randomly selected cells per isolate.
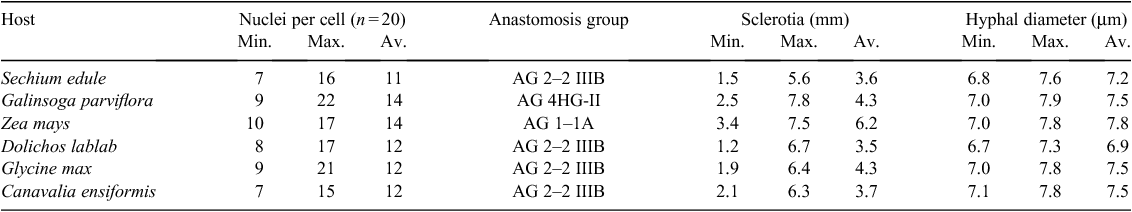
Isolations from infected tissue on PDA yielded a fungal growth with silky and fluffy, fast-growing mycelium and brown sclerotia. After ~10 days, colonies became brown because of melanisation. Hyphae were 6–8 μm wide, septate, branched at right angles, with lateral branches constricted at the junction. Young hyphae were branched near the distal septum. Sclerotia were not differentiated into cortex and medulla. Hyphal cells were multinucleate. These characters were consistent with Rhizoctonia sp. (Sneh et al. 1991) (Table 1). The number of nuclei per cell was variable (6 to 14, av. 11, n = 20), providing evidence that the isolates belonged to R. solani (Table 1).
|
Anastamosis reaction
Anastomosis reaction was determined by staining the hyphae with Safranin-O 0.03% and KOH 3% solution (Yamamoto and Uchida 1982). Criteria used for compatibility was observation of C2 and C3 type reactions from at least five points on each of four slides per isolate (Carling and Leiner 1990). A positive anastomosis reaction between the test isolates and AG tester was defined as fusion of end walls of hyphae, with subsequent plasmolysis of adjacent cells (MacNish et al. 1993). Pairing was done between test isolates and AG tester isolates of R. solani (AG 1, AG 2, AG 3, AG 4, AG 5, AG 6, AG 7, AG 8, AG 9, AG 10, AG 11, AG 12, AG 13 and AG BI), obtained from different researchers.
Pathogenicity
Koch’s postulates were demonstrated by spraying an aqueous suspension of 8-day-old culture filtrate (blended in a Waring blender: Sumeet, Mathur Micromotors & Appliances Pvt. Ltd, Vapi, India) on healthy plants of different hosts, with non-inoculated plants serving as control. Inoculated plants developed symptoms, whereas control plants remained healthy. R. solani was also reisolated from the inoculated plants.
C2 anastomosis reaction was observed between test isolates and the standard isolates. Anastomosis reactions detected AG 2–2 IIIB on Canavalia ensiformis, Sechium edule, Glycine max and Dolichos lablab; AG 1–1A was detected on Zea mays and AG 4HG-II on a weed, Galinsoga parviflora.
R. solani (AG 2–2 IIIB) on S. edule and AG 4HG-II on a weed, G. parviflora, are new records from India. Tribal communities in north-east India cultivate all other crops mentioned above except G. parviflora. G. parviflora may be acting as a reservoir and helping in multiplication and spread of the pathogen.
Carling DE, Leiner RH
(1990) Virulence of isolates of Rhizoctonia solani AG-3 collected from potato plant organs and soil. Plant Disease 74, 901–903.
| Crossref | GoogleScholarGoogle Scholar |

Carling DE,
Kuninaga S, Brainard KA
(2002) Hyphal anastomosis reactions, rDNA-internal transcribed spacer sequences, and virulence levels among subsets of Rhizoctonia solani anastomosis group-2 (AG-2) and AG-BI. Phytopathology 92, 43–50.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Cruickshank RH
(1990) Pectic zymograms as criteria in taxonomy of Rhizoctonia. Mycological Research 94, 938–946.
| Crossref | GoogleScholarGoogle Scholar |

Guillemaut C,
Edel-Hermann V,
Camporota P,
Alabouvette C,
Richard-Molard M, Steinberg C
(2003) Typing of anastomosis groups of Rhizoctonia solani by restriction analysis of ribosomal DNA. Canadian Journal of Microbiology 49, 556–568.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Kulik MM, Dery PD
(1995) Use of DAPI for anastomosis group typing of strains of the fungus Rhizoctonia solani. Biotechnic & Histochemistry 70, 95–98.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

MacNish GC,
Carling DE, Brainard KA
(1993) Characterization of Rhizoctonia solani AG-8 from bare patches by pectic isozyme (zymogram) and anastomosis techniques. Phytopathology 83, 922–927.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Priyatmojo A,
Yamauchi R,
Naito S,
Kageyama K, Hyakumachi M
(2002) Comparison of whole-cell fatty acid compositions of isolates of Rhizoctonia solani AG 2 from tobacco and tulip, AG 2–1 and AG-BI. Journal of Phytopathology 150, 283–288.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Yamamoto DT, Uchida JY
(1982) Rapid nuclear staining of Rhizoctonia solani and related fungi with acridine orange and with safranin O. Mycologia 74, 145–149.
| Crossref | GoogleScholarGoogle Scholar |



