First report of Tomato mosaic virus in Griselinia lucida, an epiphytic shrub native to New Zealand
S. Veerakone A B , B. S. M. Lebas A , J. Tang A and G. R. G. Clover AA Plant Health and Environment Laboratory, MAF Biosecurity New Zealand, PO Box 2095, Auckland 1140, New Zealand.
B Corresponding author. Email: stella.veerakone@maf.govt.nz
Australasian Plant Disease Notes 5(1) 107-109 https://doi.org/10.1071/DN10039
Submitted: 17 May 2010 Accepted: 22 September 2010 Published: 4 October 2010
Abstract
Virus-like symptoms were observed on leaves of Griselinia lucida, a New Zealand native shrub, on Rangitoto Island, New Zealand. Rod-shaped particles similar to those of tobamoviruses were observed by electron microscopy and were identified by reverse transcription polymerase chain reaction as Tomato mosaic virus (ToMV). This is the first record of a virus in G. lucida and the first definite report of ToMV in New Zealand.
Griselinia lucida (akapuka, family Griseliniaceae) grows naturally as an epiphyte in forest areas and on rocky outcrops in coastal regions of New Zealand. It is distributed throughout the North and South Islands but is more common in the former (Dawson 1966). It also grows well as a shrub in cultivation and is a popular garden plant due to the broad, glossy foliage and low maintenance requirements. The leaves are also used in the floristry industry.
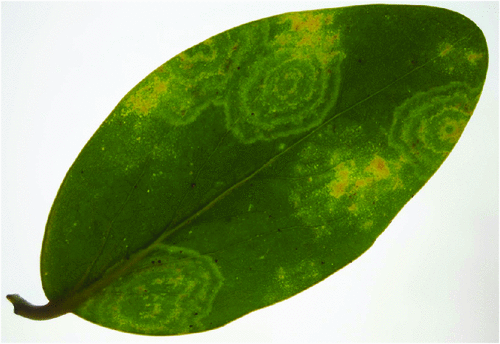
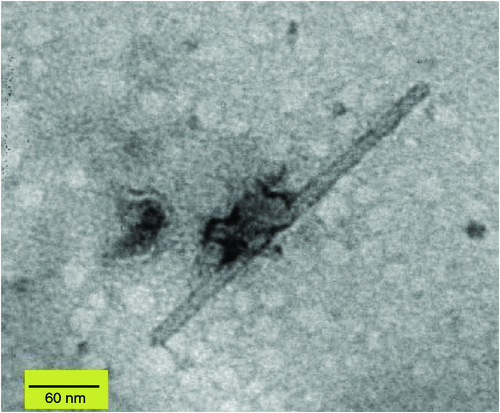
In April 2009, chlorotic mosaic and concentric ring symptoms (Fig. 1) were observed on three G. lucida shrubs from Rangitoto Island, a volcanic reserve managed by the Department of Conservation in New Zealand. Leaf samples were collected from two of the symptomatic G. lucida and sent to the laboratory for disease diagnosis. Rod-shaped particles (~18 × 300 nm) were observed by electron microscopy in crude sap preparations from symptomatic leaves negatively stained with 1% (w/v) uranyl acetate (Fig. 2).
|
|
Total RNA was extracted from the leaves of one asymptomatic and two symptomatic samples using a Qiagen® RNeasy Plant Mini Kit (Qiagen, Melbourne, Vic., Australia). An internal control reverse transcription-polymerase chain reaction (RT–PCR) was done for all the samples using nad5-s and nad5-as primers (Menzel et al. 2002) to ensure the competency of the RNA extracts (data not shown). Complementary DNA (cDNA) was synthesised using Superscript™ III reverse-transcriptase (Invitrogen, Carlsbad, CA, USA) in a final volume of 20 μL containing 4 μL of total RNA, 1 × RT buffer, 8 U RNasin Plus (Promega, Madison, WI, USA), 0.25 μg random hexamer primers, 20 μg bovine serum albumin (BSA), 100 U Superscript TM III reverse-transcriptase, 10 mM DTT, 0.5 mM dNTPs and nuclease-free water. The RNA, buffer, random primers, BSA and water were mixed and heated to 70°C for 10 min then kept on ice for 1 min. After denaturation other reagents were added and incubated for 1 h at 50°C. PCR was performed in a final volume of 20 µL using generic tobamovirus primers (Agdia Inc., Elkhart, IN, USA). The reaction mixture consisted of 10 µL 2× GoTaq® Green Master Mix (Promega), 4 µL Agdia primer mix, 2 µL cDNA and 4 µL nuclease-free water. Amplification was done according to the manufacturer’s instructions. PCR products were analysed by electrophoresis on 1.5% (w/v) TAE agarose gel stained with SYBR Safe (Invitrogen).
An amplicon of the expected size (~380 bp) was obtained from the two symptomatic samples. The amplified DNA product was purified from the gel using an extraction kit (Bio–Rad, Hercules, CA, USA) and cloned into the pCR4–TOPO vector (Invitrogen). The recombinant plasmid DNA was transformed into competent E. coli cells. After screening by PCR, four clones containing the expected size fragment were sequenced on an ABI Avant 3100 Genetic analyzer using BigDye 3.2 chemistry by EcoGene (Auckland, New Zealand). The sequences were analysed using the nucleotide Basic Local Alignment Search Tool (BLAST: National Center for Biotechnology Information: http://www.ncbi.nlm.nih.gov). The sequences were identical and had 97% nucleotide identity with the replicase gene of Tomato mosaic virus (ToMV) (GenBank Accession Nos: DQ873692, AB083196 and AJ417701) and Tobacco mosaic virus (TMV) (GenBank Accession Nos: AJ243571, Z92909 and X02144). These TMV GenBank accession numbers are now classified as ToMV by the International Committee on Taxonomy of Viruses (Lewandowski 2005).
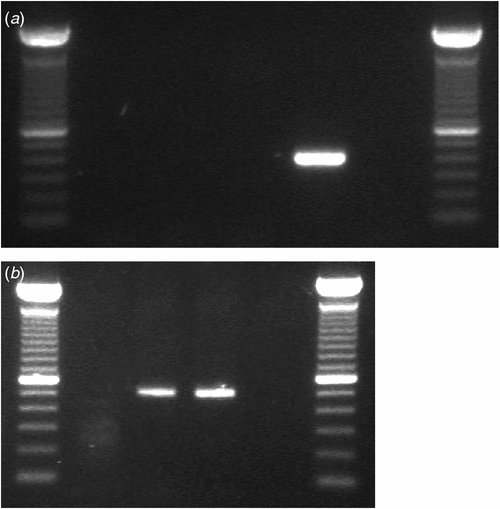
The identity of the virus was confirmed by RT–PCR using discriminative species-specific primers for TMV and ToMV (Jacobi et al. 1998). The TMV-2 reverse primer and TMV-1 forward primer are expected to amplify a 422 bp PCR product within the viral movement protein gene of TMV, while the ToMV-5 reverse primer (5458–5436 nt of TMV-L, GeneBank Accession No. X02144) and ToMV-6 forward primer (4951–4976 nt) amplify a product of 508 bp within the same gene of ToMV. PCR reactions were done in a 20 µL volume, containing 1× GoTaq® Green Master Mix, 0.5 µM of either primer pair and 2 µL of cDNA. The PCR cycle for both viruses was performed as follows: initial denaturation at 94°C for 5 min, 40 cycles of 94°C for 30s, 62°C for 45s, and 72°C for 1 min, and a final elongation at 72°C for 5 min. No PCR product was obtained using the TMV-specific primers but a ~500 bp band was obtained with the ToMV primers (Fig. 3a and 3b). The product was cloned and two clones were sequenced as described above. The consensus sequence was submitted to GenBank (GenBank Accession No. HM026496) and analysis showed 100% nucleotide identity with ToMV (GenBank Accession Nos: AF 332868 and AB355139) confirming that G. lucida was infected only with ToMV.
|
The virus isolated from G. lucida was identified as ToMV based on molecular analysis with supporting evidence from electron microscopy. ToMV has been reported previously in New Zealand but was thought to be a strain of TMV (Pennycook 1989). This is the first definite report of ToMV in New Zealand and the first report of any virus infecting the native plant G. lucida. ToMV is associated with diseases in a variety of annual and perennial plants and trees (Jacobi et al. 1992). Symptoms include leaf mottling, mosaic, necrosis and total crop loss. The virus is seed- and soil-borne but is primarily disseminated by human handling and contaminated tools (Broadbent 1976).
Acknowledgements
The authors thank Dr Eric Mckenzie (Landcare Research Institute, Auckland, New Zealand) for sample collection.
Broadbent L
(1976) Epidemiology and control of Tomato mosaic virus. Annual Review of Phytopathology 14, 75–96.
| Crossref | GoogleScholarGoogle Scholar |

Dawson JW
(1966) Vegetative features of Griselinia lucida – A New Zealand shrub epiphyte. Tuatara 14, 121–129.

Jacobi V,
Castello JD, Flachmann M
(1992) Isolation of Tomato mosaic virus from Red Spruce. Plant Disease 76, 518–522.
| Crossref | GoogleScholarGoogle Scholar |

Jacobi V,
Bachand GD,
Hamelin RC, Castello JD
(1998) Development of a multiplex immunocapture RT-PCR assay for detection and differentiation of tomato and tobacco mosaic tobamoviruses. Journal of Virological Methods 74, 167–178.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Menzel W,
Jelkmann W, Maiss E
(2002) Detection of four apple viruses by multiplex RT-PCR assays with coamplification of plant mRNA as internal control. Journal of Virological Methods 99, 81–92.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |



