A new tospovirus species infecting Solanum esculentum and Capsicum annuum in Thailand
P. Chiemsombat A , M. Sharman B C , K. Srivilai A , P. Campbell B , D. Persley B and S. Attathom AA Department of Plant Pathology, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University Kamphaeng Saen Campus, Nakhon Pathom 73140, Thailand.
B Department of Employment, Economic Development and Innovation, 80 Meiers Road, Indooroopilly, Qld 4068, Australia.
C Corresponding author. Email: Murray.Sharman@dpi.qld.gov.au
Australasian Plant Disease Notes 5(1) 75-78 https://doi.org/10.1071/DN10027
Submitted: 25 May 2010 Accepted: 1 July 2010 Published: 15 July 2010
Abstract
Based on distinct sequences, serology and host range, isolates of a new tospovirus species infecting field crops of tomato and pepper in Thailand are shown to be members of the provisionally named Tomato necrotic ringspot virus.
Tospoviruses are among the most damaging and economically important group of plant viruses, causing significant crop losses in a wide range of ornamental and food crops in many regions of the world (Mumford et al. 1996). The greatest tospovirus diversity is found in the Asian region, with at least 14 species reported (Pappu et al. 2009). At least three species occur in Thailand including Watermelon silver mottle virus (WSMoV), Melon yellow spot virus (MYSV) and Capsicum chlorosis virus (CaCV) (Chiemsombat et al. 2008). Pepper (Capsicum annuum) and tomato (Solanum esculentum) displaying symptoms of bud necrosis and necrotic ringspots on leaves, stem and fruits were collected from production areas in Chiang Rai and Ratchaburi provinces of Thailand during 2007. This paper reports the molecular and biological identification of a new tospovirus species from these samples.
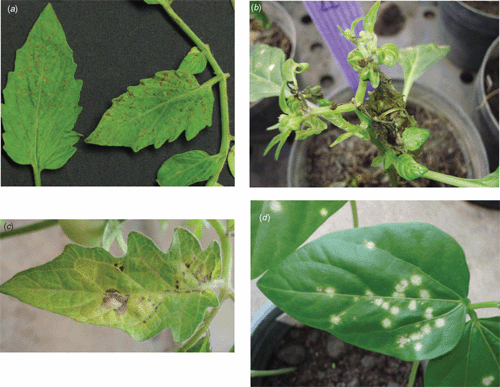
In March 2007 a symptomatic tomato sample (isolate-Q1510) was collected from the Ratchburi region, while in December 2007 several symptomatic pepper samples were collected from the Chiang Rai region, one of which is hereafter referred to as isolate-CR. Field infected Q1510-tomato displayed symptoms of fine necrotic spots and rings on leaves (Fig. 1a), while pepper and tomato (Fig. 1b and c) inoculated with isolate-CR-infected pepper leaves, showed bud necrosis with severe mosaic and necrotic ring spots on leaves. A freeze-dried field leaf sample from isolate-Q1510 was negative by enzyme-linked immunosorbent assay (ELISA) for tospovirus serogroup IV (WSMoV/GBNV: AGDIA ELISA reagent set, Cat. No. SRA61500/0500) with A405 nm values comparable to those of the healthy controls while values of the freeze-dried positive control (CaCV) were 50 times greater than the means of the healthy controls.
|
Isolate-CR was transmitted by mechanical inoculation, using a field sample of pepper to tobacco (Nicotiana tabacum cv. Xanthi NC), Physalis micrantha and Datura stramonium, all of which produced severe mosaic symptoms. Isolate-CR was maintained in tomato cv. LY514 for subsequent tests. Diagnostic indicator species for isolate-CR were shown to be Vigna sinensis cvv.Chiatai and KY Bush 693 with white local lesions at 7 days post-inoculation (Fig. 1d) while CaCV induced chlorotic concentric ring spots on the same host species (Chiemsombat et al. 2008). The test plants above were indexed by non-structural (NSs) gene-specific polymerase chain reaction (PCR), as described below, to confirm infection with isolate-CR.
Symptomatic leaf of isolate-CR-infected tomato reacted weakly in plate-trapped antigen (PTA) ELISA (Mowat and Dawson 1987) to polyclonal antibody A3 (PAb-A3) which was produced against expressed MYSV nucleoprotein (N) for detection of tospovirus serogroup IV species and MYSV (Gajanadana et al. 2006). This weak reaction against the PAb-A3 suggested the presence of tospovirus in these samples. Further field observations in 2009, with the use of PAb-A3 antibody in PTA-ELISA and inoculations to diagnostic species described below, indicated that several pepper crops in Nakhon Pathom province had significant losses due to bud necrosis disease caused by the same virus as isolate-CR (data unpublished).
For isolate-Q1510, RNA was extracted from leaf tissue using the Concert RNA Reagent (Invitrogen, Carlsbad, CA, US) before preparation of cDNA using SuperScript III reverse transciptase (Invitrogen) and PCR using native Taq (Invitrogen) essentially as per the manufacturers’ instructions. Degenerate primers gL3637 and gL4435c (Chu et al. 2001) were used to amplify an ~800 bp product of the partial L-gene which was directly sequenced. For amplification of the total medium RNA (MRNA) segment from isolate-Q1510, previously published tospovirus MRNA sequences (GenBank accessions: CaCV, NC_008303 and DQ256125; MYSV, NC_008307; WSMoV, NC_003841; Groundnut bud necrosis virus (GBNV), NC_003620; Impatiens necrotic spot virus (INSV), NC_003616 and DQ425095; and Tomato spotted wilt virus (TSWV), NC_002050, AY870390 and AY870389) were aligned and a degenerate primer TosMtotal (5′ CCC GGG AGA GCA ATC RGT GC 3′) designed to the complementary MRNA segment genome termini. The underlined sequence shown for primer TosMtotal above represents non-viral nucleotides added to increase primer stability in PCR. It was anticipated that the entire MRNA could be amplified using the TosMtotal primer. However, an internal region with homology to the primer sequence resulted in the amplification of an ~1300 bp product which was cloned into PCR 2.1 Topo vector (Invitrogen) and sequenced as above (GenBank HM136988).
For amplification of complete N-gene from the small RNA segment of isolate-Q1510, previously published sequences (GenBank accessions: WSMoV, NC_003843; GBNV, NC_003619; CaCV, DQ256123 and DQ355974; Iris yellow spot virus, AF001387; Calla lily chlorotic spot virus, AY867502; MYSV, NC_008300; Tomato yellow ring virus, DQ462163; and Tomato necrotic ringspot virus, FJ489600) were used to design primers. Primers TNRV.NPF (5′ CAC TAT CTG ATA TAA ATG AT 3′), CS2821F (5′ ACT TTG CAR ABC TGY TCA TA 3′) and CS3477R (5′ CGA CCA GAG CAA TCG AGG 3′) were used to amplify two overlapping products spanning 899 nt (GenBank HM113532) and containing the entire N-gene of 846 nt which encode a putative protein of 281 amino acids. PCR products were directly sequenced as described above.
For isolate-CR, RNA was extracted from leaf tissues using CTAB buffer followed by lithium chloride precipitation (Chang et al. 1993). Complete N-gene was amplified using the primers 3VGr4R (5′ GTA AAC ACC ATG TCT AMC GT 3′) and 5′ WSMV Forward (5′ GTG AGA GCA ACC CTG GAA CAG C 3′) and complete NSs-gene was amplified using the primers 3VGr4R and 5VGr4F (5′ TTA MAM TTC YAK MGA 3′) (Chiemsombat et al. 2008). Resulting PCR products were cloned into pGEMT-easy vector (Promega, Madison, WI, USA) and sequenced. For back-indexing of host range test plants, partial NSs gene-specific RT–PCR was used with the primers 3VGr4R and 5′ WSMV Forward. The entire NSs gene was 1356 nt long. Isolate-CR sequences have been lodged with GenBank accession numbers FN806775 and FN677988 for N and NSs genes, respectively.
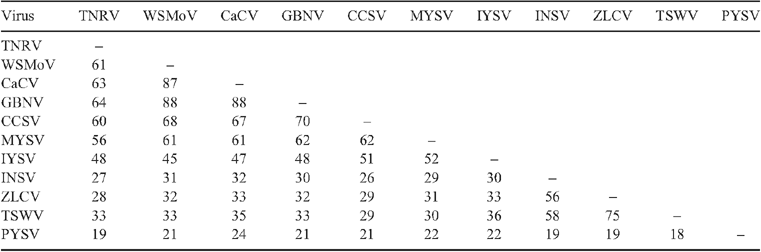
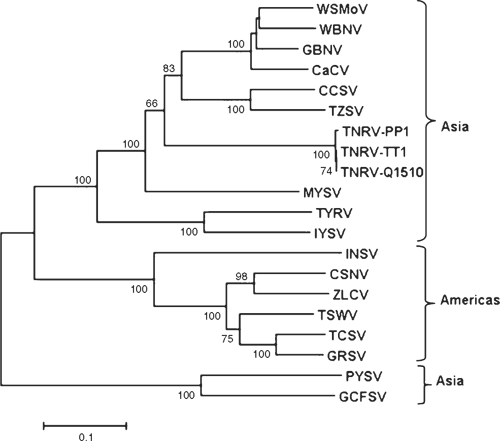
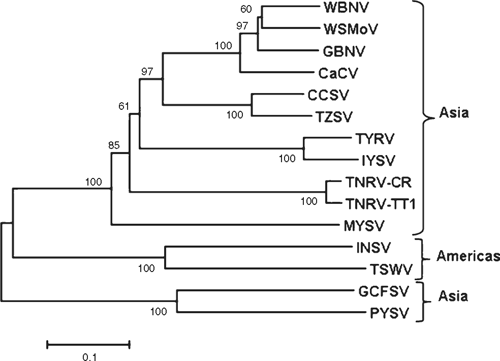
The putative partial nucleocapsid (N) protein sequence of 248 amino acids for isolate-CR (GenBank FN806775) was 98% identical to isolate-Q1510 (GenBank HM113532) and to Tomato necrotic ringspot virus (TNRV) isolate-PP1, which was first recorded from Thailand pepper samples and provisionally named by other workers (GenBank FJ947153). The complete putative N protein sequence of 281 amino acids for isolate-Q1510 was 99.6% identical to TNRV-PP1 and 100% identical to TNRV-TT1 (GenBank FJ489600). As such, isolates-Q1510 and -CR are considered to be isolates of TNRV. The next most closely related tospovirus to TNRV-Q1510 was Groundnut bud necrosis virus (GBNV) with 64% identity for the complete N protein (Table 1). These results indicate TNRV is a distinct tospovirus species which clusters with the other Asian or Old World tospoviruses when comparing complete N protein sequences (Fig. 2). The putative complete NSs-gene amino acid sequences for isolates TNRV-CR and TNRV-TT1 (GenBank FN677988 and FJ489600 respectively) were 96% identical. Comparison of these TNRV isolates with other tospoviruses confirmed that TNRV is a new distinct tospovirus species (Fig. 3). The next most closely related tospovirus species to TNRV-CR was CCSV (GenBank AY867502) with only 55% identity for the complete NSs protein.
|
|
|
Partial MRNA nucleotide and amino acid sequence for TNRV-Q1510 were 99% and 98%, respectively, identical to TNRV-TT1 (GenBank FJ947152, Seepiban et al. 2009) over a 1230 nt (410 aa) overlap. The next closest related tospovirus to TNRV-Q1510 was GBNV (GenBank U42555) with 64% amino acid identity over a 410 aa overlap. Partial L RNA nucleotide sequence for TNRV-Q1510 (GenBank GQ487713, Sharman et al. 2009) was 99% identical, over a short overlap of 96 nt, to TNRV which has been reported from another tomato isolate from Thailand (GenBank GU479877) while the next closest related tospovirus was CCSV (GenBank FJ822962) with 78% identity over a 752 nt overlap.
This paper reports that isolates of a new tospovirus infecting field crops of tomato and pepper from different regions of Thailand are members of the provisionally named Tomato necrotic ringspot virus. Based on differences from other recognised tospovirus species with respect to serology, host range and nucleoprotein (N) and non-structural protein (NSs) sequence similarity, TNRV appears to be a distinct species grouping with other Old World species from Asia. The described differences to other tospovirus species satisfy the species demarcation criteria used by the International Committee on Taxonomy of Viruses (Fauquet et al. 2005). Many aspects of TNRV biology remain unclear and further studies on the alternative hosts, thrips vectors and the screening for natural resistance in germplasm collections will be important for development of effective management strategies. Recent advances in molecular techniques have now enabled the discovery of at least 19 distinct tospovirus species with one or more tospovirus species now found on every continent except Antarctica (Pappu et al. 2009). Along with TNRV, the number of tospovirus species described is likely to continue to increase with at least two further species being recently proposed (Ciuffo et al. 2009; Hassani-Mehraban et al. 2010).
Chang S,
Puryear J, Cairney J
(1993) A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter 11, 113–116.
| Crossref | GoogleScholarGoogle Scholar |

Chiemsombat P,
Gajanandana O,
Warin N,
Hongprayoon R,
Bhunchoth A, Pongsapich P
(2008) Biological and molecular charaterization of tospoviruses in Thailand. Archives of Virology 153, 571–577.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Chu F-H,
Chao C-H,
Chung M-H,
Chen C-C, Yeh S-D
(2001) Completion of the genome sequence of Watermelon silver mottle virus and utilization of degenerate primers for detecting tospoviruses in five serogroups. Phytopathology 91, 361–368.
| Crossref | GoogleScholarGoogle Scholar | PubMed |

Ciuffo M,
Kurowski C,
Vivoda E,
Copes B,
Masenga V,
Falk BW, Turina M
(2009) A New Tospovirus sp. in Cucurbit Crops in Mexico. Plant Disease 93, 467–474.
| Crossref | GoogleScholarGoogle Scholar |

Hassani-Mehraban A,
Botermans M,
Verhoeven J,
Meekes E,
Saaijer J,
Peters D,
Goldbach R, Kormelink R
(2010) A distinct tospovirus causing necrotic streak on Alstroemeria sp. in Colombia. Archives of Virology 155, 423–428.
| Crossref | GoogleScholarGoogle Scholar | PubMed |
PubMed |

Mowat WP, Dawson S
(1987) Detection and identification of plant viruses by ELISA using crude sap extracts and unfractionated antisera. Journal of Virological Methods 15, 233–247.
| Crossref | GoogleScholarGoogle Scholar | PubMed |
PubMed |

Mumford RA,
Barker I, Wood KR
(1996) The biology of the tospoviruses. The Annals of Applied Biology 128, 159–183.
| Crossref | GoogleScholarGoogle Scholar |

Pappu HR,
Jones RAC, Jain RK
(2009) Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Research 141, 219–236.
| Crossref | GoogleScholarGoogle Scholar | PubMed |
PubMed |



