First report of Colletotrichum boninense infecting yellow passion fruit (Passiflora edulis f. flavicarpa) in Brazil
H. J. Tozze Júnior A , I. H. Fischer B , M. P. S. Câmara C and N. S. Massola Júnior A DA Universidade de São Paulo, Escola Superior de Agricultura ‘Luiz de Queiroz’, Departamento de Fitopatologia e Nematologia Agrícola, CP 09, Piracicaba, SP 13418-900, Brazil.
B Agência Paulista de Tecnologia dos Agronegócios, Av. Rodrigues Alves, 40-40 Bauru, SP 17030-000, Brazil.
C Universidade Federal Rural de Pernambuco, Departamento de Agronomia, Dois Irmãos, Recife, PE 52171-900, Brazil.
D Corresponding author. Email: nmassola@esalq.usp.br
Australasian Plant Disease Notes 5(1) 70-72 https://doi.org/10.1071/DN10025
Submitted: 5 March 2010 Accepted: 20 June 2010 Published: 12 July 2010
Abstract
In June 2006, fruits of yellow passion fruit with typical anthracnose symptoms were collected in São Paulo state, Brazil. Based on cultural, morphological characteristics and molecular analyses of internal transcribed spacer rRNA region (ITS), the causal agent of the disease was determined as Colletotrichum boninense. This is the first report of C. boninense associated with anthracnose of passion fruit in Brazil.
Passion fruit is widely consumed worldwide. Brazil is the world’s largest producer of passion fruit, represented mainly by yellow passion fruit (Passiflora edulis f. flavicarpa) (97%) and sweet passion fruit (P. alata) (3%), with an estimated cultivation area of 46 866 ha, and an annual yield of 664 286 t (FNP 2010). Anthracnose of passion fruit, previously assumed to be caused by Colletotrichum gloeosporioides, occurs in all production areas of Brazil, and can be a serious problem in some seasons (Lima et al. 2005).
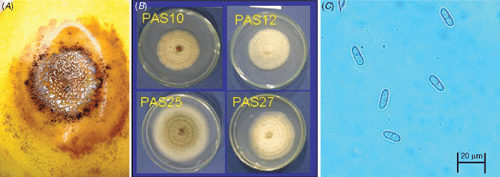
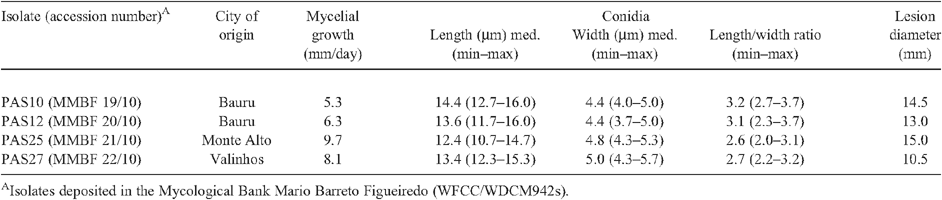
Anthracnose symptoms were observed on yellow passion fruit collected in Bauru, Monte Alto and Valinhos cities, São Paulo state, Brazil, in June 2006. Typical symptoms on fruits were circular, sunken lesions with orange spore masses in the centre (Fig. 1A). Four single-conidium isolates (PAS10, PAS12, PAS25, PAS27) were obtained from infected fruits. Colonies of these isolates grown on potato dextrose agar (PDA) at 25°C with a 12-h light photoperiod were white to cream (Fig. 1B). Mycelial growth ranged from 5.3 to 9.7 mm/day (Table 1). Conidia were cylindrical, obtuse at both ends, with a hilum-like low protuberance at the base (Fig. 1C), and measured 12.4 to 14.4 μm long × 4.4 to 5.0 μm wide. Conidial length/width ratio was 2.6 : 3.2. These morphological characters are consistent with the description of C. boninense (Moriwaki et al. 2003). These isolates were deposited in the Mycological Bank Mario Barreto Figueiredo (WFCC/WDCM942) with accession numbers (MMBF 19/10, MMBF 20/10, MMBF 21/10, MMBF 22/10) (Table 1).
|
|
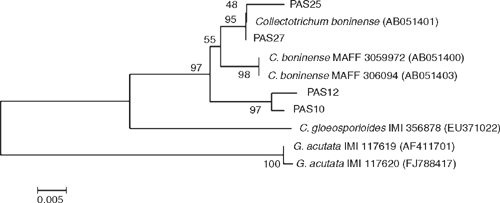
To confirm morphological identification, the internal transcribed spacer rRNA region (ITS) genes were sequenced [GenBank Accessions FJ981603 (PAS12), FJ981604 (PAS10), FJ981605 (PAS25), FJ981606 (PAS27)]. These sequences were compared with the same regions of the isolates used in the original characterisation of C. boninense (GenBank Accessions AB051401, AB051400, AB051403) (Moriwaki et al. 2003). Similarity between these sequences was 97–100%. In order to further confirm the identity of the isolates from passion fruit, ITS region sequences of closely related species C. acutatum (GenBank Accession number AF411701, FJ788417) and C. gloeosporioides (GenBank Accession number EU371022) were obtained from GenBank and a phylogenetic analysis was carried out. These sequences were edited with the Staden Package (Staden 1996) and aligned with CLUSTALW (www.ebi.ac.uk/clustalw). An unrooted tree was generated in order to illustrate the relatedness of closely related species without making assumptions about ancestry among species. Alignment was visually inspected and manually corrected using the MEGA 4.0 program (Tamura et al. 2007). The phylogenetic analysis was done with 545 bp sequences of the ITS rDNA region using the Neighbour Joining (NJ) algorithm on the Kimura 2-parameter distance, without an outgroup. The analysis was run in MEGA 4.0, with 10 000 bootstrap replicates (Fig. 2).
|
The pathogenicity of the four isolates was determined on yellow passion fruit cv. ‘Afruvec’. Inoculations were performed by depositing 40 μL of a conidial suspension (105 conidia/mL) on the fruit surfaces, followed by wounding with a sterilised hypodermic needle, adapted from Cao et al. (2008). Five replicates (fruits) were used per isolate. Inoculation of control fruits was similar in procedure and number of test fruits, except that sterile distilled water was used at the wound site instead of the conidial suspension. Incubation took place in a moist chamber at 25°C with a 12-h light photoperiod. Initial symptoms appeared 3 days after inoculation and were characterised by dark spots surrounding the inoculation points. A week after inoculation the lesions were circular, slightly sunken and measured 10.5–15.0 mm in diameter. At this stage, orange coloured spore masses were observed on the centre. No symptoms were observed on the control fruits.
The fungus was reisolated from inoculated fruits and the cultures were identical to the original isolates.
To our knowledge, this is the first report of C. boninense infecting yellow passion fruit in Brazil. Further studies are warranted to clarify the role of C. gloeosporioides in this disease in Brazil.
Cao S,
Zheng Y,
Yang Z, Tang S
(2008) Effect of methyl jasmonate on the inhibition of Colletotrichum acutatum infection in loquat fruit and the possible mechanisms. Postharvest Biology and Technology 49, 301–307.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Lima RM,
De Oliveira SMA,
Maciel MIS, De Moraes LM
(2005) Influence of temperature and wetness periods on development of anthracnose of yellow passion fruit in post-harvest. Summa Phytopathologica 31, 27–32.

Moriwaki J,
Toyozo S, Tsukiboshi T
(2003) Morphological and molecular characterization of Colletotrichum boninense sp. nov. from Japan. Mycoscience 44, 47–53.
| Crossref | GoogleScholarGoogle Scholar |
CAS |

Staden R
(1996) The Staden Sequence Analysis Package. Molecular Biotechnology 5, 233–241.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |

Tamura K,
Dudley J,
Nei M, Kumar S
(2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution 24, 1596–1599.
| Crossref | GoogleScholarGoogle Scholar |
CAS |
PubMed |



