DNA-binding, multivalent interactions and phase separation in transcriptional activation
Ngaio C. Smith A and Jacqueline M. Matthews
A and Jacqueline M. Matthews  A *
A *
A School of Life and Environmental Sciences, Building G08, The University of Sydney, Darlington, NSW 2006, Australia.

Ngaio Smith obtained her BSc (Hons) and PhD from the University of Sydney under the supervision of Professor Jacqui Matthews. Her work focusses on the interactions of transcription factors and DNA, particularly in the LIM-HD family of proteins. A key area of interest of her work has been how protein–protein interactions affect DNA-binding specificity and gene regulation. |

Jacqui Matthews is a graduate of UNSW and was awarded her PhD from the University of Cambridge, working with Sir Alan Fersht to define the principles underlying protein folding. She undertook postdoctoral work with Professor Richard Simpson at the Ludwig Institute for Cancer Research in Melbourne, where she first met and worked with Ed Nice. She developed an independent research program at The University of Sydney centered on biomolecular interactions, particularly the assembly of multiprotein transcriptional complexes involved in cell-specific transcriptional regulation. |
Australian Journal of Chemistry 76(8) 351-360 https://doi.org/10.1071/CH22269
Submitted: 21 December 2022 Accepted: 6 April 2023 Published: 5 July 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Transcription is an essential process in biology whereby gene-specific transcription factors target sites on DNA to recruit the basal transcription machinery that will produce messenger RNA (mRNA). It is a highly regulated multi-step process that involves many proteins and protein complexes. Transcription factors, the proteins that mark genes for activation, and other transcriptional regulators are highly enriched in low-complexity disordered regions, which are strongly linked to multivalent binding and phase separation. These disordered regions can form multivalent dynamic complexes that are essential for many aspects of transcription. Many of these proteins can phase separate in vitro and show evidence of phase separation in vivo. Whether these interactions represent biologically relevant phase separation in vivo is controversial. However, what these events do demonstrate is that many transcriptional proteins co-cluster with other factors in vivo, forming multivalent dynamic clusters that contribute to transcriptional events. We review some of these recently investigated events and consider how they contribute to our understanding of transcription.
Keywords: DNA binding, dynamic hubs, intrinsic disorder, liquid–liquid phase separation, multivalent interactions, protein–protein interactions, transcription factors, transcriptional activation.
Introduction
Gene regulation in eukaryotes is a complex process that involves the interplay of multiple proteins acting together to regulate chromatin structure, mark which genes are to be up- or down-regulated and modulate the recruitment or activity of RNA polymerase II (Pol II), which is responsible for transcribing most genes into messenger RNA (mRNA). For a given gene to be transcribed into mRNA, transcription factor proteins (TFs) bind specific sequences at promoter and enhancer sites. Whereas promoters lie close to the start site of transcription, enhancers can lie many thousands of base pairs away, either up- or down-stream of the start site. The bringing together of enhancers and promoters is thought to be an essential part of gene activation, usually involving interaction with Mediator and other proteins such as CTCF, condensin and the basal TFs to recruit and/or activate Pol II. Many of these basal factors, which are common to the expression of all genes, are multiprotein complexes.
The genome needs to have many layers of organisation for correct function. This includes the packaging of the genome into chromatin – the combination of DNA and proteins that makes up chromosomes within the nucleus of eukaryotic cells. Chromatin exists in two major forms, of which euchromatin is less condensed and can be transcribed, whereas heterochromatin is highly condensed and typically not transcribed. The presence of architectural proteins maintains or restricts accessibility, and the compartmentalisation of nuclear elements enhances or inhibits function.[1,2]
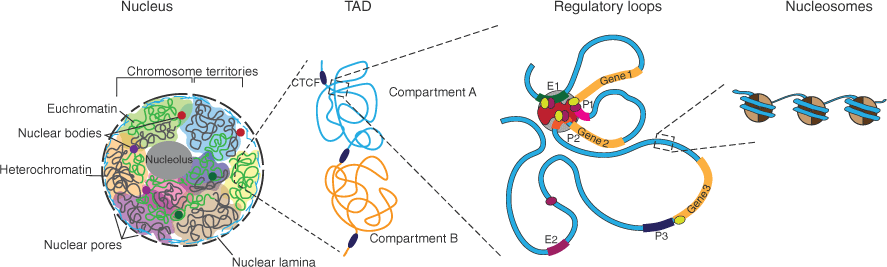
In recent decades, methods based on chromatin conformation capture (3C) techniques (which identify regions of DNA that lie close together in space), other molecular biology approaches to map the genome architecture, and super-resolution microscopy and imaging have provided considerable insight into 3D genome organisation and function (reviewed in ref. [3]). These studies confirm that chromosomes separate into active and inactive compartments. Chromatin interactions are enriched within smaller domains – the topologically associating domains (TADs), which are further divided into smaller chromatin nanodomains, which are exclusively transcriptionally active or inactive. Chromatin loops and extrusions play an additional role in both gene regulation and chromatin folding, and DNA is wrapped around histone octamers, forming nucleosomes, which give expanded chromatin a ‘beads on a string’ appearance[4] (Fig. 1).
Nuclear organisation and transcription. The nuclear membrane contains numerous nuclear pores, and the nuclear lamina lies on the inner surface of the nuclear membrane, helping to maintain stability and organise chromatin. The chromosomes lie predominantly in chromosome territories that can be made up of open euchromatin (green) and closed heterochromatin (grey). Membraneless nuclear bodies including the nucleolus and other structures are formed through liquid–liquid phase separation. Transcriptionally active domains (TADs) are separated into compartments that are transcriptionally active (Compartment A) or inactive (Compartment B). Within these regions, regulatory loops are formed, bringing enhancers and promoters of genes into close proximity through interactions between gene-specific transcription factors (green and purple) and the basal transcription machinery including Mediator, basal transcription factors and RNA polymerase II. DNA is wound around histone octets to form nucleosomes.
In this review, we look at how protein–protein and protein–nucleic acid interactions contribute to the fundamental cellular process of gene activation, with a particular focus on how transcription factors specifically target genes and transcription initiation, and how liquid–liquid phase separation (LLPS) contributes to these events.
Transcription factor binding
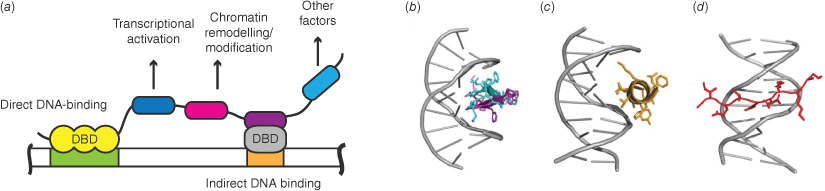
Most cell-specific eukaryotic TFs are multi-domain proteins comprising one or more DNA-binding domains (DBDs) plus other motifs or domains, including in most cases substantial regions of intrinsic disorder (Fig. 2a). DBD-DNA interactions depend on the shape complementarity of both molecules and the ability to make favourable interactions.[5,6] Notably, alpha helices and beta hairpins fit well into the major groove of DNA, whereas extended protein chains can target the minor groove (Fig. 2b, c). Most DBDs are positively charged under physiological conditions, and can bind non-specifically to DNA – predominantly through basic sidechains from proteins and the acidic sugar/phosphate backbone of DNA. In contrast, specific interactions can be classed as sequence-specific/direct readout (close contacts between the nucleobases and protein chains) or conformation-specific/indirect readout (reviewed in ref. [6]). There are some preferences of nucleobase-specific recognition by given amino acids involving the nucleobases themselves or the widths of major and minor grooves (which vary according to sequence), and some binding events distort protein structure (reviewed in ref. [7]). Although there is no simple formula for predicting efficient DNA–protein interfaces, computational tools incorporating Rosetta, machine learning and/or AlphaFold2 predictions have been developed to help predict new and redesign existing DNA–protein interfaces (e.g. refs [8–11]).
Transcription factor–DNA binding. (a) Transcription factors typically contain a DNA-binding domain (DBD; yellow) or can interact with DNA through binding to another protein that contains a DBD (grey), and usually have other protein interaction domains that will recruit proteins with various other functional domains. Many of these interaction domains are within large regions of intrinsic disorder. (b) β-Hairpin from the ARC (Activity-regulated cytoskeleton-associated protein) repressor binding to the DNA major groove.[97] (c) α-Helix, and (d) extended chain from GATA1 (GATA-binding factor 1) binding the major and minor grooves, respectively.[98]
Although specificity is required to precisely regulate the eukaryotic genome, the intrinsic ability of eukaryotic TFs to recognise cognate DNA sequences with high specificity is poor. Unlike prokaryotic TFs that bind long DNA sites with high levels of specificity, eukaryotic TFs tend to recognise shorter DNA motifs (~6–8 base pairs (bp)) with high levels of degeneracy, and so lack the necessary information to precisely target specific sites in the genome.[12] Concentrations of canonical binding sites in the genome are thought to be high (~mM), which should favour binding, and there will be many other slightly varied sites that would be bound with lower affinity. The combination of non-specific and specific binding of TFs to DNA and rapid exchange of non-optimal binding sites is thought to facilitate search strategies that enable TFs to find their target binding sites much faster than predicted from random binding events alone.[13,14]
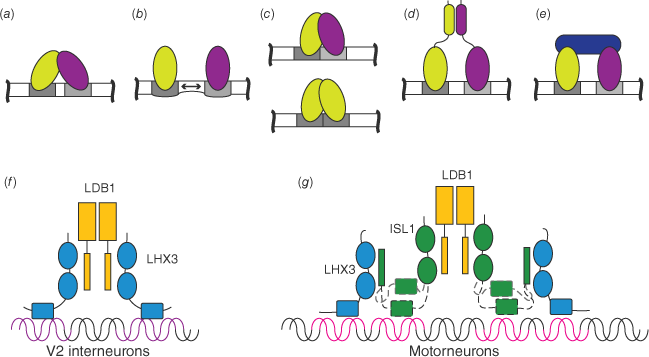
TFs have been identified as binding throughout the genome, to both canonical sites and regions that lack a canonical binding motif, but most of these sites are non-functional and binding here does not alter transcription.[15,16] Rather, it is sites co-occupied by multiple TFs that are associated with active expression of genes.[5] The non-DBD parts of TFs, and especially the disordered regions (see below), tend to mediate protein–protein interactions. This property gives them the ability to recruit other factors that either promote or repress transcription. Multiple binding events and protein–protein interactions play an important role in specificity, through a range of different mechanisms including the modification of DNA structure, enhanced binding through avidity effects and combinatorial binding, such as heterodimerisation of TFs that individually recognise half-sites (reviewed in ref. [7]) (Fig. 3a–e). An example of changing specificity through involvement in different protein complexes focuses on ISL1 (Islet-1), a LIM-homeodomain (LIM: Lin11, Isl1, Mec3) protein required for the development of many different organs including the heart and nervous system, which acts as a transcriptional switch to reduce binding of another LIM-homeodomain protein, LHX3 (LIM-homeobox protein 3), to interneuron specific genes and enhance binding to motor neuron-specific genes.[17] We showed that ISL1 replaces LHX3 as the direct binding partner for LDB1 (LIM domain-binding protein 1), providing an alternative intrinsically disordered binding site for LHX3,[18] forming lower-affinity but kinetically favoured complexes.[19] Although the ISL1-DBD has quite low binding affinity and specificity for DNA, it can modulate the binding of the LHX3-DBD, apparently though competition for binding to interneuron gene sequences, but synergistically enhances binding to motor neuron genes through dynamic binding events[20] (Fig. 3f, g). In this way, protein–protein interactions can have a major influence on DNA binding specificity and downstream transcriptional activation without directly altering DNA binding characteristics of partner proteins.
Transcription factor interactions modulate DNA binding by a range of mechanisms: (a) direct interactions between DBDs (light green and purple indicate different TFs; DNA binding sites are grey inside longer white DNA sequences); (b) DNA binding modulates DNA structure of nearby binding sites; (c) half-site binding facilitates combinatorial binding; (d) interactions of non-DBDs; (e) higher level complex formation (additional TF is dark blue); (f) schematic of LHX3 (blue) binding to V2 interneuron DNA sites (purple) within the binary LHX3-LDB1 complex (LDB1 is yellow); (g) in neighbouring motor neurons, ISL1(green) replaces LHX3 as the LDB1-binding protein binding both partners. The ISL1-DBD facilitates binding of LHX3 to these motorneuron sites (magenta) through weak apparently dynamic binding events.
Contributions of disordered regions of TFs
It was the study of TFs that spearheaded research into intrinsic disorder in proteins. Numerous investigations have demonstrated that intrinsically disordered regions are enriched in binding function, with flexible and adaptable modes of binding that can be rapidly modulated by post-translational modification (PTM), with some disordered binding domains able to bind multiple partner proteins (e.g. refs [21,22]). Some disordered binding domains fold on binding, with recent studies suggesting that the more common mechanism is binding preceding induced-fit folding.[23] However, it has become increasingly evident that folding on binding exists on a spectrum from high levels of order to highly dynamic or fuzzy complexes (e.g. ref. [24]). Indeed, while intrinsically disordered regions are well suited to facilitating binding, the same properties make them well suited to undergo LLPS.
Multivalent networks of interactions: phase separation and alternative models
It is now commonly accepted that biological molecules can undergo LLPS to form membrane-less organelles or condensates (Box 1), providing a mechanism to organise the contents of living cells and regulate a plethora of cellular functions (e.g. ref. [25]). Many proteins that have been found undergoing LLPS contain disordered or low-complexity regions that contribute to phase separation through the formation of multivalent interaction networks.[26] Proteins involved in transcription have been found to have particularly high levels of disorder,[27] and there is a growing set of data that implicates phase transitions as having important roles in transcription.[28] The likely ability of most proteins to undergo phase separation[29] and the presence of a crowded environment in the nucleus (including the presence of 30–50% chromatin) to facilitate phase separation[30–32] support these phenomena. However, there are caveats.
| Box 1. Characteristics of phase separation |
| Liquid–liquid phase separation (LLPS) involving proteins and/or nucleic acids refers to the segregation of one or more of these biopolymers into a concentrated liquid phase and a dilute surrounding phase, and distinct separate liquid states exist inside and outside the condensate; surface tension causes these to form a spherical or droplet state that can undergo fusion with droplets of the same type. LLPS is triggered by the formation of networks of multivalent interactions and segregation occurs only when a threshold concentration of biopolymers is reached (see reviews by Alberti et al.[33], Shin and Brangwynne[92], Banani et al.[93]). Whether a solution undergoes LLPS depends not only on the concentration but also on environmental conditions such as temperature, buffers and pH, co-solutes and crowding (i.e. the volume excluded by other macromolecules). |
| Biological condensates appear to have very different properties, such as differences in liquidity (noting that some condensates change over time to become more gel-like, including forming fibrils), sensitivity to hexanediols and whether they exclude other biomolecules, and can vary considerably in terms of size.[38] This is in line with proteins and nucleic acids having different spacing and sequences of binding sites and spacers giving different physical and chemical properties (e.g. ref. [36]). |
| The key functional features of LLPS that distinguish it from other multivalent networks appear to be depletion of segregated biopolymer outside the condensate, and separation of liquid states inside and outside the condensate. The latter can result in a lowering of the dielectric constant and modification of water activity, which can enhance electrostatic effects, weaken hydrophobic forces and modulate enzyme activity.[94] The concentration effect and concentration dependence of LLPS is also a property of gels, albeit at lower biopolymer concentrations (e.g. ref. [36]), so formation of high-concentration foci of enhanced binding and activity is not unique to LLPS states. Some increases of binding affinity and modulation of the dielectric constant and water activity can be brought about by crowding in the absence of phase separation.[95,96] |
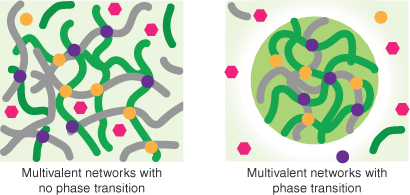
Robust data exist to demonstrate that nuclear organelles are formed by proteins in a phase-separated state. Many proteins have also been shown to undergo LLPS in vitro, but it is much more difficult to confirm LLPS in vivo when such states are small and probably transient, and observations of LLPS may be exacerbated because experiments that artificially increase target protein concentrations could artificially induce LLPS.[33–35] Furthermore, not all multivalent networks of interactions are associated with LLPS. Multivalent proteins and nucleic acids can form stable scaffolds (formed through non-exchangeable interactions, which may take part in larger structures) or networks of exchangeable contacts such as gels that occur in the absence of phase separation[34,36] (Fig. 4). Some refer to the latter as ‘hubs’ or ‘dynamic hubs’ (e.g.[37,38]), whereas others use the same terms to describe phenomena without implying mechanism.[34] Note that size is not a good discriminator of hubs versus condensates.[38] In many cases, both forms of dynamic multivalent networks are reasonable models for observable events in transcription[34,37] (Box 1).
Multivalent interaction networks. Multivalent networks involving proteins and/or nucleic acids can form dynamic hubs or gels that do not involve phase separation but can also lead to phase separation that depletes the surrounding environment of components of the dense phase. In this schematic green and grey lines represent elongated polymers (nucleic acids or disordered proteins), purple, pink and yellow shapes represent specific TFs. Light green indicates average concentrations and dark green is a concentrated liquid phase or droplet. Note the depleted (white) zone srrounding the droplet.
Simulations of LLPS often invoke the ‘stickers-and-spacers’ lattice-based model from associative polymer theory, in which multivalent proteins are represented as heteropolymers comprising stickers (LLPS-binding motifs) and spacers (regions in between stickers).[39] Such simulations can be used to predict phase behaviour and allow the modelling of complex systems, leading to the proposal that some proteins form scaffolds that drive LLPS, while other proteins can take part in LLPS only after it has been established (clients).[40]
In the next sections, we review some of the many recent investigations in transcriptional activation where LLPS of proteins has been implicated. In most cases, evidence is provided for in vitro phase separation and in vivo clustering/puncta formation, often with other experiments supporting phase separation (such as liquid behaviour/or disruption by 1,6-hexanediol). However, in most cases, physiologically relevant LLPS has not yet been unequivocally demonstrated, and it is therefore difficult to attribute roles as drivers or clients of LLPS. Further technological development will be required to make detailed studies of these systems, as current experimental techniques cannot directly observe all condensates with the required level of detail.
Genome organisation
LLPS could contribute to chromatin organisation and overall genome structure by a range of different mechanisms, including facilitating the formation of compartments of heterochromatin and generating functional nuclear compartments for targeted activation of gene expression (reviewed by Kantidze and Razin[41]). The aliphatic alcohol 1,6-hexanediol inhibits weak hydrophobic interactions and can disrupt some condensates in vitro and in vivo.[42] Ulianov et al. recently used super-resolution microscopy analysis and 3C techniques to demonstrate that the addition of 1,6-hexanediol to cells (under controlled exposure conditions to avoid disruption of membranes) was able to disrupt genome structure.[43] These cells were able to partially recover on removal of the 1,6-hexanediol. This suggests a significant role of LLPS in maintaining genome organisation. However, 1,6-hexanediol in cells has been reported to drive chromatin condensation in a manner independent of its ability to dissolve hydrophobic condensates,[44] so it remains unclear to what extent LLPS plays a role in overall genome organisation. Further, the mechanism of action of 1,6-hexanediol in cells should be more closely investigated, and interpretation of in vivo data reliant on this technique should be taken with caution until further investigations can be carried out.
CCCT-binding protein, CTCF
CTCF, together with condensin (a multiprotein complex that mediates cohesion between replicated sister chromatids in chromosome segregation in dividing cells) plays multiple roles in transcriptional regulation including defining TAD boundaries, insulating promoters/enhancers, loop extrusion and regulation of genome structure.[45–47] CTCF forms LLPS-like clusters in cells,[48–50] which in one study was shown to be dependent on interaction with RYBP (RING1 and YY1-binding protein), a largely disordered and self-associating polycomb protein.[51] Polycomb proteins typically modify histones (and other proteins) to silence target genes; however, these two proteins co-localise to puncta in embryonic stem cells associated with strongly transcriptionally active genes. CTCF was also shown to form co-clusters with Pol II in which CTCF appeared to play an important scaffolding role in generating multiprotein transcriptionally active clusters or condensates.[50] In that study, the presence of CTCF in clusters was insensitive to 1,6-hexanediol, but Pol II clusters were sensitive to and reliant on the presence of CTCF. These studies imply a role of CTCF clustering in gene activation, but it is not clear if RYBP is also involved in those Pol II clusters. Indeed, this study and many others focus on co-clustering of pairs of proteins, but it is possible that larger networks may be involved in vivo that modulate clustering and activity. Without being able to clearly see this broader context, our understanding of these systems remains limited.
RNA polymerase II
Pol II first forms a pre-initiation complex, and thereafter initiates transcription (promoter escape) and generates the mRNA transcript through elongation.[52] The C-terminal domain (CTD) of the large subunit (Rbp1) of Pol II is a low-complexity domain required for production of a variety of transcripts,[53] and is at least partially responsible for full-length Pol II clustering and in vitro phase separation.[54] The CTD itself can undergo LLPS in vitro,[54,55] both alone and when interacting with a low-complexity domain from TAF15 (TATA-box binding protein associated factor 15; a component of the basal transcription factor IID (TFIID) complex).[54,55] Phosphorylation of the CTD has been shown to disrupt binding and prevent phase separation. Bacterial RNA polymerase (RNAP) also forms dynamic LLPS-like clusters in vivo, suggesting that it is an intrinsic property of RNA polymerases.[56]
The CTD comprises multiple heptapeptide (YSPTSPS) repeats, the number of which varies across species (from 26 repeats in yeast to 52 in humans). The number of heptapeptide repeats present in the CTD influences the stability of condensates formed by the domain; lower numbers of repeats result in less stable condensates in vitro and less association with chromatin in cells.[54] All residues in the CTD heptapeptide repeats are post-translationally modified in a cyclical fashion, which has regulatory and mechanistic consequences both for basal transcription and transcription-associated processes, including the recruitment of many different regulatory proteins.[53] For example, CTD in its unmodified form binds Mediator in the initiation stage, followed by phosphorylation by CDK7 (cyclin dependent kinase 7) in TFIIH to allow promoter escape; then, hyperphosphorylation by p-TEFb (positive transcription elongation factor b) enables transcriptional elongation.[53] p-TEFb is made up of CDK9 and Cyclin T1. A low-complexity histidine-rich domain in Cyclin T1 can undergo LLPS in vivo, and can form puncta in vitro with colocalisation of the CTD.[57] CTD phosphorylation by CDK7 enhances the colocalisation effect, but subsequent hyperphosphorylation by CDK9 (cyclin dependent kinase 9) causes the puncta to dissipate. The various CTD-associated clusters have different properties, with a CTD-only non-phosphorylated state being sensitive to hexanediols whereas the CDK7-phosphorylated state is more resistant.[54,57] Thus, the CTD may form different phase-separated states regulated by phosphorylation (which in turn enables binding to different partners) throughout successive stages of transcription. This is a good example of the complex interplay between LLPS, post-translational modification and protein–protein interaction and how these mechanisms combine to precisely regulate transcription.
Mediator
Mediator is a eukaryotic multi-subunit transcriptional complex that links Pol II to activating TFs on chromatin.[58,59] It is characterised by the ability to interact with many regulatory factors, high levels of intrinsic disorder and conformational flexibility, and forms LLPS-like clusters in vivo.[59] Many TFs that colocalise with Mediator in vivo can phase-separate in vitro with mammalian Mediator, or components thereof, via their intrinsically disordered activation domains, suggesting a common mechanism for TF activation domains.[60] Residues required for phase separation were also necessary for gene activation.[60] Activation domains from the yeast transcription factors GAL4 (Regulatory protein GAL4) and GCN4 (General Control Nondepressible protein 4) form dynamic or fuzzy complexes with the yeast homologue of Med15 (a Mediator subunit[61]).
Mediator has been observed to form condensates or clusters with Pol II both in vivo and in cells (for example, refs [50,62,63]). These clusters are likely associated with chromatin and facilitate transcription. Hypophosphorylated CTD colocalises with Mediator in vivo but hyperphosphorylated CTD does not,[62] supporting the model of differently phosphorylated states of Pol II making different networks of interactions throughout its activity cycle.
Super-enhancers
Large clusters of enhancers, referred to as super-enhancers, control the transcription of genes with prominent roles in cell-type specific processes.[64,65] Some of these assemblies were previously referred to as transcriptional factories.[66] They are formed through networks of interactions between chromatin and TFs.[67] ChIP-seq (chromatin immunoprecipitation with sequencing) data indicate the presence of Mediator and hypophosphorylated Pol II at super-enhancers, suggesting that they are setting up the local environment for transcriptional initiation.[62] Characterising super-enhancers has been difficult because of their complex and transient nature. However, there are now several databases that catalogue where super-enhancers occur in the genome and their functions.[68–70] The identities of the protein components of super-enhancers remain poorly described owing to technical limitations. Several candidate TFs have been implicated in phase separation at super-enhancers, including BRD4 (Bromodomain-containing protein 4).[71]
BRD4
BET (bromodomain and extra-terminal domain, ET) TFs are epigenetic readers that bind to acetylated lysine residues in histones and other TFs and recruit transcriptional regulators.[72] BRD4 is a broadly expressed BET protein with two isoforms, the longer of which contains a low-complexity CTD (LCD). It regulates the transcription of numerous genes, particularly at super-enhancers.[73] BRD4 interacts with components of Mediator, as well as p-TEFb.[73–75] BRD4 can phase-separate in vitro and colocalises with MED1 from Mediator, as well as localising to H3K27 acetylation sites on chromatin (which are linked to transcriptionally active regions), and binding DNA directly.[76] Although phase separation can be mediated by the LCD,[71] a short isoform of BRD4 that lacks the LCD also drives cluster formation in vivo.[76,77] Both inhibition of BRD4 to block binding to acetyllysine residues and pharmacological degradation have been shown to inhibit transcription, phase separation and clustering of Mediator while maintaining promoter–enhancer contacts.[63,78,79] Although BDR4 is generally considered to be a transcriptional activator,[80–82] it can also act as a repressor through the recruitment of the repressive NuRD (Nucleosome remodelling and deacetylase) complex,[83] highlighting that many transcriptional regulators can either activate or repress transcription depending on their binding partners.
Nucleic acids
Although proteins have probably been the major focus of recent research in LLPS, nucleic acids also play a key role. Initial studies found that RNA binding partners could be drivers of protein-based LLPS.[84] It was later shown that DNA could play a similar role, and that the sequence of the DNA can influence the likelihood of LLPS occurring.[85–87] The multivalent nature of many protein:DNA complexes provides the opportunity to form networks of interactions within the nucleus, adding an additional layer of complexity in the regulatory landscape.
Multivalent dynamic networks and LLPS in transcription
Overall, it is clear that many proteins that are involved with transcription can undergo LLPS in vitro and form clusters or puncta in vivo. In many cases, it is the formation of local high protein concentration clusters that explains experimental data. LLPS may be involved but does not necessarily provide a clear mechanistic advantage over other forms of multivalent network formation.[34] For example, a simplified model that accounts for high levels of transcriptional activity appears to capture the multivalent networks of interactions at these sites rather than other properties of phase separation.[88] Given the links between multivalent interactions, crowding and sensitivity to concentrations, phase separation could be a consequence of, rather than a driving force for, clustering in many cases – particularly when considering small transient multivalent networks. Higher-resolution techniques for investigating these events in vitro, along with simulation and mutagenic experiments to specifically modify phase separation while maintaining key networks (or vice versa) may help to resolve these possibilities.
The formation of compartments in the nucleus through phase separation should make it easier for TFs to find their target sites if they are in the desired compartments. Separating highly condensed heterochromatin from accessible euchromatin means that most TFs will not be searching unproductively through heterochromatin, increasing the efficiency of their action. Regardless of the possible involvement of phase separation, multiple TFs binding at enhancers and promoters would synergistically recruit Mediator to those sites via their activation domains – note that Mediator possesses many different subunits that individually bind different TFs.[89,90] The ability of Mediator to interact with multiple different TFs via their activation domains suggests that it could act as a scaffold to drive LLPS. Simultaneously, the formation of condensates with gene-specific TFs could exclude other TFs from taking part in transcriptional activation complexes.
Palacio and Taatjes argue that both condensate and hub models associated with Pol II are likely simultaneously in play during Pol II transcription.[37] As domains of Pol II cycle through different phosphorylated states, phase separation could help to modulate the activity of kinases and phosphatases to enable very rapid changes (Box 1). Phase separation to deplete surrounding regions of Pol II or other essential factors could partly account for transcriptional bursting – the phenomenon of non-stochastic gene expression where genes show short periods of active gene expression followed by variable periods of inactivity (reviewed in ref. [91]). Burst size and duration are proposed to be predominantly limited by the stability of initiation complexes and activators to help regulate the short-term occupancy of Pol II at active gene promoters;[91] stability regulated by post-translational modification or local depletion of substrates within condensates could trigger dissociation.
Conclusions
Transcriptional activation has many layers, but fundamentally, it involves multiple gene-specific TFs binding to promoters and enhancers inside areas of euchromatin that are primed for activity and separated from parts of the genome that are not. Combinatorial binding often includes interactions between TFs to increase binding affinity and specificity at target sites, and many of these factors come with other partners, such as chromatin remodellers, to make conditions even more favourable for transcription. These factors recruit the basal transcription machinery, probably predominantly through interactions with Mediator, which in turn brings Pol II and the basal TFs to promoter sites to initiate transcription and facilitate elongation in small bursts of activity before moving to other genes. All these steps rely on a combination of multiprotein and nucleic acid complexes, which lie on a spectrum of very stable to very dynamic networks of rapidly exchangeable components. The varying stability of these interactions provides an additional layer of precise regulation to gene expression. Hopefully, technological advances will soon resolve some of the current controversies about the underlying mechanisms of small transient multivalent networks, but at this stage, while acknowledging that many of them may be truly phase-separating, it is worth being aware of alternative models and that many aspects of the alternative models, such as multivalency, crowding and concentration dependence, are interrelated.
References
1 Pombo A, Branco MR. Functional organisation of the genome during interphase. Curr Opin Genet Dev 2007; 17(5): 451-5.
| Crossref | Google Scholar |
2 Ing-Simmons E, Rigau M, Vaquerizas JM. Emerging mechanisms and dynamics of three-dimensional genome organisation at zygotic genome activation. Curr Opin Cell Biol 2022; 74: 37-46.
| Crossref | Google Scholar |
3 Jerkovic´ I, Cavalli G. Understanding 3D genome organization by multidisciplinary methods. Nat Rev Mol Cell Biol 2021; 22(8): 511-28.
| Crossref | Google Scholar |
4 Zhou K, Gaullier G, Luger K. Nucleosome structure and dynamics are coming of age. Nat Struct Mol Biol 2019; 26(1): 3-13.
| Crossref | Google Scholar |
5 Slattery M, Zhou T, Yang L, Dantas Machado AC, Gordân R, Rohs R. Absence of a simple code: how transcription factors read the genome. Trends Biochem Sci 2014; 39(9): 381-99.
| Crossref | Google Scholar |
6 Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of Specificity in Protein–DNA Recognition. Annu Rev Biochem 2010; 79(1): 233-69.
| Crossref | Google Scholar |
7 Smith NC, Matthews JM. Mechanisms of DNA-binding specificity and functional gene regulation by transcription factors. Curr Opin Struct Biol 2016; 38: 68-74.
| Crossref | Google Scholar |
8 Qiu J, Bernhofer M, Heinzinger M, Kemper S, Norambuena T, Melo F, et al. ProNA2020 predicts protein–DNA, protein–RNA, and protein–protein binding proteins and residues from sequence. J Mol Biol 2020; 432(7): 2428-43.
| Crossref | Google Scholar |
9 Yuan Q, Chen S, Rao J, Zheng S, Zhao H, Yang Y. AlphaFold2-aware protein–DNA binding site prediction using graph transformer. Brief Bioinform 2022; 23(2): bbab564.
| Crossref | Google Scholar |
10 Baek M, McHugh R, Anishchenko I, Baker D, DiMaio F. Accurate prediction of nucleic acid and protein-nucleic acid complexes using RoseTTAFoldNA. bioRxiv 2022;
| Crossref | Google Scholar |
12 Wunderlich Z, Mirny LA. Different gene regulation strategies revealed by analysis of binding motifs. Trends Genet 2009; 25(10): 434-40.
| Crossref | Google Scholar |
13 Gerland U, Moroz JD, Hwa T. Physical constraints and functional characteristics of transcription factor–DNA interaction. Proc Natl Acad Sci 2002; 99(19): 12015-20.
| Crossref | Google Scholar |
14 Rajewsky N, Vergassola M, Gaul U, Siggia ED. Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo. BMC Bioinformatics 2002; 3: 30.
| Crossref | Google Scholar |
15 Cusanovich DA, Pavlovic B, Pritchard JK, Gilad Y. The functional consequences of variation in transcription factor binding. PLoS Genet 2014; 10(3): e1004226.
| Crossref | Google Scholar |
16 Ballester B, Medina-Rivera A, Schmidt D, Gonzàlez-Porta M, Carlucci M, Chen X, et al. Multi-species, multi-transcription factor binding highlights conserved control of tissue-specific biological pathways. Elife 2014; 3: e02626.
| Crossref | Google Scholar |
17 Thaler JP, Lee S-K, Jurata LW, Gill GN, Pfaff SL. LIM Factor LHX3 Contributes to the Specification of Motor Neuron and Interneuron Identity through Cell-Type-Specific Protein–Protein Interactions. Cell 2002; 110(2): 237-49.
| Crossref | Google Scholar |
18 Bhati M, Lee C, Nancarrow AL, Lee M, Craig VJ, Bach I, et al. Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes. EMBO J 2008; 27(14): 2018-29.
| Crossref | Google Scholar |
19 Robertson NO, Smith NC, Manakas A, Mahjoub M, McDonald G, Kwan AH, et al. Disparate binding kinetics by an intrinsically disordered domain enables temporal regulation of transcriptional complex formation. Proc Natl Acad Sci 2018; 115(18): 4643-8.
| Crossref | Google Scholar |
20 Smith NC, Wilkinson-White LE, Kwan AHY, Trewhella J, Matthews JM. Contrasting DNA-binding behaviour by ISL1 and LHX3 underpins differential gene targeting in neuronal cell specification. J Struct Biol X 2021; 5: 100043.
| Crossref | Google Scholar |
21 Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005; 6(3): 197-208.
| Crossref | Google Scholar |
22 Shammas SL. Mechanistic roles of protein disorder within transcription. Curr Opin Struct Biol 2017; 42: 155-61.
| Crossref | Google Scholar |
23 Smith NC, Kuravsky M, Shammas SL, Matthews JM. Binding and folding in transcriptional complexes. Curr Opin Struct Biol 2021; 66: 156-62.
| Crossref | Google Scholar |
24 Miskei M, Antal C, Fuxreiter M. FuzDB: database of fuzzy complexes, a tool to develop stochastic structure–function relationships for protein complexes and higher-order assemblies. Nucleic Acids Res 2016; 45(D1): gkw1019.
| Crossref | Google Scholar |
25 Strom AR, Brangwynne CP. The liquid nucleome – phase transitions in the nucleus at a glance. J Cell Sci 2019; 132(22): jcs235093.
| Crossref | Google Scholar |
26 Hardenberg M, Horvath A, Ambrus V, Fuxreiter M, Vendruscolo M. Widespread occurrence of the droplet state of proteins in the human proteome. Proc Natl Acad Sci 2020; 117(52): 33254-62.
| Crossref | Google Scholar |
27 Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry 2006; 45(22): 6873-88.
| Crossref | Google Scholar |
28 Wagh K, Garcia DA, Upadhyaya A. Phase separation in transcription factor dynamics and chromatin organization. Curr Opin Struct Biol 2021; 71: 148-55.
| Crossref | Google Scholar |
29 Fuxreiter M, Vendruscolo M. Generic nature of the condensed states of proteins. Nat Cell Biol 2021; 23(6): 587-94.
| Crossref | Google Scholar |
30 Raghunath M, Zeugolis DI. Transforming eukaryotic cell culture with macromolecular crowding. Trends Biochem Sci 2021; 46(10): 805-11.
| Crossref | Google Scholar |
31 André AAM, Spruijt E. Liquid–liquid phase separation in crowded environments. Int J Mol Sci 2020; 21(16): 5908.
| Crossref | Google Scholar |
32 Zhang Y, Lee DSW, Meir Y, Brangwynne CP, Wingreen NS. Mechanical Frustration of Phase Separation in the Cell Nucleus by Chromatin. Phys Rev Lett 2021; 126(25): 258102.
| Crossref | Google Scholar |
33 Alberti S, Gladfelter A, Mittag T. Considerations and Challenges in Studying Liquid–Liquid Phase Separation and Biomolecular Condensates. Cell 2019; 176(3): 419-34.
| Crossref | Google Scholar |
34 McSwiggen DT, Mir M, Darzacq X, Tjian R. Evaluating phase separation in live cells: diagnosis, caveats, and functional consequences. Genes Dev 2019; 33(23–24): 1619-34.
| Crossref | Google Scholar |
35 Mir M, Bickmore W, Furlong EEM, Narlikar G. Chromatin topology, condensates and gene regulation: shifting paradigms or just a phase? Development 2019; 146(19): dev182766.
| Crossref | Google Scholar |
36 Harmon TS, Holehouse AS, Rosen MK, Pappu RV. Intrinsically disordered linkers determine the interplay between phase separation and gelation in multivalent proteins. Elife 2017; 6: e30294.
| Crossref | Google Scholar |
37 Palacio M, Taatjes DJ. Merging Established Mechanisms with New Insights: Condensates, Hubs, and the Regulation of RNA Polymerase II Transcription. J Mol Biol 2022; 434(1): 167216.
| Crossref | Google Scholar |
38 Forman-Kay JD, Ditlev JA, Nosella ML, Lee HO. What are the distinguishing features and size requirements of biomolecular condensates and their implications for RNA-containing condensates? RNA 2022; 28(1): 36-47.
| Crossref | Google Scholar |
39 Martin EW, Holehouse AS, Peran I, Farag M, Incicco JJ, Bremer A, et al. Valence and patterning of aromatic residues determine the phase behavior of prion-like domains. Science 2020; 367(6478): 694-9.
| Crossref | Google Scholar |
40 Espinosa JR, Joseph JA, Sanchez-Burgos I, Garaizar A, Frenkel D, Collepardo-Guevara R. Liquid network connectivity regulates the stability and composition of biomolecular condensates with many components. Proc Natl Acad Sci 2020; 117(24): 13238-47.
| Crossref | Google Scholar |
41 Kantidze OL, Razin SV. Weak interactions in higher-order chromatin organization. Nucleic Acids Res 2020; 48(9): 4614-26.
| Crossref | Google Scholar |
42 Kroschwald S, Maharana S, Simon A. Hexanediol: a chemical probe to investigate the material properties of membrane-less compartments. Matters 2017; 3(5):.
| Crossref | Google Scholar |
43 Ulianov SV, Velichko AK, Magnitov MD, Luzhin AV, Golov AK, Ovsyannikova N, et al. Suppression of liquid–liquid phase separation by 1,6-hexanediol partially compromises the 3D genome organization in living cells. Nucleic Acids Res 2021; 49(18): 10524-41.
| Crossref | Google Scholar |
44 Itoh Y, Iida S, Tamura S, Nagashima R, Shiraki K, Goto T, et al. 1,6-Hexanediol rapidly immobilizes and condenses chromatin in living human cells. Life Sci Alliance 2021; 4(4): e202001005.
| Crossref | Google Scholar |
45 Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell 2009; 137(7): 1194-211.
| Crossref | Google Scholar |
46 Braccioli L, de Wit E. CTCF: a Swiss-army knife for genome organization and transcription regulation. Essays Biochem 2019; 63(1): 157-65.
| Crossref | Google Scholar |
47 Stam M, Tark-Dame M, Fransz P. 3D genome organization: a role for phase separation and loop extrusion? Curr Opin Plant Biol 2019; 48: 36-46.
| Crossref | Google Scholar |
48 Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 2017; 6: e25776.
| Crossref | Google Scholar |
49 Gu B, Comerci CJ, McCarthy DG, Saurabh S, Moerner WE, Wysocka J. Opposing Effects of Cohesin and Transcription on CTCF Organization Revealed by Super-resolution Imaging. Mol Cell 2020; 80(4): 699-711.e7.
| Crossref | Google Scholar |
50 Lee R, Kang MK, Kim YJ, Yang B, Shim H, Kim S, et al. CTCF-mediated chromatin looping provides a topological framework for the formation of phase-separated transcriptional condensates. Nucleic Acids Res 2022; 50(1): 207-26.
| Crossref | Google Scholar |
51 Wei C, Jia L, Huang X, Tan J, Wang M, Niu J, et al. CTCF organizes inter-A compartment interactions through RYBP-dependent phase separation. Cell Res 2022; 32(8): 744-60.
| Crossref | Google Scholar |
52 Sentenac A. Eukaryotic RNA polymerase. CRC Crit Rev Biochem 1985; 18(1): 31-90.
| Crossref | Google Scholar |
53 Harlen KM, Churchman LS. The code and beyond: transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat Rev Mol Cell Biol 2017; 18(4): 263-73.
| Crossref | Google Scholar |
54 Boehning M, Dugast-Darzacq C, Rankovic M, Hansen AS, Yu T, Marie-Nelly H, et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat Struct Mol Biol 2018; 25(9): 833-40.
| Crossref | Google Scholar |
55 Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, et al. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 2013; 155(5): 1049-60.
| Crossref | Google Scholar |
56 Ladouceur A-M, Parmar BS, Biedzinski S, Wall J, Tope SG, Cohn D, et al. Clusters of bacterial RNA polymerase are biomolecular condensates that assemble through liquid–liquid phase separation. Proc Natl Acad Sci 2020; 117(31): 18540-9.
| Crossref | Google Scholar |
57 Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, et al. Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 2018; 558(7709): 318-23.
| Crossref | Google Scholar |
58 Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 2015; 16(3): 155-66.
| Crossref | Google Scholar |
59 Richter WF, Nayak S, Iwasa J, Taatjes DJ. The Mediator complex as a master regulator of transcription by RNA polymerase II. Nat Rev Mol Cell Biol 2022; 23: 732-49.
| Crossref | Google Scholar |
60 Boija A, Klein IA, Sabari BR, Dall’Agnese A, Coffey EL, Zamudio AV, et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 2018; 175(7): 1842-55.e16.
| Crossref | Google Scholar |
61 Tuttle LM, Pacheco D, Warfield L, Wilburn DB, Hahn S, Klevit RE. Mediator subunit Med15 dictates the conserved ‘fuzzy’ binding mechanism of yeast transcription activators Gal4 and Gcn4. Nat Commun 2021; 12(1): 2220.
| Crossref | Google Scholar |
62 Guo YE, Manteiga JC, Henninger JE, Sabari BR, Dall’Agnese A, Hannett NM, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature 2019; 572(7770): 543-8.
| Crossref | Google Scholar |
63 Cho W-K, Spille J-H, Hecht M, Lee C, Li C, Grube V, et al. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018; 361(6400): 412-5.
| Crossref | Google Scholar |
64 Sur I, Taipale J. The role of enhancers in cancer. Nat Rev Cancer 2016; 16(8): 483-93.
| Crossref | Google Scholar |
65 Sengupta S, George RE. Super-enhancer-driven transcriptional dependencies in cancer. Trends Cancer 2017; 3(4): 269-81.
| Crossref | Google Scholar |
66 Rippe K. Liquid–liquid phase separation in chromatin. Cold Spring Harb Perspect Biol 2022; 14(2): a040683.
| Crossref | Google Scholar |
67 Hnisz D, Day DS, Young RA. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 2016; 167(5): 1188-200.
| Crossref | Google Scholar |
68 Wei Y, Zhang S, Shang S, Zhang B, Li S, Wang X, et al. SEA: a super-enhancer archive. Nucleic Acids Res 2016; 44(D1): D172-9.
| Crossref | Google Scholar |
69 Jiang Y, Qian F, Bai X, Liu Y, Wang Q, Ai B, et al. SEdb: a comprehensive human super-enhancer database. Nucleic Acids Res 2019; 47(D1): D235-43.
| Crossref | Google Scholar |
70 Khan A, Zhang X. dbSUPER: a database of super-enhancers in mouse and human genome. Nucleic Acids Res 2016; 44(D1): D164-71.
| Crossref | Google Scholar |
71 Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018; 361(6400): eaar3958.
| Crossref | Google Scholar |
72 Cheung KL, Kim C, Zhou MM. The Functions of BET Proteins in Gene Transcription of Biology and Diseases. Front Mol Biosci 2021; 8: 728777.
| Crossref | Google Scholar |
73 Wu S-Y, Chiang C-M. The Double Bromodomain-containing Chromatin Adaptor Brd4 and Transcriptional Regulation. J Biol Chem 2007; 282(18): 13141-5.
| Crossref | Google Scholar |
74 Bhagwat AS, Roe JS, Mok BYL, Hohmann AF, Shi J, Vakoc CR. BET Bromodomain Inhibition Releases the Mediator Complex from Select cis-Regulatory Elements. Cell Rep 2016; 15(3): 519-30.
| Crossref | Google Scholar |
75 Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol Cancer 2018; 17(1): 164.
| Crossref | Google Scholar |
76 Han X, Yu D, Gu R, Jia Y, Wang Q, Jaganathan A, et al. Roles of the BRD4 short isoform in phase separation and active gene transcription. Nat Struct Mol Biol 2020; 27(4): 333-41.
| Crossref | Google Scholar |
77 Wang C, Zhang E, Wu F, Sun Y, Wu Y, Tao B, et al. The C-terminal low-complexity domain involved in liquid–liquid phase separation is required for BRD4 function in vivo. J Mol Cell Biol 2019; 11(9): 807-9.
| Crossref | Google Scholar |
78 Wang C, Lu H, Liu X, Gao X, Tian W, Chen H, et al. A natural product targets BRD4 to inhibit phase separation and gene transcription. iScience 2022; 25(1): 103719.
| Crossref | Google Scholar |
79 Crump NT, Ballabio E, Godfrey L, Thorne R, Repapi E, Kerry J, et al. BET inhibition disrupts transcription but retains enhancer–promoter contact. Nat Commun 2021; 12(1): 223.
| Crossref | Google Scholar |
80 Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013; 153(2): 320-34.
| Crossref | Google Scholar |
81 Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. NURD, a Novel Complex with Both ATP-Dependent Chromatin-Remodeling and Histone Deacetylase Activities. Mol Cell 1998; 2(6): 851-61.
| Crossref | Google Scholar |
82 Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 2005; 19(4): 523-34.
| Crossref | Google Scholar |
83 Liu B, Liu X, Han L, Chen X, Wu X, Wu J, et al. BRD4-directed super-enhancer organization of transcription repression programs links to chemotherapeutic efficacy in breast cancer. Proc Natl Acad Sci 2022; 119(6): e2109133119.
| Crossref | Google Scholar |
84 Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, et al. RNA Controls PolyQ Protein Phase Transitions. Mol Cell 2015; 60(2): 220-30.
| Crossref | Google Scholar |
85 Zhou H, Song Z, Zhong S, Zuo L, Qi Z, Qu L-J, et al. Mechanism of DNA-Induced Phase Separation for Transcriptional Repressor VRN1. Angew Chem Int Ed Engl 2019; 58(15): 4858-62.
| Crossref | Google Scholar |
86 Shakya A, King JT. DNA Local-Flexibility-Dependent Assembly of Phase-Separated Liquid Droplets. Biophys J 2018; 115(10): 1840-7.
| Crossref | Google Scholar |
87 Sato Y, Sakamoto T, Takinoue M. Sequence-based engineering of dynamic functions of micrometer-sized DNA droplets. Sci Adv 2020; 6(23): eaba3471.
| Crossref | Google Scholar |
88 Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell 2017; 169(1): 13-23.
| Crossref | Google Scholar |
89 Quevedo M, Meert L, Dekker MR, Dekkers DHW, Brandsma JH, van den Berg DLC, et al. Mediator complex interaction partners organize the transcriptional network that defines neural stem cells. Nat Commun 2019; 10(1): 2669.
| Crossref | Google Scholar |
90 Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153(2): 307-19.
| Crossref | Google Scholar |
91 Rodriguez J, Larson DR. Transcription in living cells: molecular mechanisms of bursting. Annu Rev Biochem 2020; 89(1): 189-212.
| Crossref | Google Scholar |
92 Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Science 2017; 357(6357): eaaf4382.
| Crossref | Google Scholar |
93 Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 2017; 18(5): 285-98.
| Crossref | Google Scholar |
94 O’Flynn BG, Mittag T. The role of liquid–liquid phase separation in regulating enzyme activity. Curr Opin Cell Biol 2021; 69: 70-9.
| Crossref | Google Scholar |
95 Harada R, Sugita Y, Feig M. Protein crowding affects hydration structure and dynamics. J Am Chem Soc 2012; 134(10): 4842-9.
| Crossref | Google Scholar |
96 Kim YC, Best RB, Mittal J. Macromolecular crowding effects on protein–protein binding affinity and specificity. J Chem Phys 2010; 133(20): 11B608.
| Crossref | Google Scholar |
97 Raumann BE, Rould MA, Pabo CO, Sauer RT. DNA recognition by β-sheets in the Arc represser–operator crystal structure. Nature 1994; 367(6465): 754-7.
| Crossref | Google Scholar |
98 Wilkinson-White L, Lester KL, Ripin N, Jacques DA, Mitchell Guss J, Matthews JM. GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA. Protein Sci 2015; 24(10): 1649-59.
| Crossref | Google Scholar |


