Fluorescence Quenching of Quantum Dots by DNA Nucleotides and Amino Acids1
Daniel Siegberg A and Dirk-Peter Herten A BA Cellnetworks Cluster and Institute for Physical Chemistry, Heidelberg University, Im Neuenheimer Feld 267, D-69120, Heidelberg, Germany.
B Corresponding author. Email: dirk-peter.herten@urz.uni-hd.de
Australian Journal of Chemistry 64(5) 512-516 https://doi.org/10.1071/CH10293
Submitted: 7 August 2010 Accepted: 31 October 2010 Published: 30 May 2011
Abstract
Quantum dots found widespread application in the biosciences as bright and highly photo-stable fluorescent probes, i.e. for single-particle tracking. In this work we used ensemble spectroscopy and single-molecule techniques to study the quenching of quantum dots by various biochemical compounds that are usually present in living cells and might thus influence the experiments. We found not only nucleotides such as cytosine, guanine, and thymine can significantly influence the fluorescence emission of CdSe and CdTe quantum dots, but also amino acids, like asparagine and tryptophan. Bulk studies on fluorescence quenching indicated a static quenching mechanism. Interestingly, we could also show by single-molecule fluorescence spectroscopy that quenching of the quantum dots can be irreversible, suggesting either a redox-reaction between quantum dot and quencher or strong binding of the quencher to the surface of the bio-conjugated quantum dots.
Introduction
The development of novel labels and labelling strategies is driven by growing interest for in vivo imaging and life-cell microscopy to investigate complex biological networks in living cells and organisms. Recent advances in the synthesis and coating of quantum dots (QD) enhanced and broadened their applicability for in vivo experiments; which is of great interest because of their unique spectroscopic features.[1–4] Fluorescent QD usually have high absorbance, increasing with shorter wavelengths. At the same time they have a narrow emission band, with the emission maximum depending on constitution and size of the QD. Both features together make them highly suitable for multiplexing by excitation with a single laser wavelength. Being made of semi-conductor material, the QD are at the same time extremely photo-stable. It is therefore not astonishing that QD have attracted researchers from many disciplines of sciences since their discovery in the 1970s.[1–3,5,6] However, known photo-physical features, such as ‘blinking’ and ‘blueing’, sometimes render interpretation of the corresponding experiment difficult.[7–9] Furthermore, their application and use in cell biology demands not only detailed knowledge about toxicity but also about potential interactions with various biochemical compounds that might influence fluorescence emission. Whereas such influences on organic dyes have been described in literature,[10] to our knowledge, systematic investigations of the influence of nucleotides and amino acids on the fluorescence emission of QD are yet to be studied. In current literature, quenching of QD by metal ions, such as Cu2+ and Ag+, and by organic molecules containing amino-groups have been reported.[11–13] Influence of various compounds on the fluorescence emission properties of QD raised a strong interest in their use as fluorescent sensors, like the quenching by metal ions,[12,14,15] gold nanoparticles,[16,17] and various other compounds.[18–23] Further studies focussed on the oxidation of QD,[24] or photo-induced charge transfer,[11] and also on the influence of experimental conditions, such as temperature or pH.[7,25,26]
Herein we present a systematic study on the interaction of nucleotides and amino acids with QD, which occasionally leads to a significant quenching of fluorescence intensity. For our studies we used two different commercially available core shell QD, QD655 and QD705, with a ZnS shell and a CdSe and CdTe core respectively. QD were purchased bio-functionalized with streptavidin for labelling of biotinylated compounds. Fluorescence quenching of QD has been investigated by means of steady-state as well as fluorescence lifetime spectroscopy as dependent on quencher concentration. Additionally, single-molecule fluorescence spectroscopy has been used to study reversibility of quenching.
Results and Discussion
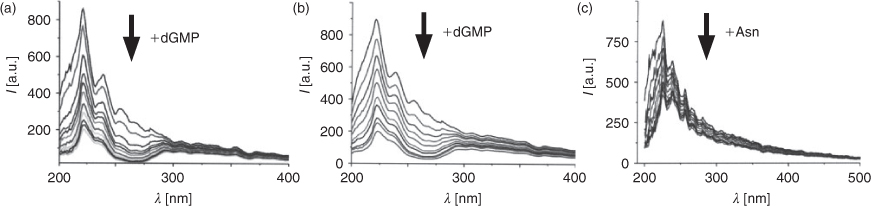
Among the numerous biochemical compounds present in living cells, nucleotides are the most prominent, located anywhere within the cell. Therefore, we were interested in examining the interaction between QD and nucleotides; as measured by the change in the fluorescence intensity upon addition of the nucleotide. With increasing amounts of dGMP to a solution of QD655, we observed a strong attenuation of the fluorescence emission (Fig. 1a). Concurrently, the emission maximum and shape of the emission spectra of QD655 remained constant. Attenuation of the fluorescence emission spectra at different concentrations of dGMP was used to create Stern-Volmer (SV) plots, shown in Figs 1b (QD655) and 1c (QD705), to quantify quenching. According to Eqn 1 (cf. Experimental Methods below) the initial fluorescence intensity (F0) in absence of nucleotide was divided by the fluorescence intensity (F) of the nucleotide containing sample and plotted against the respective nucleotide concentration. An initial visual inspection of the SV-plots, immediately reveals interesting differences in the quenching of the two different types of QD. QD655 is strongly quenched by dGMP, and to a lesser degree by dTMP and dCMP. In contrast, QD705 shows greatest sensitivity to the addition of dTMP, with the other three nucleotides affecting its emission weakly.

|
The initial linear portion of the SV-plots was used for fitting a linear model (Eqn 1) according to Stern–Volmer theory of collisional quenching (dotted lines in Fig. 1b and 1c).[27] The regression factor of all fits was of 0.99 or better. The results summarized in Table 1 show clearly that fluorescence emission of both QD is attenuated upon addition of any of the four nucleotides. Additionally, differences in the behaviour of the two types of QD can easily be identified. Quenching of QD655 by the three nucleotides (dCMP, dGMP, and dTMP) is approximately 2–3 times stronger, as compared with dAMP. QD705, however, is most strongly quenched by dTMP, with the corresponding fluorescence quenching by dAMP, dCMP, and GMP approximately a factor of 3–4 times weaker. It is noteworthy that in all cases we found deviations from linearity at higher concentration in the SV-plots (data not shown); showing mostly a trend to saturation, which could be interpreted as direct surface interaction of the quencher with the respective QD.

|
These observations first focussed our interest towards possible interactions with other biochemical compounds. To this end, similar experiments were carried out with both QD655 and QD705 and various amino acids summarized in Table 2. Interestingly, the strongest quenching of QD705 does not occur, as we had expected, with any of the aromatic amino acids, but with asparagine. The aromatic amino acids also show quenching of the fluorescence of QD705, but this is only about half of that observed for asparagine. While most of the amino acids do not show any effect on the fluorescence emission of QD705, some amino acids quench the fluorescence weakly.

|
We also became interested in whether, or to what extent, quenching by these compounds is reversible. We subsequently conducted experiments with QD655 immobilized on glass cover slides. Prior to immobilization, the glass surface was covered with bovine serum albumin (BSA) and then doped with biotinylated BSA to attach streptavidin coated QD. The non-covalent biotin/streptavidin binding allows removal of quencher containing supernatant and subsequent washing of the immobilized QD. Immobilized QD were then imaged with a confocal microscope capable of spectrally-resolved fluorescence lifetime imaging microscopy, by excitation with at 635 nm.[28] From the resulting fluorescence intensity image (Fig. 2a), a density of about less than one QD per µm2 was estimated. After the first image had been taken, concentration of dGMP was gradually increased by adding increasing amounts from a 200 µM stock solution of dGMP. After each addition of dGMP another image was taken by scanning the sample to follow attenuation of the fluorescence emission of the QD (Figs 2b–2e). The image series shows the strong influence of dGMP on the fluorescence emission of the QD, which are barely visible at a concentration of ∼6 mM of dGMP (Fig. 2e). After removal of supernatant the sample was washed three times using PBS-buffered aqueous solution. However, the final image (Fig. 2f), taken after washing, shows barely any recovery of the fluorescence emission.

|
We also measured the influence of nucleotides and amino acids on the fluorescence lifetimes of the QD; as previous experiments indicated an irreversible quenching mechanism for dGMP. However, the changes in fluorescence lifetime were rather small, ruling out any dynamic quenching mechanism, such as collisional quenching (Table 1). Additionally, we found that quenching of QD655 (Fig. 3a) and QD705 (Fig. 3b) with dGMP depends on the excitation wavelength. In both cases quenching is most prominent in a band around 270 nm. In contrast, quenching of QD705 with asparagine leads to a more equally distributed quenching over the whole excitation range (Fig. 3c). Therefore, the observed static fluorescence quenching by dGMP might also be explained by a redox-reaction of the QD with dGMP which is further supported by absorption measurements showing a comparable behaviour (data not shown).
Discussion
We systematically studied quenching of fluorescence emission of semi-conductor QD, QD655 and QD705, by different nucleotides and amino acids. In our studies, we found that quenching was in some cases very specific, i.e. QD655 was strongly quenched by dGMP, while QD705 was strongly quenched by dTMP. We also found, that of the amino acids, asparagine quenches QD705 to the greatest degree. These finding might be of interest for future developments of analytical assays.
For all quenchers we observed saturation of quenching. This indicates that quenching is limited by the number of quencher molecules that can interact with the QD. This notion is supported by recent reports of aliphatic amines and mercaptanes effecting the fluorescence emission of CdSe cores.[13,18,22] Although the QD are shielded not only by a ZnS shell, but also by polymer coating and biofunctionalization, one might imagine small molecules, like dNTP, penetrating the streptavidin and eventually also the polymer coating. Recently, non-uniformity of ZnS shells was reported, suggesting partially thin ZnS layers could even contain pores that could form potential binding sites for quenchers.[29] At the moment we can only speculate about the mechanism of quenching. Interestingly, there are some reports in the literature where similar compounds have altered the emission of QD. Amines, for example, have been found to quench fluorescence in some cases, suggesting interactions with surface holes.[13] However, many other publications report fluorescence enhancement, instead suggesting a completely different kind of interaction.[30] This contradiction might be solved by the observation that properties of QD depend not only on the material but also on the particle synthesis and surface coating.[31] Indeed, we also found variations between batches of QD of the same type. Nevertheless, the idea of direct binding of the quencher to the QD surface supports our finding that quenching is mostly static, i.e. either formation of non-fluorescent complexes or due to chemical reaction of the QD. The latter is supported by the observation that dGMP specifically quenches an excitation band of QD655 and QD705 around 270 nm. In contrast, quenching by asparagine does not directly impact specific features in the excitation spectra, pointing more towards formation of a non-fluorescent complex.
Conclusions
In summary, emission of QD is quenched by various nucleotides and amino acids. Our data indicates that quenching mechanisms depends on the particular compound; with quenching found to be specific for certain compounds which may influence the future development of analytical assays. Furthermore, the provided information regarding the quenching of QD may be useful to explain and avoid side effects when working in with biological samples or in living cells containing nucleotides and/or amino acids. It is well known that the properties of QD depend not only on the material itself, but may vary considerably on particle synthesis and surface preparation. Therefore, the described observations should not be generalized but instead considered and recognised as potential side-effects when QD are used for assay development in biological samples.
Experimental
Materials and Reagents
The semi-conductor QD, QD655 and QD705, were obtained from Quantum Dot Corporation (Hayward, USA; now Invitrogen). QD655 has a CdSe core and emits at 655 nm, while the emission of QD705 is red-shifted at 705 nm due to the CdTe core. Both QD were obtained as coated core/shell QD with a shell of ZnS that was coated with a polymer which was already biofunctionalized with streptavidin by the vendor. Suspensions of the QD were used as delivered without any further modification and diluted 1:800 in phosphate buffered saline (PBS, 0.1 mM, pH 7.3).
Spectroscopy
All experiments were conducted in standard quartz cuvettes (Hellma, Müllheim, Germany) with a maximum volume of 1.5 mL. For UV/Vis and fluorescence spectroscopy the concentration of QD was chosen well below an optical density of 0.1, to avoid inner filter effects. Optical density at excitation wavelength was verified with a Cary Scan 500 UV/Vis Photo spectrometer (Varian, Darmstadt, Germany). Steady-state fluorescence spectra were measured on a Cary Eclipse fluorescence spectrometer (Varian, Darmstadt, Germany). Ensemble fluorescence lifetime measurements were performed on a FluoTime 100 (PicoQuant, Berlin, Germany) using time-correlated single-photon counting (TCSPC). For excitation, we used a pulsed LED emitting at 370 nm with a pulse width <600 ps (fwhm) operating at 1 MHz. The measurements were completed in standard quartz cuvettes (d = 0.3 cm). To exclude polarization effects, fluorescence was observed under magic angle conditions (54.7°). Typically 3000–5000 photons were collected in the maximum channel of a total of 4096 channels. The lifetime was determined with the software provided by the manufacturer (FluoFit Ver. 4.1, PicoQuant, Berlin, Germany) by least-squares deconvolution, using the instrument response function acquired with LUDOX and their quality judged by the reduced χ2 values and the randomness of the weighted residuals.
Quenching Experiments
For quenching experiments, solutions of the four monophosphate nucleotides dAMP, dCMP, dGMP, and dTMP, as well as the amino acids tryptophane, threonine, histidine, methionine, proline, asparagine, leucine, glutamine, arginine, serine, and glycine, were used. All nucleotides and amino acids were purchased from Sigma–Aldrich (Stockelsdorf, Germany) in pro analysi quality.
Quenching of the QD by the different bio-compounds was investigated at 25°C by adding increasing amounts of bio-compound stock solution (1 mM, PBS buffered, 0.1 mM, pH 7.3) to QD containing solution (PBS buffered, 0.1 mM, pH 7.3) and measuring the fluorescence emission spectrum/fluorescence lifetime upon excitation at 400/370 nm, if not stated otherwise. Addition of the respective bio-compound was continued until quenching reached saturation, which usually occurred at ∼20–50 mM of the bio-compound depending on the particular bio-compound and the QD, respectively. Measured fluorescence intensities were corrected for dilution of QD upon addition of quencher. The ratio of initial fluorescence intensity I0/lifetime τ0 to fluorescence intensity I/lifetime τ at a given quencher concentration was plotted against quencher concentration c(Q) in a SV plot. The quenching or Stern–Volmer constant KSV was then determined by fitting a straight line to the initial slope (usually 5–10 data points) of the SV plot according to Eqn 1:[27]
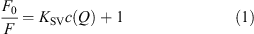
Microscopy
The experimental setup and principle of fluorescence lifetime imaging microscopy have been described elsewhere. Briefly a pulsed laser diode emitting at 635 nm (PicoQuant GmbH, Berlin, Germany) was driven by a pulsed laser driver (PDL 800B PicoQuant) at a repetition rate of 5 MHz and was directed into an inverted microscope (Axiovert 100TV, Zeiss, Jena, Germany) equipped with a xyz-piezoscanning table (PI Physik Instrumente, Karlsruhe, Germany). The collimated laser beam was directed into an oil immersion objective (100×, NA = 1.4, Olympus, Japan). Fluorescence emission was collected by the same microscope lens and was detected by focusing on two avalanche photo diodes (SPCM-AQR 15, Perkin–Elmer). For data recording and analysis a TCSPC PC-card (SPC-630 Becker&Hickl, Berlin, Germany) and custom written LabView-based software were used.
Acknowledgements
We gratefully acknowledge the Landesstiftung Baden-Württemberg (Promotionskolleg Bioquant) and the Deutsche Forschungsgemeinschaft (DFG, EXC 81) for financial support.
References
[1] A. P. Alivisatos, W. W. Gu, C. Larabell, Annu. Rev. Biomed. Eng. 2005, 7, 55.| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXps12lt7c%3D&md5=dacdb3241e3e3b9e1ccd06a64df10bbaCAS | 16004566PubMed |
[2] X. H. Gao, L. L. Yang, J. A. Petros, F. F. Marshal, J. W. Simons, S. M. Nie, Curr. Opin. Biotechnol. 2005, 16, 63.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhsFShsL4%3D&md5=7ed753a456099920b2ffe9f6d8d3186eCAS | 15722017PubMed |
[3] I. L. Medintz, H. T. Uyeda, E. R. Goldman, H. Mattoussi, Nat. Mater. 2005, 4, 435.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXks1Cit7k%3D&md5=3a55eb443c0a989d0049862257c15ad6CAS | 15928695PubMed |
[4] X. Michalet, F. F. Pinaud, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Sundaresan, A. M. Wu, S. S. Gambhir, S. Weiss, Science 2005, 307, 538.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmslOhtw%3D%3D&md5=be5fc27e060de79d88469c92ddadd0e3CAS | 15681376PubMed |
[5] E. A. Jares-Erijman, T. M. Jovin, Nat. Biotechnol. 2003, 21, 1387.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXosFOksbo%3D&md5=17287a750da7b858899e3c429f68ef52CAS | 14595367PubMed |
[6] P. Mulvaney, L. M. Liz-Marzan, M. Giersig, T. Ung, J. Mater. Chem. 2000, 10, 1259.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXjtlyjtrg%3D&md5=a75746448834906f9226427505e9dcf2CAS |
[7] J. A. Kloepfer, S. E. Bradforth, J. L. Nadeau, J. Phys. Chem. B 2005, 109, 9996.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXjsVGgt78%3D&md5=1700ecf84a738186c4765b163a121f08CAS | 16852208PubMed |
[8] W. G. J. H. M. van Sark, P. L. T. M. Frederix, A. A. Bol, H. C. Gerritsen, A. Meijerink, ChemPhysChem 2002, 3, 871.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD38XosVaksLw%3D&md5=1a5f16c246a7ed82a4308c16f28d7a8fCAS |
[9] M. Kuno, D. P. Fromm, H. F. Hamann, A. Gallagher, D. J. Nesbitt, J. Chem. Phys. 2000, 112, 3117.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3cXptFWlsA%3D%3D&md5=b63a38ef24836a163c687780b1977f03CAS |
[10] S. Doose, H. Neuweiler, M. Sauer, ChemPhysChem 2009, 10, 1389.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1MXptVGhu7s%3D&md5=a619fda1453c07aeb07827057e421246CAS | 19475638PubMed |
[11] S. N. Sharma, Z. S. Pillai, P. V. Kamat, J. Phys. Chem. B 2003, 107, 10088.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXmsFamtrg%3D&md5=0b0a42b34dd1c106d3c8e16d30eace51CAS |
[12] K. A. Gattas-Asfura, R. M. Leblanc, Chem. Commun. 2003, 2684.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3sXot1ersr4%3D&md5=4d2d94f169afda4299bdeeee860fa2e5CAS |
[13] C. Landes, C. Burda, M. Braun, M. A. El-Sayed, J. Phys. Chem. B 2001, 105, 2981.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXhvFGru70%3D&md5=e47041c8c1ef1e713fc1eb594e612362CAS |
[14] C. Q. Dong, H. F. Qian, N. H. Fang, J. C. Ren, J. Phys. Chem. B 2006, 110, 11069.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XkvVWhsLg%3D&md5=9433e5b223b738bca5fce7a87134e6a7CAS | 16771367PubMed |
[15] J. G. Liang, X. P. Ai, Z. K. He, D. W. Pang, Analyst 2004, 129, 619.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2cXltVahsbs%3D&md5=f83581ab155fe2910bf3c9410641f891CAS | 15213829PubMed |
[16] Z. Gueroui, A. Libchaber, Phys. Rev. Lett. 2004, 93, 166108.
| Crossref | GoogleScholarGoogle Scholar | 15525013PubMed |
[17] L. Dyadyusha, H. Yin, S. Jaiswal, T. Brown, J. J. Baumberg, F. P. Booy, T. Melvin, Chem. Commun. 2005, 3201.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXltlKhsb4%3D&md5=77bc415160fd3dae970572a6f79c95f8CAS |
[18] S. Hohng, T. Ha, ChemPhysChem 2005, 6, 956.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXksFajsrY%3D&md5=2ef4b99ebfbb96656ece3856e30a3c12CAS | 15884082PubMed |
[19] J. A. Kloepfer, R. E. Mielke, J. L. Nadeau, Appl. Environ. Microbiol. 2005, 71, 2548.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXktFKmurg%3D&md5=8c5a13b96f2b49ff93f9f635a6e1d7edCAS | 15870345PubMed |
[20] A. Priyam, A. Chatterjee, S. K. Das, A. Saha, Chem. Commun. 2005, 4122.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXntlejsLo%3D&md5=7ea5763c2834428afb35f6e5de361365CAS |
[21] D. Selmarten, M. Jones, G. Rumbles, P. Yu, J. Nedeljkovic, S. Shaheen, J. Phys. Chem. B 2005, 109, 15927.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXmvFSrsLY%3D&md5=56ea0dbfef38e484618c55378ccd5aa2CAS | 16853021PubMed |
[22] C. Bullen, P. Mulvaney, Langmuir 2006, 22, 3007.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xhs1Cisr8%3D&md5=9a15e49934617a505e42d8a5f777079dCAS | 16548550PubMed |
[23] M. G. Sandros, V. Shete, D. E. Benson, Analyst 2006, 131, 229.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xot1WhtQ%3D%3D&md5=e07255b24eef3580fc4af56e79d66692CAS | 16440087PubMed |
[24] Y. Zhang, J. He, P. N. Wang, J. Y. Chen, Z. J. Lu, D. R. Lu, J. Guo, C. C. Wang, W. L. Yang, J. Am. Chem. Soc. 2006, 128, 13396.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28XpvVChsLo%3D&md5=45362693cca5450302e9a961e799f613CAS | 17031951PubMed |
[25] V. Biju, Y. Makita, A. Sonoda, H. Yokoyama, Y. Baba, M. Ishikawa, J. Phys. Chem. B 2005, 109, 13899.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXltFCgu7k%3D&md5=fefaaaf292c5427f3c6f4a6d33cebd9eCAS | 16852744PubMed |
[26] M. Califano, A. Franceschetti, A. Zunger, Nano Lett. 2005, 5, 2360.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXhtFKhtbbJ&md5=a1fcf51ec281badd52e7bccd78e0340dCAS | 16351178PubMed |
[27] J. R. Lakowicz, Principles of Fluorescence Spectroscopy, 3rd edn 2006 (Springer: Berlin).
[28] P. Tinnefeld, D. P. Herten, M. Sauer, J. Phys. Chem. A 2001, 105, 7989.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD3MXlsFygur0%3D&md5=682a89a557e987b708b8046729a5595fCAS |
[29] Z. Yu, L. Guo, H. Du, T. Krauss, J. Silcox, Nano Lett. 2005, 5, 565.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD2MXitVCktb8%3D&md5=f54e3a64d7014f516a26e98747f9804dCAS | 15826088PubMed |
[30] C. Bullen, P. Mulvany, Langmuir 2006, 22, 3007.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD28Xhs1Cisr8%3D&md5=9a15e49934617a505e42d8a5f777079dCAS | 16548550PubMed |
[31] U. Resch-Genger, M. Grabolle, S. Cavaliere-Jaricot, R. Nitschke, T. Nann, Nat. Methods 2008, 5, 763.
| Crossref | GoogleScholarGoogle Scholar | 1:CAS:528:DC%2BD1cXhtVGgsbrM&md5=dcfa76003c77ac090b7154c8b1046cebCAS | 18756197PubMed |
1 This Paper is published alongside a Review by Sauer and coworkers as part of a proposed Research Front on Single Molecule Spectroscopy which did not reach fruition.