An autoantigen-ome from HS-Sultan B-Lymphoblasts offers a molecular map for investigating autoimmune sequelae of COVID-19
Julia Y. Wang A * , Wei Zhang B , Victor B. Roehrl A , Michael W. Roehrl A and Michael H. Roehrl
A * , Wei Zhang B , Victor B. Roehrl A , Michael W. Roehrl A and Michael H. Roehrl  C D *
C D *
A Curandis, Boston, USA.
B Department of Gastroenterology, Affiliated Hospital of Guizhou Medical University, Guizhou, China.
C Department of Pathology, Beth Israel Deaconess Medical Center, Boston, USA.
D Harvard Medical School, Boston, USA.
Handling Editor: Mibel Aguilar
Australian Journal of Chemistry 76(8) 525-557 https://doi.org/10.1071/CH22267
Submitted: 20 December 2022 Accepted: 8 June 2023 Published: 19 July 2023
Abstract
To understand how COVID-19 may induce autoimmune diseases, we have been compiling an atlas of COVID autoantigens (autoAgs). Using dermatan sulfate (DS) affinity enrichment of autoantigenic proteins extracted from HS-Sultan lymphoblasts, we identified 362 DS-affinity proteins, of which at least 201 (56%) are confirmed autoAgs. Comparison with available multi-omic COVID data shows that 315 (87%) of the 362 proteins are affected in SARS-CoV-2 infection via altered expression, interaction with viral components, or modification by phosphorylation or ubiquitination, at least 186 (59%) of which are known autoAgs. These proteins are associated with gene expression, mRNA processing, mRNA splicing, translation, protein folding, vesicles, and chromosome organization. Numerous nuclear autoAgs were identified, including both classical antinuclear antibodies (ANAs) and extractable nuclear antigens (ENAs) of systemic autoimmune diseases and unique autoAgs involved in the DNA replication fork, mitotic cell cycle, or telomerase maintenance. We also identified many uncommon autoAgs involved in nucleic acid and peptide biosynthesis and nucleocytoplasmic transport, such as aminoacyl-tRNA synthetases. In addition, this study found autoAgs that potentially interact with multiple SARS-CoV-2 Nsp and Orf components, including CCT/TriC chaperonin, insulin degrading enzyme, platelet-activating factor acetylhydrolase, and the ezrin-moesin-radixin family. Furthermore, B-cell-specific IgM-associated endoplasmic reticulum (ER) complex (including MBZ1, BiP, heat shock proteins, and protein disulfide-isomerases) is enriched by DS-affinity and up-regulated in B-cells of COVID-19 patients, and a similar IgH-associated ER complex was also identified in autoreactive pre-B1 cells in our previous study, which suggests a role of autoreactive B1 cells in COVID-19 that merits further investigation. In summary, this study demonstrates that virally infected cells are characterized by alterations of proteins with propensity to become autoAgs, thereby providing a possible explanation for infection-induced autoimmunity. The COVID autoantigen-ome provides a valuable molecular resource and map for investigation of COVID-related autoimmune sequelae and considerations for vaccine design.
Keywords: autoantibodies, autoantigens, autoimmunity, COVID, dermatan sulfate, Epstein-Barr virus, long COVID, SARS-CoV-2.
Introduction
The novel coronavirus SARS-CoV-2 has caused the worldwide COVID-19 pandemic with hundreds of millions infected and high morbidity and mortality. A significant proportion of patients who have recovered from the acute viral infection of COVID-19 continue to suffer from lingering health problems, so called ‘long COVID’ syndrome. It is yet unknown how long the COVID aftereffects will persist, and more importantly, what the underlying causative mechanisms of long COVID syndrome are. The acute phase of COVID-19 is accompanied by various autoimmune responses, and autoimmune diseases, which tend to be chronic and debilitating, are major concerns of COVID-19 sequelae. To understand how SARS-CoV-2 infection may induce autoimmunity and how diverse the autoimmune disorders could be, we have started to compile a comprehensive atlas of COVID autoantigens (autoAgs) and autoantibodies (autoAbs), the root elements of autoimmunity.[1,2]
We have developed a unique DS-affinity enrichment strategy for autoAg discovery.[1–8] We discovered that dermatan sulfate (DS), a glycosaminoglycan that is abundant in skin and soft connective tissues and that is involved in wound healing and tissue repair, has affinity to autoAgs.[3,4] Because of this affinity, DS binds autoAgs to form non-covalent DS–autoAg complexes, which transform non-antigenic singular self-molecules into antigenic non-self-complexes.[3] DS–autoAg complexes are capable of engaging autoreactive B-cell receptors (autoBCRs) via a two-step process: (i) DS–autoAg complexes bind autoBCRs on autoreactive B1-cells via autoAg–autoBCR specificity; (ii) DS enters cells by (DS–autoAg)–autoBCR complex internalization and recruits a cascade of molecules to stimulate autoreactive B1-cells.[3,5] In particular, DS recruits GTF2I that is required for IGH gene expression and IgH-associated multiprotein complexes in the endoplasmic reticulum (ER) to facilitate autoAb production.[5] Therefore, any self-molecule with DS-affinity has a propensity to be transformed by DS into an autoantigenic DS–autoAg complex.[5] Based on this unifying principle of DS–autoAg affinity, we have discovered and catalogued known and putative autoAgs from different cells and organs.[1,2,6–8]
AutoAbs, which target autoAgs, have been found in a significant portion of COVID-19 patients. In a cohort study of 147 hospitalized COVID-19 patients, autoAbs were detected in ~50% of patients, and antinuclear autoAbs were detected in ~25% of patients, with the target autoAgs associated with myositis, systemic sclerosis, and connective tissue disease overlap syndromes.[9] In another study of 987 COVID-19 patients with life-threatening pneumonia, over 10% developed autoAbs against interferons, which likely neutralized their ability to block SARS-CoV-2 infection.[10] Although COVID-19 is typically mild or asymptomatic in children, multisystem inflammatory syndrome and multiple autoAbs developed in a portion of infected children.[11,12] In a study of COVID-19 patients with unexplained neurological symptoms, anti-neuronal autoAbs were detected in sera or cerebrospinal fluid of all patients.[13] Antinuclear autoAbs, the most frequently tested autoAbs in clinical screening for autoimmune diseases such as lupus, Sjögren syndrome, scleroderma, or polymyositis, were found in 20–50% of COVID-19 patients.[14–16]
Viral infections have long been regarded as culprits of autoimmune diseases. However, it has remained unclear how infections induce autoimmune diseases. In this study, we investigated HS-Sultan cells, a B-cell lymphoblast line isolated from Burkitt’s lymphoma of a 7-year-old boy. HS-Sultan cells are infected with and immortalized by Epstein-Barr virus (EBV) and carry the viral DNA sequence. From the proteome extracts of HS-Sultan cells, we identified a putative DS-affinity autoantigen-ome (i.e. the totality of proteins with known autoantigenic properties) of 362 proteins, of which 201 are confirmed autoAgs by means of corresponding specific autoAbs reported in the literature. By comparing this autoantigen-ome with proteins affected by SARS-CoV-2 infection derived from multi-omic studies compiled in Coronascape,[17–38] we identified 315 DS-affinity proteins and 186 confirmed autoAgs. When host cells are infected, numerous molecules undergo significant changes via altered expression, modification, or degradation. When the infected cells die, these altered molecules are released, and those with DS-affinity may become associated with DS and transform into immunogenic autoAg–DS complexes. This study illustrates that viral infections can alter the host cell autoantigen-ome, result in a large repertoire of potential autoAgs, and may consequently lead to autoimmune disease.
Results and discussion
DS-affinity autoantigen-ome of HS-Sultan cells
HS-Sultan cells were cultured, harvested, and lysed. Total proteins were extracted from lysates and fractionated on DS-Sepharose affinity resins. Proteins with no or weak DS-affinity were removed from the resins with 0.2 M NaCl, and those with intermediate to strong DS-affinity were eluted first with 0.5 M and then with 1.0 M NaCl. Proteins in the DS-affinity fractions were collected, desalted, digested, and sequenced by mass spectrometry. A total of 362 proteins were identified, with the majority present in the 0.5 M NaCl elution. Proteins that eluted with 1.0 M NaCl possess very strong DS-affinity and include some of the classical nuclear autoAgs, e.g. histones, TOP1, Sm-D3, and 60S acidic ribosomal protein P0. Other proteins with strong DS-affinity include both known autoAgs (vimentin, ATP synthetase ATP5B, and PABPC1) and unknown ones (RPL10A, L15, RPS27A, and mitochondrial single-stranded DNA binding protein SSBP1).
Of the 362 DS-affinity proteins identified from HS-Sultan cells, 201 (55.5%) are confirmed humoral autoAgs based on prior literature reports of specific autoAbs (see references in Table 1). These autoAgs and their respective autoAbs are found in a wide spectrum of autoimmune diseases and cancer. The number of actual autoAgs is likely much greater, as most of the unconfirmed proteins have structural resemblance to known autoAgs. For example, SSBP1 is structurally and functionally similar to the classical lupus autoAg SSB, but it has not formally been identified as an autoAg. As another example, nucleosome assembly protein 1-like 1 and 4 (NAP1L1 and NAP1L4) are identified by DS-affinity but unconfirmed as autoAgs, whereas their close relative NAP1L3 has been reported as an autoAg. Due to the structural similarity of many DS-affinity proteins with known autoAgs, it is likely that there are additional yet-to-be discovered (considered putative) autoAgs in this group.
| Number of Peptides | Protein | Alteration in COVID (up and/or down) | SARS-CoV-2 interaction | DS-affinity (1.0 M) | DS-affinity (0.5 M) | Ref. | |||
|---|---|---|---|---|---|---|---|---|---|
| 6 | AARS | Alanine-tRNA ligase, cytoplasmic | u | d | + | [39] | |||
| 15 | ACLY | ATP-citrate synthase | u | d | + | [40] | |||
| 9 | ACTA2 | Actin, aortic smooth muscle | u | d | + | [41] | |||
| 6 | ACTB | Actin, cytoplasmic 1 | u | d | + | [42] | |||
| 7 | ACTBL2 | Beta-actin-like protein 2 | u | d | + | [42] | |||
| 8 | ACTN1 | Alpha-actinin-1 (f-actin cross linking protein) | u | d | + | [43] | |||
| 7 | ACTN4 | Alpha-actinin-4 | u | d | + | [43] | |||
| 2 | ACTR2 | Actin-related protein 2 | u | d | + | [44] | |||
| 2 | ACTR3 | Actin-related protein 3 | u | + | [45] | ||||
| 3 | AFP | Alpha-fetoprotein | d | + | [46] | ||||
| 15 | AHCY | Adenosylhomocysteinase | d | + | |||||
| 4 | AHSA1 | Activator of 90 kDa heat shock protein ATPase homolog 1 | d | + | |||||
| 5 | ALDH18A1 | Delta-1-pyrroline-5-carboxylate synthetase | d | + | |||||
| 3 | ALDOA | Fructose-bisphosphate aldolase A | u | d | + | [47] | |||
| 4 | ALDOC | Fructose-bisphosphate aldolase C | u | d | + | [48] | |||
| 2 | ANP32A | Acidic leucine-rich nuclear phosphoprotein 32 family member A | u | d | + | ||||
| 4 | ANP32B | Acidic leucine-rich nuclear phosphoprotein 32 family member B | d | N | + | [49] | |||
| 2 | ANP32C | Acidic leucine-rich nuclear phosphoprotein 32 family member C | + | ||||||
| 8 | ANXA5 | Annexin A5 | u | d | + | [50] | |||
| 5 | ANXA6 | Annexin A6 | u | d | + | [51] | |||
| 4 | APEH | Acylamino-acid-releasing enzyme | + | ||||||
| 2 | APEX1 | DNA-(apurinic or apyrimidinic site) lyase | u | d | + | [52] | |||
| 2 | API5 | Apoptosis inhibitor 5 | d | + | |||||
| 8 | ARF1 | ADP-ribosylation factor 1 | Nsp6 | + | |||||
| 6 | ARHGDIA | Rho GDP-dissociation inhibitor 1 | u | d | + | ||||
| 8 | ARHGDIB | Rho GDP-dissociation inhibitor 2 | d | + | [53] | ||||
| 3 | ARPC2 | Actin-related protein 2/3 complex subunit 2 | d | + | |||||
| 2 | ASNS | Asparagine synthetase [glutamine-hydrolyzing] | u | + | |||||
| 14 | ATIC | Bifunctional purine biosynthesis protein, PURH | + | [54] | |||||
| 13 | ATP5F1B | ATP synthase subunit beta, mitochondrial, ATP5B | u | d | Nsp6 | + | [55] | ||
| 3 | ATXN10 | Ataxin-10, spinocerebellar ataxia type 10 protein | u | d | + | ||||
| 2 | BZW1 | Basic leucine zipper and W2 domain-containing protein 1 | u | + | |||||
| 3 | BZW2 | Basic leucine zipper and W2 domain-containing protein 2 | N | + | |||||
| 2 | C1QBP | Complement component 1 Q subcomponent-binding protein | d | + | [56] | ||||
| 8 | CALM1 | Calmodulin | u | d | + | [12] | |||
| 13 | CALR | Calreticulin | u | d | + | [57] | |||
| 15 | CAND1 | Cullin-associated NEDD8-dissociated protein 1 | + | ||||||
| 6 | CAP1 | Adenylyl cyclase-associated protein 1 | u | d | + | ||||
| 3 | CAPN1 | Calpain-1 catalytic subunit | + | ||||||
| 2 | CAPRIN1 | Caprin-1, cell cycle associated protein 1 | d | + | |||||
| 3 | CAPZA1 | F-actin capping protein subunit alpha-1 (capz alpha-1) | d | + | [58] | ||||
| 3 | CAPZB | F-actin-capping protein subunit beta | d | + | [59] | ||||
| 10 | CCT2 | T-complex protein 1 subunit beta (tcp-1-beta) (cct-beta) | d | Nsp12 | + | [60] | |||
| Orf8 | |||||||||
| Orf9b | |||||||||
| Orf10 | |||||||||
| 6 | CCT3 | T-complex protein 1 subunit gamma (chaperonin containing TCP1, subunit 3 isoform) | u | Orf8 | + | [61] | |||
| Orf10 | |||||||||
| 6 | CCT4 | T-complex protein 1 subunit delta (tcp-1-delta) (cct-delta) (stimulator of tar rna-binding) | u | + | [62] | ||||
| 10 | CCT5 | T-complex protein 1 subunit epsilon (tcp-1-epsilon) (cct-epsilon) | u | d | Nsp1 | + | [60] | ||
| Nsp12 | |||||||||
| Orf8 | |||||||||
| Orf10 | |||||||||
| 7 | CCT6A | T-complex protein 1 subunit zeta (acute morphine dependence-related protein 2) | u | d | Nsp1 | + | [60] | ||
| Nsp12 | |||||||||
| Orf10 | |||||||||
| 5 | CCT7 | T-complex protein 1 subunit eta (hiv-1 nef-interacting protein) | Orf10 | + | [60] | ||||
| 20 | CCT8 | T-complex protein 1 subunit theta | u | d | Nsp1 | + | [61] | ||
| Nsp12 | |||||||||
| Nsp14 | |||||||||
| 4 | CDC37 | Hsp90 co-chaperone, hsp90 chaperone protein kinase-targeting | u | d | Nsp16 | + | |||
| 6 | CLIC1 | Chloride intracellular channel protein 1 | u | d | Nsp16 | + | [62] | ||
| 12 | CLTC | Clathrin heavy chain 1 | u | d | + | [4] | |||
| 4 | CLTCL1 | Clathrin heavy chain 2 | + | ||||||
| 4 | CLUH | Clustered mitochondria protein homolog (mRNA-binding) | d | Nsp7 | + | ||||
| Nsp16 | |||||||||
| 2 | CMPK1 | UMP-CMP kinase | d | + | |||||
| 3 | CNDP2 | Cytosolic non-specific dipeptidase | u | Orf3 | + | ||||
| Orf10 | |||||||||
| 2 | CNPY2 | Protein canopy homolog 2, MSAP, TMEM4, Zsig9 | d | Orf3a | + | ||||
| 2 | COPZ1 | Coatomer subunit zeta-1 | d | + | |||||
| 12 | CORO1A | Coronin-1A | u | + | [63] | ||||
| 3 | CPNE1 | Copine-1 | + | ||||||
| 4 | CPNE3 | Copine-3 | u | d | + | ||||
| 3 | CS | Citrate synthase, mitochondrial | u | d | + | [40] | |||
| 10 | CSE1L | Exportin-2 | d | + | |||||
| 4 | CSK | Tyrosine-protein kinase CSK | d | + | [64] | ||||
| 6 | DARS | Aspartate-tRNA ligase, cytoplasmic, DARS1 | + | [65] | |||||
| 3 | DCK | Deoxycytidine kinase | u | + | |||||
| 3 | DCTN2 | Dynactin subunit 2 | Orf6 | + | |||||
| 3 | DCTPP1 | dCTP pyrophosphatase 1 | d | + | |||||
| 7 | DDB1 | DNA damage-binding protein 1 | u | d | + | [4] | |||
| 4 | DDX39A | ATP-dependent RNA helicase DDX39A | u | d | + | ||||
| 3 | DDX39B | Spliceosome RNA helicase DDX39B | d | + | |||||
| 2 | DDX5 | Probable ATP-dependent RNA helicase, RNA helicase p68 | u | d | + | [66] | |||
| 2 | DHX15 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 | d | + | |||||
| 3 | DHX9 | ATP-dependent RNA helicase A | + | [67] | |||||
| 3 | DIABLO | Diablo homolog, mitochondrial | u | d | Nsp6 | + | |||
| Nsp15 | |||||||||
| 4 | DLD | Dihydrolipoyl dehydrogenase, mitochondrial | + | [68] | |||||
| 2 | DNAJC8 | DnaJ homolog subfamily C member 8 | u | + | |||||
| 4 | DNPH1 | 2′-deoxynucleoside 5′-phosphate N-hydrolase 1 | u | + | |||||
| 6 | DPP3 | Dipeptidyl-peptidase 3 | d | + | |||||
| 5 | DUT | Deoxyuridine 5′-triphosphate nucleotidohydrolase, mitochondrial | u | d | + | ||||
| 2 | EEF1A1 | Elongation factor 1-alpha | u | d | + | [69] | |||
| 4 | EEF1A2 | Elongation factor 1-alpha 2 | u | Orf3 | + | [70] | |||
| 2 | EEF1B2 | Elongation factor 1-beta | d | + | |||||
| 5 | EEF1D | Elongation factor 1-delta | d | + | [71] | ||||
| 6 | EEF1G | Elongation factor 1-gamma | u | d | + | ||||
| 17 | EEF2 | Elongation factor 2 | u | d | + | [72] | |||
| 2 | EFTUD2 | U5 small nuclear ribonucleoprotein component 116 kDa | d | + | [73] | ||||
| 3 | EIF2A | Eukaryotic translation initiation factor 2 subunit alpha, EIF2S1 | + | ||||||
| 5 | EIF3A | Eukaryotic translation initiation factor 3 subunit A | u | d | Nsp1 | + | [74] | ||
| 5 | EIF3B | Eukaryotic translation initiation factor 3 subunit B | u | d | Nsp1 | + | |||
| 2 | EIF3C | Eukaryotic translation initiation factor 3 subunit C | d | Nsp1 | [75] | ||||
| 3 | EIF3E | Eukaryotic translation initiation factor 3 subunit E (viral integration site protein int-6 homolog) | u | d | Nsp1 | + | [76] | ||
| 4 | EIF3F | Eukaryotic translation initiation factor 3 subunit F | u | d | Nsp1 | + | |||
| 2 | EIF3I | Eukaryotic translation initiation factor 3 subunit I | d | Nsp1 | + | [75] | |||
| 10 | EIF3L | Eukaryotic translation initiation factor 3 subunit L (subunit E interacting protein) | d | Nsp1 | + | ||||
| 19 | EIF4A1 | Eukaryotic initiation factor 4A-I | u | d | + | ||||
| 4 | EIF4A3 | Eukaryotic initiation factor 4A-III | + | [77] | |||||
| 2 | EIF5 | Eukaryotic translation initiation factor 5 | u | d | + | [78] | |||
| 5 | EIF5A | Eukaryotic translation initiation factor 5A-1 | u | d | + | [78] | |||
| 2 | EIF6 | Eukaryotic translation initiation factor 6 | u | + | |||||
| 3 | ELAVL1 | ELAV-like protein 1 | d | + | [79] | ||||
| 3 | ENO2 | Gamma-enolase | u | d | + | [80] | |||
| 2 | EPRS | Bifunctional glutamate/proline-tRNA ligase | u | + | [81] | ||||
| 4 | ERO1L | ERO1-like protein alpha | d | Orf3a | + | ||||
| 4 | ETF1 | Eukaryotic peptide chain release factor subunit 1 | u | + | |||||
| 14 | EZR | Ezrin | u | d | S | + | [82] | ||
| 3 | FARSB | Phenylalanine-tRNA ligase beta subunit | + | [83] | |||||
| 19 | FASN | Fatty acid synthase | u | d | + | [84] | |||
| 2 | FDPS | Farnesyl pyrophosphate synthetase like-4 protein | d | + | |||||
| 2 | FEN1 | Flap endonuclease 1 | u | d | + | ||||
| 2 | FERMT3 | Fermitin family homolog 3 | u | + | |||||
| 11 | FKBP4 | Peptidyl-prolyl cis-trans isomerase FKBP4, FKBP-52 | Nsp12 | + | [85] | ||||
| 2 | FKBP5 | Peptidyl-prolyl cis-trans isomerase FKBP5 (FK506-binding protein) | u | + | |||||
| 2 | FUBP1 | Far upstream element-binding protein 1 | u | d | + | ||||
| 2 | G6PD | Glucose-6-phosphate 1-dehydrogenase | u | d | + | [86] | |||
| 5 | GANAB | Neutral alpha-glucosidase AB | d | Orf6 | + | [87] | |||
| Orf8 | |||||||||
| Orf9c | |||||||||
| 6 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | u | d | Orf8 | + | [88] | ||
| 4 | GARS | Glycine-tRNA ligase, GARS1 | u | + | [89] | ||||
| 2 | GART | Trifunctional purine biosynthetic protein adenosine-3 | d | Nsp15 | + | ||||
| 2 | GBE1 | 1,4-alpha-glucan-branching enzyme | u | + | |||||
| 8 | GDI1 | Rab GDP dissociation inhibitor alpha | u | d | + | [90] | |||
| 10 | GDI2 | Rab GDP dissociation inhibitor beta | u | d | Nsp12 | + | [91] | ||
| Orf9b | |||||||||
| 2 | GGCT | Gamma-glutamylcyclotransferase, cytochrome c-releasing factor 21 | u | + | |||||
| 3 | GLO1 | Lactoylglutathione lyase | d | Orf3 | + | [92] | |||
| 3 | GLRX3 | Glutaredoxin-3 | d | + | |||||
| 10 | GLUD1 | Glutamate dehydrogenase 1, mitochondrial | + | [93] | |||||
| 2 | GMFB | Glia maturation factor, beta | u | + | |||||
| 3 | GSPT1 | Eukaryotic peptide chain release factor GTP-binding subunit ERF3A | + | ||||||
| 3 | GSS | Glutathione synthetase | d | + | |||||
| 2 | GTF2I | General transcription factor II-I | u | d | + | ||||
| 3 | H2AFV | Histone H2A.V, H2AZ2 | u | d | + | [94] | |||
| 2 | H2AFY2 | Core histone marco-H2A.1, MARCOH2A1 | + | [95] | |||||
| 3 | HARS | Histidine-tRNA ligase, cytoplasmic | + | [12] | |||||
| 3 | HDGF | Hepatoma-derived growth factor, hmg-1l2 | u | d | + | [96] | |||
| 5 | HIST1H1C | Histone 1.2, H1-2 | u | d | Nsp8 | + | [97] | ||
| 2 | HIST1H2AB | Histone H2A type 1-B/E, H2AC4 | d | + | [98] | ||||
| 2 | HIST1H2BL | Histone H2B type 1-L, H2BC13 | u | + | [99] | ||||
| 14 | HIST1H4J | Histone H4, H4C1 | + | [100] | |||||
| 9 | HIST2H2BE | Histone H2B type 2-E, H2BC21 | u | d | + | [101] | |||
| 5 | HIST2H3A | Histone H3.2, H3C15 | u | d | + | [102] | |||
| 4 | HMGB1 | High mobility group protein 1-like 10 (hmg-1l10) | d | + | [96] | ||||
| 3 | HMGCS1 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic | u | d | + | ||||
| 4 | HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | u | d | + | [103] | |||
| 8 | HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | u | d | + | [104] | |||
| 2 | HNRNPA3 | Heterogeneous nuclear ribonucleoprotein A3 | u | d | + | [105] | |||
| 2 | HNRNPAB | Heterogeneous nuclear ribonucleoprotein A/B | d | + | [105] | ||||
| 3 | HNRNPC | Heterogeneous nuclear ribonucleoprotein C1/C2 | u | d | + | [106] | |||
| 5 | HNRNPCL1 | Heterogeneous nuclear ribonucleoprotein C-like 1 | + | [106] | |||||
| 5 | HNRNPD | Heterogeneous nuclear ribonucleoprotein D | + | [107] | |||||
| 2 | HNRNPDL | Heterogeneous nuclear ribonucleoprotein D-like | u | d | + | [108] | |||
| 3 | HNRNPF | Heterogeneous nuclear ribonucleoprotein F | d | + | [109] | ||||
| 2 | HNRNPH1 | Heterogeneous nuclear ribonucleoprotein H (hnrnp h) | u | d | + | [110] | |||
| 9 | HNRNPK | Heterogeneous nuclear ribonucleoprotein K (transformation up-regulated nuclear protein) | u | + | [111] | ||||
| 5 | HNRNPR | Heterogeneous nuclear ribonucleoprotein R | u | d | + | [112] | |||
| 2 | HNRNPU | Heterogeneous nuclear ribonucleoprotein U | u | d | + | [113] | |||
| 6 | HPRT1 | Hypoxanthine-guanine phosphoribosyltransferase | + | ||||||
| 16 | HSP90AA1 | Heat shock protein HSP 90-alpha (hsp 86) (ny-ren-38 antigen) | u | d | [114] | ||||
| 6 | HSP90AA2 | Heat shock protein HSP 90-alpha A2 | u | + | [115] | ||||
| 16 | HSP90AB1 | Heat shock protein HSP 90-beta (hsp 84) (hsp 90) | u | d | Nsp12 | + | [116] | ||
| 19 | HSP90B1 | Endoplasmin, GRP94, tumor rejection antigen 1 | u | d | Orf3a | + | [115] | ||
| Orf9c | |||||||||
| 4 | HSPA4 | Heat shock 70 kDa protein 4 | u | d | + | [117] | |||
| 15 | HSPA5 | Endoplasmic reticulum chaperone, GRP78, BiP | u | d | S | + | [118] | ||
| E | |||||||||
| M | |||||||||
| Nsp2 | |||||||||
| Nsp4 | |||||||||
| Nsp6 | |||||||||
| Orf3a | |||||||||
| Orf7a | |||||||||
| Orf7b | |||||||||
| 24 | HSPA8 | Heat shock cognate 71 kDa protein | u | d | Nsp2 | + | [119] | ||
| Nsp12 | |||||||||
| 12 | HSPA9 | Stress-70 protein, mitochondrial (grp 75) | u | d | N | + | [120] | ||
| 19 | HSPD1 | 60 kDa heat shock protein, mitochondrial matrix protein p1 | u | d | + | [121] | |||
| 4 | HSPH1 | Heat shock protein 105 kDa | u | + | [122] | ||||
| 2 | IDE | Insulin-degrading enzyme | Nsp4 | + | |||||
| Nsp12 | |||||||||
| Nsp14 | |||||||||
| Nsp15 | |||||||||
| Nsp16 | |||||||||
| Orf3b | |||||||||
| 2 | IDH3A | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | + | ||||||
| 6 | ILF2 | Interleukin enhancer-binding factor 2 | u | + | [123] | ||||
| 6 | IPO5 | Importin-5, KPNB3, RANBP5 | + | [124] | |||||
| 3 | IPO7 | Importin-7, RANBP7 | Nsp6 | + | [125] | ||||
| Orf9c | |||||||||
| 8 | IQGAP1 | Ras GTPase-activating-like protein IQGAP1 | u | + | [126] | ||||
| 4 | KARS | Lysine-tRNA ligase, KARS1 | Nsp7 | + | [81] | ||||
| 3 | KHSRP | Far upsteam element-binding protein 2 (KH-type splicing regulatory protein) | u | d | + | ||||
| 2 | KPNA2 | Importin subunit alpha-1 | d | Orf6 | + | ||||
| 11 | KPNB1 | Importin subunit beta-1 | + | [124] | |||||
| 30 | LCP1 | Plastin-2 (l-plastin) (lymphocyte cytosolic protein 1) | u | d | + | [127] | |||
| 7 | LDHA | l-lactate dehydrogenase A chain (proliferation-inducing gene 19 protein) | u | d | Nsp12 | + | [128] | ||
| 10 | LDHB | l-lactate dehydrogenase B chain | u | d | Nsp12 | + | [129] | ||
| Nsp7 | |||||||||
| 16 | LRPPRC | Leucine-rich PPR motif-containing protein | d | + | [130] | ||||
| 7 | MCM2 | DNA replication licensing factor MCM2 | d | + | [131] | ||||
| 3 | MCM3 | DNA replication licensing factor mcm3 (dna polymerase alpha holoenzyme-associated protein p1)(p102 protein) (p1-mcm3) | u | d | + | [131] | |||
| 3 | MCM4 | DNA replication licensing factor mcm4 (cdc21 homolog) (p1-cdc21) | u | d | + | [131] | |||
| 2 | MCM6 | DNA replication licensing factor mcm6 (p105mcm) | u | d | + | [131] | |||
| 2 | ME2 | NAD-dependent malic enzyme, mitochondrial | u | d | Nsp15 | + | |||
| 6 | MSN | Moesin | u | S | + | [132] | |||
| Nsp6 | |||||||||
| Orf3 | |||||||||
| 2 | MYG1 | UPF0160 protein MYG1, mitochondrial, C12orf10 | + | ||||||
| 4 | MZB1 | Marginal zone B- and B1-cell-specifc protein (Proapoptotic caspase adapter protein, plasma cell-induced resident protein) | + | ||||||
| 3 | NAA15 | N-alpha-acetyltransferase 15, NatA auxiliary subunit (NMDA receptor-regulated protein, NARG1) | d | + | |||||
| 2 | NAA25 | N-alpha-acetyltransferase 25, NatB auxiliary subunit (TPR repeat-containing protein C12orf30) | + | ||||||
| 3 | NACA | Nascent polypeptide-associated complex subunit alpha (nac-alpha) | u | d | + | [133] | |||
| 7 | NAP1L1 | Nucleosome assembly protein 1-like 1 (nap-1-related protein) (hnrp) | d | + | |||||
| 4 | NAP1L4 | Nucleosome assembly protein 1-like 4 (nucleosome assembly protein 2) | u | d | + | ||||
| 5 | NARS | Asparagine-tRNA ligase, cytoplasmic, NARS1 | d | + | [134] | ||||
| 4 | NASP | Nuclear autoantigenic sperm protein (histone binding protein) | u | d | + | [135] | |||
| 12 | NCL | Nucleolin | u | d | + | [136] | |||
| 8 | NME1 | Nucleoside diphosphate kinase A | d | + | [137] | ||||
| 15 | NPEPPS | Puromycin-sensitive aminopeptidase | + | ||||||
| 4 | NPM1 | Nucleophosmin | u | d | Orf9c | + | [138] | ||
| 3 | NSFL1C | NSFL1 cofactor p47 | u | + | |||||
| 8 | NUDC | Nuclear migration protein nudC (nuclear distribution protein c homolog) | d | Nsp12 | + | ||||
| 2 | NUDT21 | Cleavage and polyadenylation specificity factor subunit 5 | d | + | |||||
| 2 | NUDT3 | Diphosphoinositol polyphosphate phosphohydrolase | + | ||||||
| 4 | NUDT5 | ADP-sugar pyrophosphatase (nucleoside diphosphate-linked moiety × motif 5) (nudix motif 5) | d | + | |||||
| 2 | OTUB1 | Ubiquitin thioesterase protein OTUB1 | + | ||||||
| 5 | P4HB | Protein disulfide-isomerase (cellular thyroid hormone-binding protein) (p55) | u | d | Nsp7 | + | [139] | ||
| Orf3b | |||||||||
| 14 | PA2G4 | Proliferation-associated protein 2G4 | u | + | |||||
| 9 | PABPC1 | Polyadenylate-binding protein 1 | d | N | + | [140] | |||
| 2 | PAFAH1B2 | Platelet-activating factor acetylhydrolase IB subunit beta | u | d | + | ||||
| 2 | PAFAH1B3 | Platelet-activating factor acetylhydrolase IB subunit gamma | u | Nsp12 | + | ||||
| Nsp5 | |||||||||
| Orf3b | |||||||||
| 6 | PAICS | Multifunctional protein ADE2 | d | + | |||||
| 2 | PARP1 | Poly [ADP-ribose] polymerase 1 | u | d | + | ||||
| 8 | PCNA | Proliferating cell nuclear antigen (cyclin) | u | d | + | [88] | |||
| 2 | PDCD10 | Programmed cell death protein 10 | + | ||||||
| 21 | PDIA3 | Protein disulfide-isomerase A3 | u | d | M | + | [141] | ||
| Orf3a | |||||||||
| Orf3b | |||||||||
| Orf8 | |||||||||
| Orf9c | |||||||||
| 8 | PDIA4 | Protein disulfide-isomerase A4 (protein erp-72) (erp72) | u | d | Nsp16 | + | [142] | ||
| Nsp7 | |||||||||
| Orf3b | |||||||||
| 5 | PDIA6 | Protein disulfide-isomerase A6 (protein disulfide isomerase p5) (thioredoxin domain-containing protein 7) | u | d | + | [139] | |||
| 7 | PFAS | Phosphoribosylformylglycinamidine synthase | Nsp12 | + | |||||
| Nsp15 | |||||||||
| Nsp16 | |||||||||
| Nsp7 | |||||||||
| 3 | PFDN2 | Prefoldin subunit 2 | u | Nsp12 | + | [143] | |||
| Nsp15 | |||||||||
| Orf10 | |||||||||
| 2 | PFDN3 | Prefoldin subunit 3, von hippel-lindau-binding protein 1, VBP1 | d | Nsp12 | + | ||||
| Nsp15 | |||||||||
| 9 | PFN1 | Profilin-1 (profilin i) | u | d | + | [131] | |||
| 4 | PGAM1 | Phosphoglycerate mutase 1 | u | d | + | [144] | |||
| 4 | PGAM2 | Phosphoglycerate mutase 2 | + | ||||||
| 3 | PGLS | 6-phosphogluconolactonase | u | + | |||||
| 3 | PHGDH | d-3-phosphoglycerate dehydrogenase | u | d | + | [145] | |||
| 10 | PLCG2 | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase gamma-2 | u | + | |||||
| 2 | PMPCB | Mitochondrial-processing peptidase subunit beta | d | M | + | ||||
| 8 | PPA1 | Inorganic pyrophosphatase | u | Orf3 | + | [146] | |||
| 3 | PPAT | Amidophosphoribosyltransferase | d | + | |||||
| 6 | PPIB | Peptidyl-prolyl cis-trans isomerase B | u | d | Orf8 | + | [147] | ||
| 2 | PPP1R7 | Protein phosphatase 1 regulatory subunit 7 (protein phosphatase 1 regulatory subunit 7) (protein phosphatase 1 regulatory subunit 22) | u | + | |||||
| 3 | PPP2R1A | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | d | + | |||||
| 3 | PRDX1 | Peroxiredoxin-1 | u | d | + | [148] | |||
| 5 | PRDX3 | Thioredoxin-dependent peroxide reductase, mitochondrial | u | d | + | [149] | |||
| 2 | PRKCSH | Glucosidase 2 subunit beta (protein kinase c substrate) | d | S | + | ||||
| Nsp6 | |||||||||
| Orf3 | |||||||||
| Orf3a | |||||||||
| 2 | PRMT1 | Protein arginine n-methyltransferase 1 (hnRNP methyltransferase-like 2 isoform (ec 2.1.1.-) (interferon receptor 1-bound protein 4) | d | + | [150] | ||||
| 2 | PRPSAP2 | Phosphoribosyl pyrophosphate synthetase-associated protein 2 | u | + | |||||
| 3 | PSMA1 | Proteasome subunit alpha type-1 | d | Orf3b | + | [151] | |||
| 2 | PSMA2 | Proteasome subunit alpha type-2 | d | + | |||||
| 6 | PSMA3 | Proteasome subunit alpha type-3 (macropain subunit c8) | u | d | Nsp2 | + | [152] | ||
| Nsp4 | |||||||||
| Nsp7 | |||||||||
| Nsp10 | |||||||||
| Nsp12 | |||||||||
| 5 | PSMA4 | Proteasome subunit alpha type-4 (C9) | u | + | [153] | ||||
| 5 | PSMA5 | Proteasome subunit alpha type-5 (macropain zeta chain) | u | Orf3b | + | [154] | |||
| 8 | PSMA6 | Proteasome subunit alpha type-6 | u | d | Orf3b | + | |||
| 4 | PSMA7 | Proteasome subunit alpha type-7 | u | d | + | [155] | |||
| 5 | PSMB1 | Proteasome subunit beta type-1 | + | [156] | |||||
| 3 | PSMB3 | Proteasome subunit beta type-3 (proteasome theta chain) | d | Orf3b | + | [152] | |||
| 7 | PSMB4 | Proteasome subunit beta type-4 (proteasome beta chain) (macropain beta chain) (multicatalytic endopeptidase complex beta chain) | Orf3b | + | |||||
| 2 | PSMB6 | Proteasome subunit beta type-6 (proteasome delta chain)(multicatalytic endopeptidase complex delta chain) | d | Orf3b | + | ||||
| 3 | PSMB7 | Proteasome subunit beta type-7 (proteasome subunit z) | d | + | [157] | ||||
| 3 | PSMB8 | Proteasome subunit beta type-8 | u | d | + | ||||
| 4 | PSMB9 | Proteasome subunit beta type-9 | u | d | + | ||||
| 2 | PSMC3 | 26S protease regulatory subunit 6A | d | Orf6 | + | ||||
| 5 | PSMD1 | 26S proteasome non-ATPase regulatory subunit 1 | u | d | Nsp7 | + | |||
| Orf6 | |||||||||
| Orf8 | |||||||||
| 9 | PSMD11 | Proteasome 26S non-ATPase regulatory subunit 11 | u | + | |||||
| 3 | PSMD12 | 26S proteasome non-ATPase regulatory subunit 12 | d | + | |||||
| 2 | PSMD14 | 26S proteasome non-ATPase regulatory subunit 14 | + | ||||||
| 8 | PSMD3 | 26S proteasome non-ATPase regulatory subunit 3 | d | + | |||||
| 3 | PSMD6 | 26S proteasome non-ATPase regulatory subunit 6 | + | ||||||
| 11 | PSME1 | Proteasome activator complex subunit 1 | u | Nsp15 | + | ||||
| 8 | PSME2 | Proteasome activator complex subunit 2 | u | + | |||||
| 4 | PSME3 | Proteasome activator complex subunit 3 | d | Nsp16 | + | [158] | |||
| 2 | PSPH | Phosphoserine phosphatase | + | ||||||
| 3 | PTBP1 | Polypyrimidine tract-binding protein 1 | u | d | + | [159] | |||
| 2 | PTBP3 | Polypyrimidine tract-binding protein (cDNA FLJ51619, highly similar to regulator of differentiation) ROD1 | u | d | + | ||||
| 2 | PTGES3 | Prostaglandin E synthase 3 (telomerase-binding protein p23) (hsp90 co- chaperone) (progesterone rec) | d | + | |||||
| 2 | PTPN6 | Tyrosine-protein phosphatase non-receptor type 6 | u | d | + | ||||
| 2 | PUS1 | tRNA pseudouridine synthase A | + | ||||||
| 5 | RAB7A | Ras-related protein Rab-7a | u | d | Nsp7 | + | |||
| Orf3 | |||||||||
| Orf7b | |||||||||
| 2 | RAD23A | UV excision repair protein RAD23 homolog A | d | + | [160] | ||||
| 3 | RANBP1 | RanBP | u | d | + | ||||
| 2 | RANBP6 | Ran-binding protein 6 | d | Orf7a | + | ||||
| 7 | RDX | Radixin, isoform CRA (actin binding to plasma membrane) | u | d | Nsp13 | + | [161] | ||
| 2 | RPL10A | 60S ribosomal protein L10a | + | ||||||
| 2 | RPL11 | 60S ribosomal protein L11 | u | + | |||||
| 2 | RPL15 | 60S ribosomal protein L15 | d | + | |||||
| 2 | RPL22 | 60s ribosomal protein L22 (Epstein-Barr virus small rna-associated protein) (heparin-binding protein hbp15) | d | + | [157] | ||||
| 4 | RPLP0 | 60s acidic ribosomal protein P0 | u | d | + | [162] | |||
| 2 | RPS15A | 40s ribosomal protein S15a | u | ||||||
| 2 | RPS25 | 40S ribosomal protein S25 | u | d | Orf8 | + | |||
| 3 | RPS27A | Ubiquitin-40S ribosomal protein S27a | u | d | S | + | [157] | ||
| Nsp4 | |||||||||
| 6 | RPS3 | 40S ribosomal protein S3 | u | d | Orf8 | + | [163] | ||
| 3 | RPS3A | 40S ribosomal protein S3a | u | d | Orf8 | + | |||
| 3 | RPS7 | 40S ribosomal protein S7 | u | d | + | ||||
| 5 | RPSA | Similar to 40S ribosomal protein SA (p40) (34/67 kDa laminin receptor) (colon carcinoma laminin-binding protein) (NEM/1CHD4) (multidrug resistance-associated protein MGr1-Ag), RPSAP12 | + | ||||||
| 6 | RUVBL1 | RuvB-like 1, tata box-binding protein-interacting protein, nuclear matrix protein 238 | Nsp1 | + | [164] | ||||
| Nsp7 | |||||||||
| Nsp12 | |||||||||
| Orf9b | |||||||||
| 5 | RUVBL2 | RuvB-like 2 | d | Nsp1 | + | [165] | |||
| Nsp7 | |||||||||
| Nsp12 | |||||||||
| 2 | SARS | Serine-tRNA ligase, cytoplasmic, SARS1 | Nsp15 | + | |||||
| 2 | SEPT11 | Septin-11 | d | + | |||||
| 2 | SEPT2 | Septin-2, NEDD5, DIFF6 | + | [166] | |||||
| 3 | SEPT7 | Septin-7 | d | + | [167] | ||||
| 8 | SERPINB9 | Serpin B9 | u | d | + | ||||
| 2 | SERPINC1 | Antithrombin-III | u | + | |||||
| 6 | SET | Protein SET (phosphatase 2a inhibitor i2pp2a) (i-2pp2a) (template- activating factor i) (taf-i) (hla-dr-associated protein ii) (phapii) | u | d | + | [168] | |||
| 3 | SF3B3 | Splicing factor 3B subunit 3 | + | [169] | |||||
| 2 | SFPQ | Splicing factor, proline- and glutamine-rich | u | d | + | [170] | |||
| 3 | SGTA | Small glutamine-rich tetratricopeptide repeat-containing protein alpha | u | d | M | + | |||
| 2 | SHMT1 | Serine hydroxymethyltransferase, cytosolic | d | + | |||||
| 9 | SHMT2 | Serine hydroxymethyltransferase, mitochondrial | d | + | |||||
| 2 | SLIRP | SRA stem-loop-interacting RNA-binding protein, mitochondrial | u | d | + | ||||
| 8 | SND1 | Staphylococcal nuclease domain-containing protein 1 | u | d | + | ||||
| 2 | SNRNP70 | U1 small nuclear ribonucleoprotein 70 kDa | u | d | + | [171] | |||
| 3 | SNRPA | U1 small nuclear ribonucleoprotein A | u | + | [172] | ||||
| 3 | SNRPA1 | U2 small nuclear ribonucleoprotein A′ | + | [173] | |||||
| 2 | SNRPD1 | Small nuclear ribonucleoprotein Sm D1 | u | + | [174] | ||||
| 2 | SNRPD2 | Small nuclear ribonucleoprotein Sm D2 | d | + | [175] | ||||
| 2 | SNRPD3 | Small nuclear ribonucleoprotein Sm d3 (snrnp core protein d3) (sm-d3) | d | + | [174] | ||||
| 2 | SNRPE | Small nuclear ribonucleoprotein E | d | + | [176] | ||||
| 2 | SPTAN1 | Spectrin alpha chain, non-erythrocytic 1 | u | d | + | [177] | |||
| 3 | SRM | Spermidine synthase | d | + | |||||
| 3 | SRP14 | Signal recognition particle 14 kDa protein | u | d | Nsp13 | + | |||
| Orf8 | |||||||||
| 3 | SRSF1 | Serine/argine-rich splicing factor 1 (Isoform ASF-1 of splicing factor, arginine/serine-rich | u | d | + | [178] | |||
| 3 | SRSF2 | Arginine/serine-rich splicing factor 2, SFRS2 | u | d | + | [179] | |||
| 3 | SSB | Lupus La protein (Sjoegren syndrome type b antigen) (La/SSB) | u | d | + | [180] | |||
| 4 | SSBP1 | Single-stranded DNA-binding protein, mitochondrial | N | + | |||||
| 4 | ST13 | Hsc70-interacting protein (hip) (suppression of tumorigenicity protein 13) (putative tumor suppressor st13) (protein fam10a1) (progesterone receptor-associate) | u | Nsp12 | + | [181] | |||
| Orf3b | |||||||||
| Orf6 | |||||||||
| Orf8 | |||||||||
| Orf10 | |||||||||
| 3 | STIP1 | Stress-induced-phosphoprotein 1 | u | d | Nsp12 | + | |||
| Orf3a | |||||||||
| Orf8 | |||||||||
| E | |||||||||
| 2 | SUGT1 | Protein SGT1 homolog (suppressor of G2 allele of SKP1 homolog) | u | Nsp12 | + | ||||
| Nsp15 | |||||||||
| 2 | SWAP70 | Switch-associated protein 70 | d | Nsp2 | + | ||||
| 5 | SYNCRIP | Heterogeneous nuclear ribonucleoprotein Q | d | + | |||||
| 2 | TALDO1 | Transaldolase | u | d | + | [182] | |||
| 3 | TBCA | Tubulin-specific chaperone A | Nsp11 | + | |||||
| 3 | TCL1A | T-cell leukemia/lymphoma protein 1A | u | d | + | ||||
| 7 | TCP1 | T-complex protein 1 subunit alpha (tcp-1-alpha) (cct-alpha) | d | Orf10 | + | [60] | |||
| 10 | TLN1 | Talin-1 | u | d | + | [183] | |||
| 5 | TNPO1 | Transportin-1 | + | ||||||
| 2 | TOP1 | DNA topoisomerase 1 (Scl 70) | u | + | [184] | ||||
| 5 | TPM1 | Tropomyosin alpha-1 chain isoform | u | d | Nsp9 | + | [185] | ||
| 6 | TPM3 | Tropomyosin alpha-3 chain (tropomyosin gamma) | u | d | + | [186] | |||
| 5 | TPM4 | Tropomyosin alpha-4 chain | u | d | + | [187] | |||
| 4 | TPP2 | Tripeptidyl-peptidase 2 | + | ||||||
| 3 | TTLL12 | Tubulin-tyrosine ligase-like protein 12 | d | + | [188] | ||||
| 2 | TTLL3 | Tubulin monoglycylase TTLL3 | u | + | |||||
| 4 | TUBA1C | TUBA1C protein | u | d | + | [189] | |||
| 12 | TUBA4A | Tubulin alpha-4A chain | u | d | + | [190] | |||
| 5 | TUBB | Tubulin beta chain | u | d | + | [191] | |||
| 4 | TUBB1 | Tubulin beta-1 chain | + | [192] | |||||
| 2 | TUBB4 | Tubulin beta-3 chain, beta-4 chain | u | d | + | [193] | |||
| 12 | TUBB4B | Tubulin beta-4B chain (Tubulin beta-2C chain) | u | d | + | [192] | |||
| 2 | TXNDC17 | Thioredoxin domain-containing protein 17 | u | d | + | ||||
| 4 | TXNRD1 | Thioredoxin reductase 1, cytoplasmic | u | d | + | [194] | |||
| 2 | U2AF2 | Splicing factor U2AF 65 kDa subunit | d | + | |||||
| 7 | UBA1 | Ubiquitin-like modifier-activating enzyme 1 (Ubiquitin-activating enzyme E1) | u | d | + | [195] | |||
| 2 | UBA6 | Ubiquitin-like modifier-activating enzyme 6 | Nsp7 | + | |||||
| 2 | UBE2K | Ubiquitin-conjugating enzyme E2 K | + | ||||||
| 3 | USP5 | Ubiquitin carboxyl-terminal hydrolase 5 | u | d | + | ||||
| 3 | VARS1 | Valine-tRNA ligase | + | ||||||
| 12 | VCP | Transitional endoplasmic reticulum ATPase (valosin-containing protein) | u | d | + | [196] | |||
| 10 | VIM | Vimentin | u | d | + | [197] | |||
| 6 | WARS | Tryptophan-tRNA ligase, cytoplasmic, WARS1 | u | d | + | [198] | |||
| 2 | XPNPEP1 | Xaa-Pro aminopeptidase 1 | d | + | |||||
| 4 | XPO1 | Exportin-1 | Nsp4 | + | |||||
| Orf7a | |||||||||
| 5 | XPOT | Exportin-T (trna exportin) (exportin(trna)) | u | Orf7a | + | ||||
| 3 | XRCC5 | X5-ray repair cross-complementing protein 5, lupus ku86 ku80 autoantigen | d | + | [199] | ||||
| 9 | XRCC6 | X-ray repair cross-complementing protein 6 (ku70) | u | d | + | [200] | |||
| 6 | YARS | Tyrosine-tRNA ligase, cytoplasmic, YARS1 | u | d | + | [201] | |||
| 5 | YWHAB | 14-3-3 protein beta/alpha (protein kinase c inhibitor protein 1) | u | d | + | ||||
| 12 | YWHAE | 14-3-3 protein epsilon | u | d | + | [202] | |||
| 3 | YWHAG | 14-3-3 protein gamma | u | d | + | [202] | |||
| 3 | YWHAH | 14-3-3 protein eta (protein as1) | d | + | [203] | ||||
| 3 | YWHAQ | 14-3-3 protein theta | u | d | + | [204] | |||
| 7 | YWHAZ | 14-3-3 protein zeta/delta | u | d | + | [205] | |||
| 2 | ZPR1 | Zinc finger protein ZPR1 | u | d | + | [206] | |||
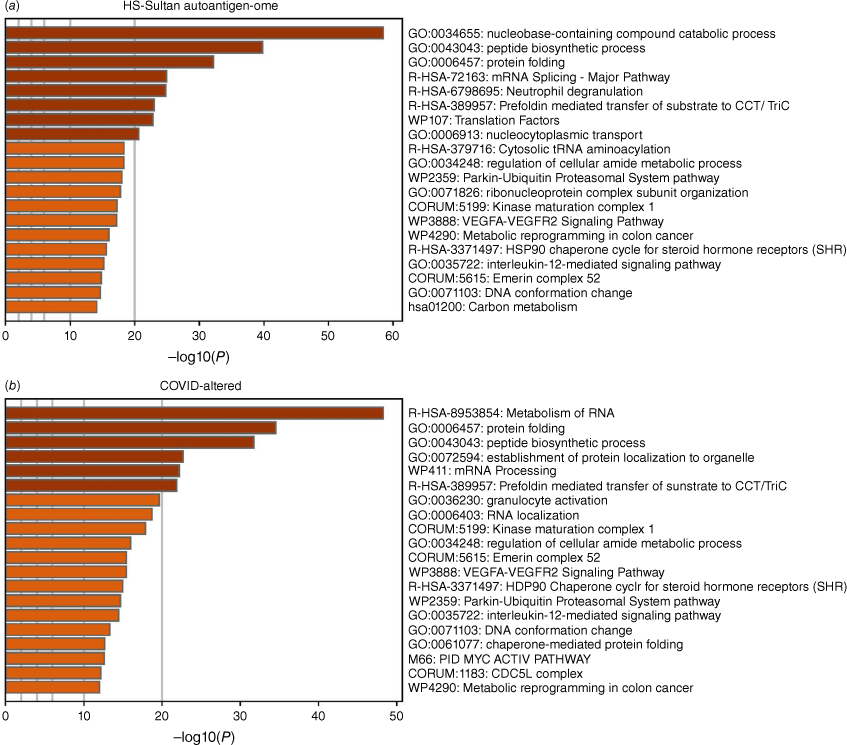
The DS-affinity autoantigen-ome, which includes confirmed and putative autoAgs, is not a random collection of proteins but functionally highly connected. As shown by protein–protein interaction analysis, the DS-affinity autoantigen-ome possesses significantly more interactions than expected (3105 interactions at high confidence level vs 1249 expected; protein–protein interaction (PPI) enrichment P-value < 1.0 × 10−16). Based on Gene Ontology (GO) biological process analysis, the DS-affinity autoantigen-ome of HS-Sultan cells is significantly associated with RNA slicing, translation, peptide biosynthesis, protein folding, proteolysis, biosynthesis and metabolism of nucleobase-containing small molecules (e.g. nucleobase, nucleoside, and nucleotide phosphate), cytoskeleton organization, and chromosome organization (Fig. 1). Pathway and process enrichment analysis reveals that it is also significantly associated with neutrophil degranulation, nucleocytoplasmic transport, kinase maturation complex, metabolic reprogramming, and IL-12 mediated signaling (Fig. 2a).
The autoantigen-ome of HS-Sultan cells identified by DS-affinity. Marked proteins are associated with peptide biosynthesis and catabolic process (58 proteins, yellow), protein folding (34 proteins, light green), RNA splicing (41 proteins, red), nucleobase-containing small molecule metabolic process (39 proteins, blue), proteolysis (55 proteins, pink), import into nuclear (13 proteins, dark green), cytoskeleton organization (39 proteins, amber), and chromosome organization (40 proteins, aqua).
Top 20 enriched pathways and processes among COVID-altered autoAgs. (a) Pathways of 362 proteins identified by DS-affinity from HS-Sultan cells. (b) Pathways of 316 DS-affinity proteins that are altered by COVID infection. The x-axes show the negative decadic logarithm of the respective pathway’s enrichment P-value.
The DS-affinity autoantigen-ome is dominated by several families of proteins. There are 24 proteasomal proteins, 22 spliceosome proteins, 14 hnRNPs, 13 aminoacyl-tRNA synthases (ligases), 13 translation initiation factors subunits, 12 ribosomal proteins, 10 heat shock proteins (HSPs), nine actin and actin-related proteins, nine tubulins, eight histones, eight snRNPs, seven T-complex proteins (CCT/TriC), six elongation factor subunits, and six 14-3-3 proteins. The majority of the proteins in these families have been reported as autoAgs (Table 1). For example, all hnRNP and snRNP proteins identified by DS-affinity in this study are among the best-known nuclear autoAgs. Interestingly, autoAgs included in clinical diagnostic autoimmune disease ANA screening panels, such as SSB (lupus La), SNRPD1 (Sm D1), SNRPD3 (Sm D3), histones, and TOP1, are all identified in this study by DS-affinity enrichment from HS-Sultan cells.
In addition to proteins, such as ribosomal and ribonucleoproteins, that can be consistently identified from a variety of cell types, HS-Sultan B lymphoblast cells give rise to a large number of unique DS-affinity protein categories. In particular, many proteins associated with biomolecule biosynthesis are identified. Among them are proteins involved in inosine monophosphate and purine nucleotide biosynthesis (ATIC, GART, HPRT1, PAICS, PFAS, PPAT, and SHMT1), amino acid biosynthesis (CS, IDH3A, PHGDH, PGAM1, PGAM2, and PSPH), and carbohydrate biosynthesis and catabolic processes (ALDOA, ALDOC, ENO2, G6PD, GBE1, LDHA, and TALDO1). There are also proteins involved in protein transport (ARF1, CSE1L, GDI1, GDI2, HMGB1, IPO5, KPNA2, RAB7A, RANBP1, RANBP6, SRP14, TNPO1, and XPOT), dephosphorylation (NSF1C, PPP1R7, PPP2R1A, SET, and SWAP70), and ubiquitination and de-ubiquitination (OTUB1, SHMT2, UBA1, UBA6, UBE2K, and USP5). Seventeen of these 44 proteins are currently known autoAgs, while the remainder await further investigation. Overall, HS-Sultan cells appear to be especially rich in biosynthetic protein machinery, which may explain the rapid proliferation of these cells in Burkitt lymphoma.
Thirteen aminoacyl-tRNA synthetases were identified by DS affinity from HS-Sultan cells, including AARS, DARS, ERPS, FARSB, GARS, HARS, KARS, NARS, PUS1, SARS, VARS, WARS, and YARS. Ten of these are already known autoAgs (Table 1), although we suspect that the remainder will also likely be autoAgs. This group of proteins are the culprits of antisynthetase syndrome, an autoimmune disease characterized by autoAbs against one or multiple tRNA synthetases. Antisynthetase syndrome is a chronic disorder that can affect many parts of the body, with common symptoms including myositis, interstitial lung disease, polyarthritis, skin thickening and cracking of fingers and toes, or Raynaud disease. Antisynthetase syndrome has been reported in a case report of COVID-19 interstitial lung disease.[207]
HS-Sultan cells are B lymphoblasts immortalized by EBV infection and carry viral DNA sequences. Using DS affinity, we identified numerous proteins involved in DNA repair and the mitotic cell cycle, including CLTC, DCTN2, MCM2, MCM3, MCM4, MCM6, NSF1C, PNCA, PPAT, and SUGT1. Using DS affinity, we also identified many proteins associated with telomerase maintenance, including TCP1, CCT2, CCT4, CCT5, CCT7, HNRNPA1, HNRPNA2B1, HNRNPC, HNRNPD, HNRNPU, HSP90AA1, HSP90AB1, PARP1, PTGES3, and XRCC5. Telomerase maintenance, which counteracts DNA damage response, cell cycle arrest, and apoptosis, is crucial for immortalization of cells with unlimited proliferative potential. Of these 25 proteins, 19 are known autoAgs, which indicates that proteins involved in telomerase maintenance, DNA repair, and cell cycle may be affected by EBV infection and become autoantigenic.
DS-affinity autoantigen-ome related to SARS-CoV-2 infection
To find out whether DS-affinity autoAgs are affected in SARS-CoV-2 infection, we conducted similarity searches with currently available multi-omic COVID-19 data compiled in Coronascape (as of 22 February 2021).[17–38] Among our 362 DS-affinity proteins, 315 (87.0%) are affected by SARS-CoV-2 infection (Table 1). Of these 315 proteins, 209 are up-regulated and 248 are down-regulated at protein and/or mRNA levels, and 95 are in the interactomes of individual SARS-CoV-2 viral proteins. Because the COVID-19 multi-omics data have been obtained with various methods from different infected cells or patients, there are proteins found up-regulated in some studies but down-regulated in other studies, but nevertheless, these proteins are affected by the infection and thus considered COVID-affected in our analysis (Supplementary Table S1). Of the 315 COVID-affected DS-affinity proteins, 186 (59.0%) are thus far confirmed autoAgs, while 129 are putative autoAgs (Table 1).
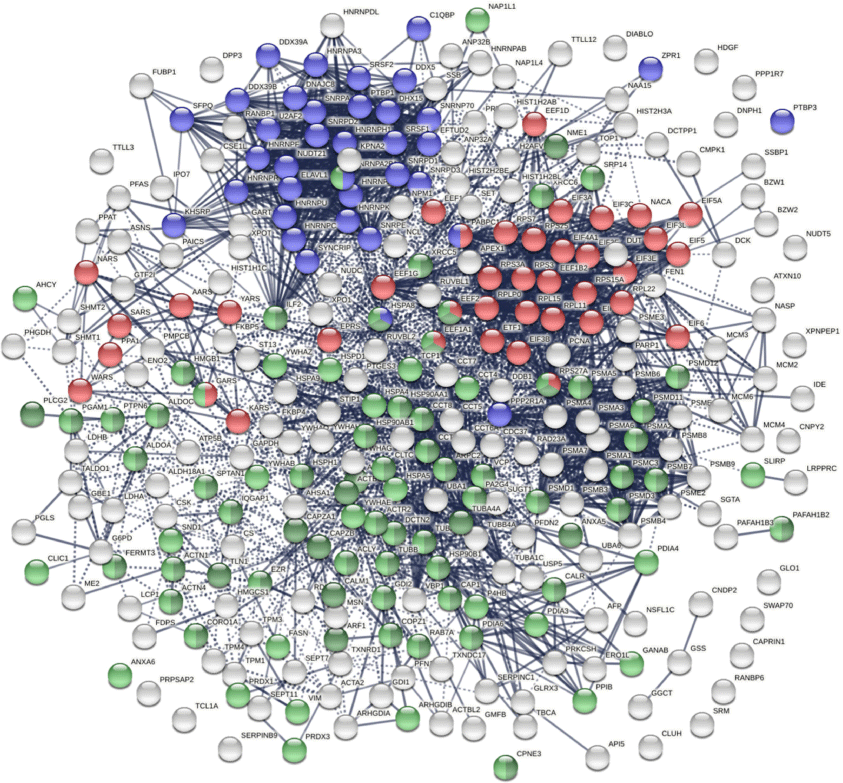
The COVID-affected DS-affinity proteins are highly connected (Fig. 3). By STRING analysis, these 315 proteins exhibit 2507 interactions at high confidence level, which is significantly higher than randomly expected (1002 interactions) with a PPI enrichment P-value <1.0 × 10−16. The proteins are primarily associated with RNA and mRNA processing, translation, vesicles, and vesicle-mediated transport (Fig. 3), which is consistent with our findings in other cell types.[1,2,8] In addition, these proteins are enriched in protein folding, peptide biosynthesis, granulocyte activation, emerin complex, IL-12 mediated signaling pathway, CDC5L complex, and metabolic reprogramming (Fig. 2b).
COVID-affected proteins shared with the HS-Sultan autoantigen-ome. Marked proteins are associated with RNA splicing (36 proteins, blue), translation (39 proteins, red), vesicle (77 proteins, green), and vesicle-mediated transport (62 proteins, dark green).
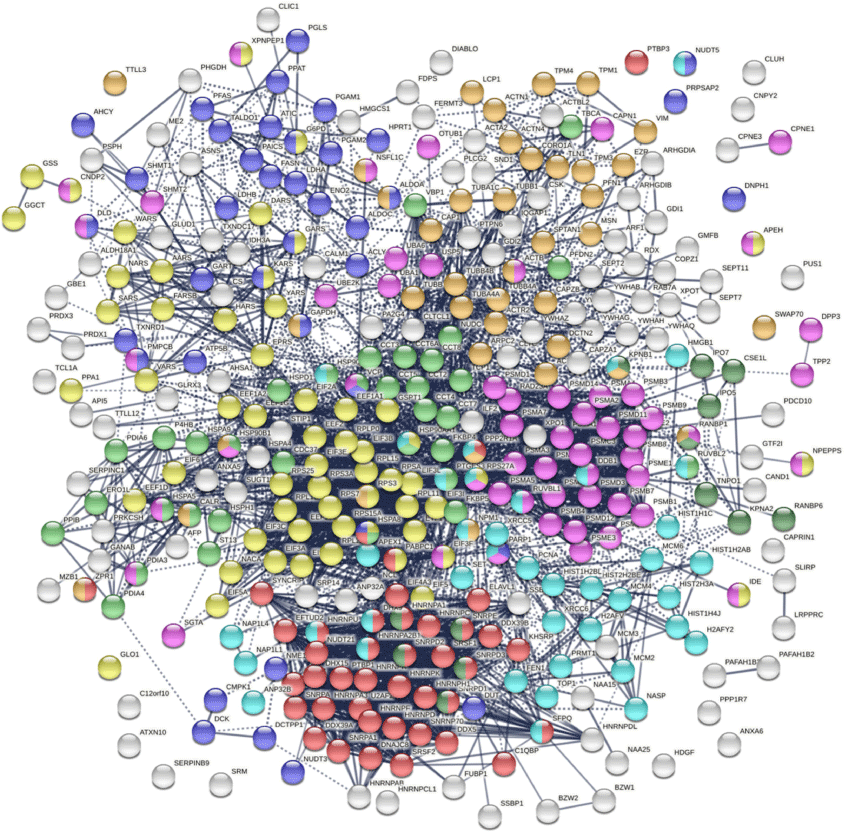
Twenty-one COVID-affected DS-affinity proteins are associated with mRNA splicing, including heterogenous nuclear ribonucleoproteins (hnRNP A1, A2B1, A3, AB, C, DL, F, H1, K, Q, R, and U), small nuclear ribonucleoproteins (SNRNP70, SNRPA, SNRPE, SNRPD1, SNRRPD2, and SNRPD3), and splicing factors (SRSF1, SRSF2, and SFPQ), all of which are well-known autoAgs.
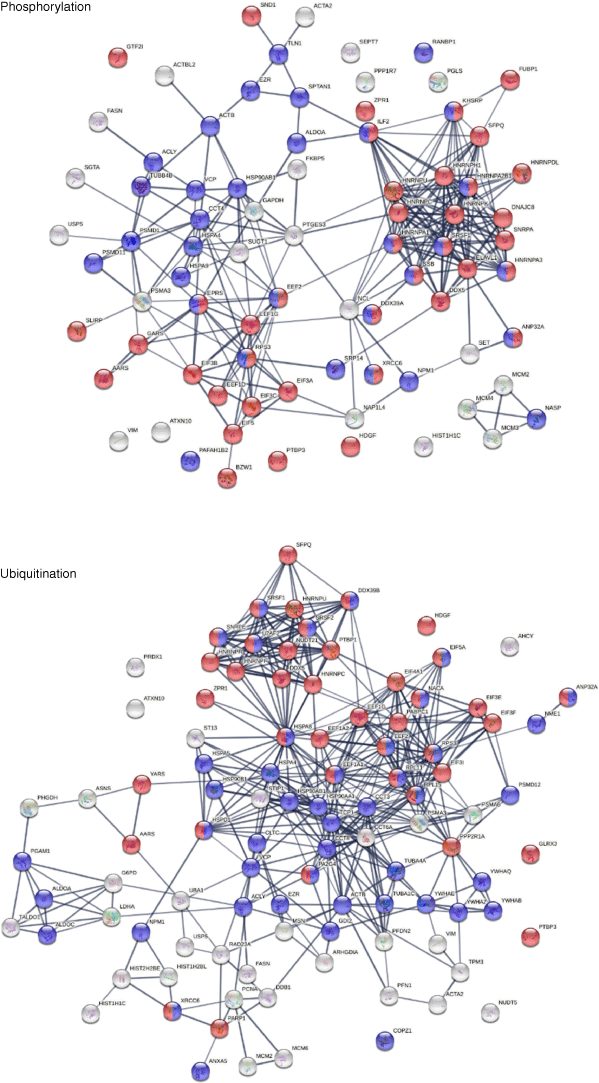
Phosphorylation and ubiquitination changes induced by SARS-CoV-2 infection are posttranslational molecular alterations that may transform native proteins into potential autoAgs (Fig. 4), which is consistent with our previous findings.[2] Phosphorylation changes affected 80 COVID-altered DS-affinity proteins, including eight hnRNPs, four initiation factors (EIF3A, 3B, 3C, and 5), three elongation factors, three replication licensing factors (MCM2, 3, and 4), SSB, XRCC6, and GTF2I. These phosphoproteins are associated with mRNA splicing, translation, telomere maintenance, DNA conformation change, and pre-replicative complex assembly. Ubiquitination changes affected 101 COVID-altered DS-affinity proteins, including eight HSPs, five initiation factors (EIF3E, 3F, 3I, 4A1, and 5A), four CCT units, four 14-3-3 proteins, three elongation factors, three histones, and two MCMS. These ubiquitinated proteins are associated with the nucleobase-containing compound catabolic process, RNA metabolism, cellular response to stress, prefoldin mediated transfer of substrate to CCT/TriC, and axon guidance.
AutoAgs that interact with SARS-CoV-2 components
There are 95 DS-affinity proteins found in the interactomes of various SARS-CoV-2 proteins (Fig. 5), meaning that these proteins can interact directly or indirectly with viral components. They appear to be intimately involved in protein metabolism, including 17 proteins related to peptide biosynthesis, 25 related to protein folding, 29 related to protein localization, and 22 related to proteolysis. In addition, these proteins are associated with the symbiont viral process, translational initiation, protein deubiquitination, protein stabilization, and posttranslational protein modification.
DS-affinity proteins in the SARS-CoV-2 interactomes. Total: marked proteins are involved in protein folding (25 proteins, red), peptide biosynthetic process (17 proteins, green), protein localization (29 proteins, blue), or proteolysis (22 proteins, pink). Orf3b: proteolysis (pink). Orf3a: endoplasmic reticulum (dark purple). Orf9b: nuclear function of prefoldin (amber), AAA+ ATPase domain or P-loop containing nucleoside triphosphate hydrolase (dark green). Nsp15: prefoldin-mediated transfer of substrate to CCT/TriC (yellow), nucleotide binding (dark green). Orf10: protein folding (red), CCT chaperonin (yellow). Orf8: protein folding (red), SRP-dependent cotranslational protein targeting to membrane (blue), CCT chaperonin (yellow). Nsp1: translation initiation (green), CCT chaperonin (yellow). Nsp12: protein folding (red), multi-organism process (aqua), CCT chaperonin (yellow).
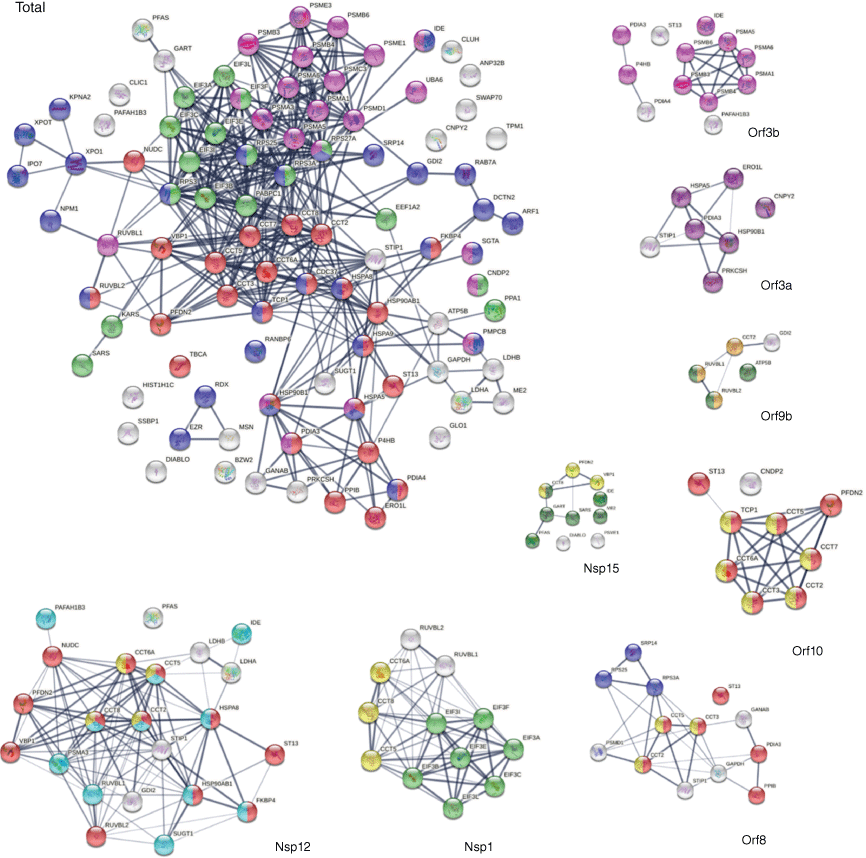
The chaperonin-containing T-complex (CCT), also known as T-complex protein ring complex (TriC), is the chaperonin of eukaryotic cells. The human CCT/TriC complex is a two-ring barrel-like complex formed by eight similar but distinct subunits. Remarkably, all eight CCT subunits are identified by DS affinity, and seven of them are found in the SARS-CoV-2 interactomes: CCT2 (interacts with Nsp12, Orf8, Orf9b, and Orf10), CCT5 (with Nsp1, Nsp12, Orf8, and Orf10), CCT6A (with Nsp1, Nsp12, and Orf10), CCT8 (with Nsp1, Nsp12, and Nsp14), CCT3 (with Orf8 and Orf10), TCP1 (with Orf10), and CCT7 (with Orf10). In total, six SARS-CoV-2 proteins interact with the host cell CCT chaperonin, with Orf10 interacting with six CCT subunits (Fig. 5). Furthermore, CCT subunits and PDFN2 (prefoldin subunit 2) are found together in the interactomes of Orf10, Nsp12, and Nsp15. CCTs assist the folding of proteins upon ATP hydrolysis, aiding in the folding of ~10% of the proteome. PDFN2 binds the nascent polypeptide chain and promotes folding in an environment in which there are many competing pathways for non-native proteins. Therefore, these findings suggest that SARS-CoV-2 exploits the CCT complex and PDFN2 to ensure competitive folding of non-native viral proteins in the host cells.
In addition to CCT/TriC, HSPs are another group of chaperones frequently identified by DS affinity. Ten HSPs are identified in this study, including HSPA4, HSPA5, HSPA8, HSPA9, HSPD1, HSPH1, HSP90AA1, HSP90AA2, HSP90AB1, and HSP9B1. All 10 are known autoAgs (Table 1). HSP8 interacts with Nsp2 and Nsp12. HSP90B1 (endoplasmin) interacts with Orf3a and Orf9c. HSPA9 interacts with N protein. HSP90AB1 interacts with Nsp12. Most strikingly, HSPA5 (GRP78, BiP) interacts with nine SARS-CoV-2 proteins, including S, E, M, Nsp2, Nsp4, Nsp6, Orf3a, Orf7a, and Orf7b. In addition, CDC37 (Hsp90 co-chaperone, hsp90 chaperone protein kinase-targeting) interacts with Nsp16. ST13 (Hsc70-interacting protein) interacts with five SARS-CoV-2 proteins (Nsp12, Orf3b, Orf6, Orf8, and Orf10). STIP1 (stress induced phosphoprotein 1, HSP90AA1 co-chaperone) interacts with four viral proteins (Nsp12, Orf3a, Orf8, and E).
The replication machinery of SARS-CoV-2 interacts with 41 different DS-affinity proteins. Nsp12, an RNA-dependent RNA polymerase and the central component of the replication machinery, interacts with the largest number (i.e. 22) of DS-affinity proteins (Fig. 5). Its cofactor Nsp7 interacts with 12 proteins and Nsp8 interacts with only one. The replication machine also includes a helicase (Nsp13), two ribonucleases (Nsp14 and Nsp15), two RNA-cap methyltransferases (Nsp14 and Nsp16), and cofactor Nsp10. Nsp15 interacts with 10 DS-affinity proteins, Nsp16 interacts with eight proteins, Nsp13 interacts with SRP14 and RDX, Nsp14 interacts with IDE and CCT8, and Nsp10 interacts with PSMA3. Nsp12-interacting DS-affinity proteins are strongly associated with protein folding, particularly prefoldin mediated transfer of substrates to the CCT complex and cooperation of prefoldin and CCT in protein folding (Fig. 5). Nsp15-interacting proteins are also associated with prefoldin-mediated substrate transfer to CCT. DS-affinity proteins interacting with other individual viral replication components have no clear biological associations.
Orf3b of SARS-CoV-2 interacts with 12 DS-affinity proteins, including six proteasomal proteins, three protein disulfide-isomerases, IDE, ST13, and PAFAH1B3 (Fig. 5). Orf3a interacts with seven proteins, including STIP1 (stress-induced-phosphoprotein 1) and six ER proteins (HSPA5, HSP90B1, CNPY2, ERO1L, PRKCSH, and PDIA3). CANPY2 prevents MIR-mediated MRLC ubiquitination and its subsequent proteasomal degradation. ERO1L (or ERO1A) is an oxidoreductase in disulfide bond formation in the ER. PRKCSH (glucosidase II subunit beta) cleaves sequentially the two innermost glucose residues from the Glc2Man9GlcNAc2 oligosaccharide precursor of immature glycoproteins. Based on the normal functions of their interacting proteins, Orf3a and Orf3b appear to affect host stress response and protein processing in the ER.
The S protein of SARS-CoV-2 is found to interact with HSPA5 (GRP78/BiP), PRKCSH, PRS27A (ubiquitin-40S ribosomal protein), MSN, and EZR. EZR and MSN are members of the ezrin-moesin-radixin (ERM) family, and its third member RDX is found to interact with Nsp13 of the virus. Moesin is localized to filopodia and other membranous protrusions that are important for cell–cell recognition, and ERM proteins connect the plasma membranes to the actin-based cytoskeleton. Actin and cytoskeleton proteins have been consistently found to be altered in SARS-CoV-2 infection in our previous studies,[1,2] and this finding suggests that ERM proteins facilitate the viral trafficking from host cell membrane to the cytoskeleton. All three ERM proteins are confirmed autoAgs.
Nsp1 is a major virulence factor of coronavirus. COVID-19 patients with autoAbs are found to have higher levels of antibodies against SARS-CoV-2 Nsp1 protein.[9] Nsp1 has been reported to hijack the host 40S ribosome by inserting its C terminus into the mRNA entry tunnel, which effectively blocks RGI-dependent innate immune responses.[208] In this study, we found that Nsp1 interacts with seven subunits of the translation initiation factor 3 complex (EIF3 A, B, C, E, F, I, L). The EIF3 complex binds the 40S ribosome and serves as a scaffold for other initiation factors, auxiliary factors, and mRNA. Hence, our study extends previous reported activities of Nsp1 and shows that Nsp1 engages both the 40S ribosome and EIF3 to manipulate host protein translation.
A few interesting SARS-CoV-2-interacting DS-affinity proteins may provide clues to potential COVID-19 symptoms. PAFAH1B3 is a catalytic unit of the platelet-activating factor acetylhydrolase complex and plays important roles in platelet activation regulation and brain development, and it interacts with Nsp12, Nsp5, and Orf3b. Another subunit, PAFAH1B2, is altered in SARS-CoV-2 infection. Both this and our previous studies[2] have identified PAFAH1B2 and B3 as potential COVID-altered autoAgs, and their roles in COVID coagulopathy merit further investigation.
IDE (insulin degrading enzyme) is a ubiquitously expressed metalloprotease that degrades insulin, beta amyloid, and others. IDE interacts with six SARS-CoV-2 proteins (Nsp4, Nsp12, Nsp14, Nsp15, Nsp16, and Orf3b). Although its role in COVID remains unknown, IDE has been partially characterized in other viral infections. It is one of the host factors of hepatitis C virus,[209] and it degrades HIV-1 p6 Gap protein and regulates virus replication in an Env-dependent manner.[210] In varicella zoster virus infection, the viral gE protein precursor associates with IDE, HSPA5, HSPA8, HSPD1, and PPIA in the ER of infected cells.[211] Interestingly, this group of ER proteins is also identified in this study, although we identified PPIB instead of PPIA. Although IDE has not yet formally been described as an autoAg, we have identified IDE in this and another study,[2] and its importance for COVID-19 and autoimmunity merits further investigation.
DS-affinity and B-cell-specific IgH-ER complex
Because HS-Sultan cells are derived from B lymphoblasts infected by EBV, we compared the DS-affinity autoantigen-ome with single-cell mRNA expression profiles of B-cells from seven patients hospitalized with COVID-19.[23] We identified 39 DS-affinity proteins that are up-regulated at mRNA level in COVID B-cells, which include seven HSPs, six proteasomal proteins, four protein disulfide-isomerases, HDGF (heparin binding growth factor), CLIC1, CPNE3, SND1, TALDO1, TCL1A, and others (Fig. 6). These up-regulated proteins are primarily associated with protein processing in the ER and the proteasome. We also identified 21 DS-affinity proteins that are down-regulated in COVID B-cells, including four translation elongation factors, two translation initiation factors, two hnRNPs, two aminoacyl-tRNA synthetases, NACA, NAP1L1, and PABPC1. These down-regulated proteins are primarily associated with gene expression (Fig. 6).
DS-affinity proteins from HS-Sultan cells that are up-regulated or down-regulated in B-cells of COVID-19 patients. Top: 39 up-regulated proteins. Red: protein processing in ER. Blue: proteasome. Bottom: 21 down-regulated proteins. Aqua: proteins involved in gene expression.
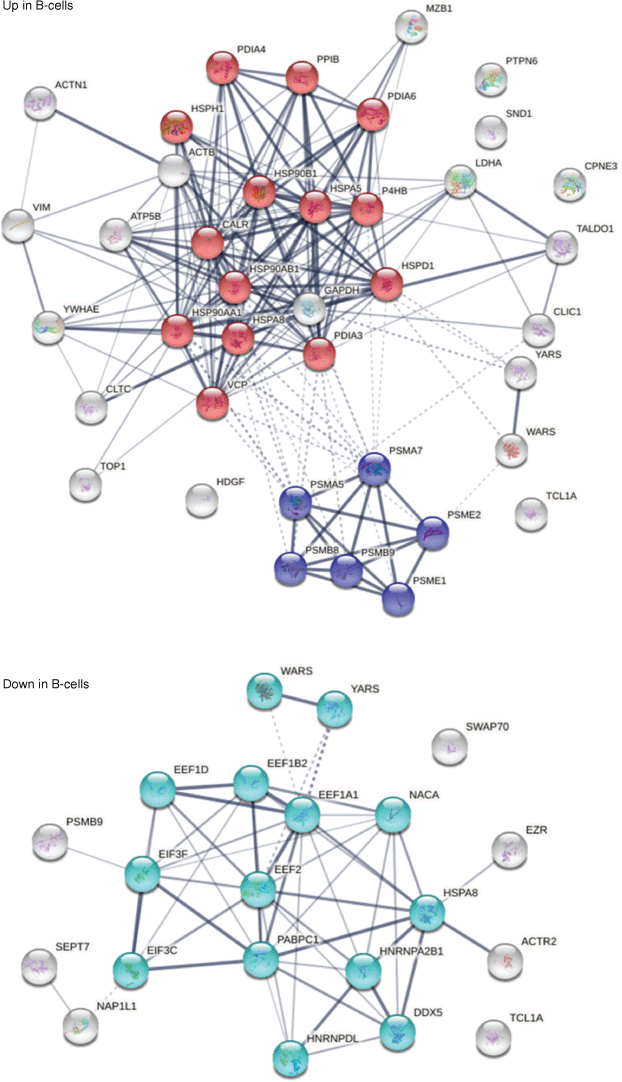
In particular, MZB1 (marginal zone B- and B1-cell-specific protein) is found up-regulated in B-cells from five of the seven COVID-19 patients. MZB1 is a B-cell-specific ER-localized protein that is most abundantly expressed in marginal zone B-cells and B1-cells.[212] These cells are also termed innate-like B cells. They differ from follicular B-cells by their attenuated Ca2+ mobilization, fast antibody secretion, and increased cell adhesion. MZB1 plays important roles in humoral immunity and helps diversify peripheral B-cell functions by regulating calcium stores, antibody secretion, and integrin activation. MZB1 mRNA expression was found increased by >2-fold in B-cells of SLE patients with active disease.[213] High MZB1 gene expression predicted adverse prognosis in chronic lymphocytic leukemia, follicular lymphoma, and diffuse large B-cell lymphoma.[214] High prevalence of MZB1-positive plasma B-cells in tissue fibrosis was found in human lung and skin fibrosis, and MZB1 levels correlated positively with tissue IgG and negatively with diffusion capacity of the lung.[215]
MZB1 is part of a B-cell-specific ER chaperone complex, associates with IgM heavy chain (IgH) and HSP90B1 (Grp94), and augments IgM assembly and secretion.[216] MZB1 is also found to augment the function of PDIA3 (ERp57).[212] In this study, MZB1, HSP90B1, HSPA5 (Grp78, BiP), PDIA4, PDIA6, and CALR are found jointly up-regulated in the B-cells of the same five COVID-19 patients. This finding suggests that these six proteins are in the same IgH-associated ER complex. In addition, several other up-regulated ER proteins are identified, including HSP90AB1, HSPD1, HSPA8, PDIA3, P4HB, and PPIB.
Interestingly, in our study of murine pre-B1 lymphoblasts, we also found that DS interacts with the same IgH-associated multiprotein complex in the ER.[5] In addition, we had observed that DS interacts directly with GTF2I in murine pre-B1 cells, and GTF2I is also identified by DS affinity in human B lymphoblast HS-Sultan cells in this study. GTF2I is a required gene transcription factor at the IgH gene locus. Pre-B1 cells, which express precursor B-cell receptors (preBCRs) that are polyreactive and autoreactive, are a critical check point in the development of mature autoreactive B cells. The Ig heavy chain (IgH) repertoire of autoAbs is determined at the pre-B stage. Our previous findings from pre-B1 cells suggested that DS is a potential master regulator of IgH at both the gene and protein expression levels, i.e. DS recruits GTFI for IgH gene expression and engages the IgH-associated ER complex for autoantibody production. The findings from this study provide further support for a key role of DS in regulating autoAb production and autoreactive B1-cell development. Furthermore, the finding from B-cells of COVID-19 patients point to a potential significance of autoreactive B1 cells in COVID-induced autoimmunity.
Conclusion
Exploiting the affinity between autoAgs and DS glycosaminoglycan, we identified 362 DS-affinity proteins from EBV-immortalized HS-Sultan cells. Of these DS-affinity proteins, 201 are already known autoAgs in a wide variety of autoimmune diseases and cancer. Of the 362, 315 DS-affinity proteins are affected by SARS-CoV-2 infection, and 186 COVID-affected DS-affinity proteins are known autoAgs. These COVID-altered proteins are largely affected by phosphorylation, ubiquitination, or interaction with viral protein components. They are associated with gene expression, mRNA processing, mRNA splicing, translation, protein folding, DNA replication fork, telomerase maintenance, chromosome organization, biosynthesis and catabolism of nucleobase-containing molecules and proteins, vesicles, and nucleocytoplasmic transport. CCT/TriC chaperonin, insulin degrading enzyme, and platelet-activating factor acetylhydrolase are found in the interactomes of multiple viral Nsp and Orf proteins. By integrating DS-affinity autoAgs with multi-omic data from COVID, our study suggests that viral infections can cause significant proteomic alterations, give rise to a diverse pool of autoAgs, and may lead to infection-induced autoimmune diseases. The COVID autoantigen-ome provided in this paper may serve as a molecular map and resource for investigating autoimmune phenomena of SARS-CoV-2 infection and its long-term sequelae. Understanding immunogenic proteins of COVID may also enhance vaccine target design.
Experimental
HS-Sultan cell culture
The human B lymphoblast HS-Sultan cell line was obtained from the ATCC (Manassas, VA) and cultured in complete RPMI-1640 medium. The growth medium was supplemented with 10% fetal bovine serum and a penicillin–streptomycin–glutamine mixture (Thermo Fisher). The cells were grown at 37°C in a CO2 incubator, and about 300 million cells were harvested for the study.
Protein extraction
Protein extraction was performed as previously described.[4] In brief, HS-Sultan cells were lysed with 50 mM phosphate buffer (pH 7.4) containing the Roche Complete Mini protease inhibitor cocktail and then homogenized on ice with a microprobe sonicator until the turbid mixture turned nearly clear with no visible cells left. The homogenate was centrifuged at 10 000g at 4°C for 20 min, and the total protein extract in the supernatant was collected. Protein concentration was measured by absorbance at 280 nm using a NanoDrop UV-Vis spectrometer (Thermo Fisher).
DS-Sepharose resin preparation
The DS-affinity resins were synthesized as previously described.[4,8] In brief, 20 mL of EAH Sepharose 4B resins (GE Healthcare Life Sciences) were washed with distilled water three times and mixed with 100 mg of DS (Sigma-Aldrich) in 10 mL of 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) buffer, pH 5.0. About 100 mg of N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (Sigma-Aldrich) powder was added, and another 100 mg was added after 8 h of reaction. The reaction proceeded by mixing on a rocker at 25°C for 16 h. The coupled resins were washed with water and equilibrated with 0.5 M NaCl in 0.1 M acetate (pH 5.0) and 0.5 M NaCl in 0.1 M Tris (pH 8.0).
DS-affinity fractionation
The total proteomes extracted from HS-Sultan cells were fractionated in a DS-Sepharose column in a manner similar to that previously described.[4] About 40 mg of proteins in 40 mL of 10 mM phosphate buffer (pH 7.4; buffer A) was loaded onto the DS-affinity column at a rate of 1 mL/min. Unbound and weakly bound proteins were removed with 60 mL of buffer A and then 40 mL of 0.2 M NaCl in buffer A. The remaining bound proteins were eluted in steps with 40 mL of 0.5 M NaCl and then with 40 mL of 1.0 M NaCl in buffer A. Fractions were desalted and concentrated with 5 kDa cut-off Vivaspin centrifugal filters (Sartorius). Fractionated proteins were separated in 1-D SDS-PAGE in 4–12% Bis-Tris gels, and the gel lanes were divided into two or three sections for mass spectrometric sequencing.
Mass spectrometry sequencing
Mass spectrometry was performed at Harvard Medical School (Taplin Biological Mass Spectrometry Facility). Protein digestion was carried out with sequencing-grade trypsin (Promega) at 4°C for 45 min. Tryptic peptides were separated by HPLC in a nanoscale C18 capillary column and analyzed in an LTQ linear ion-trap mass spectrometer (Thermo Fisher). Peptide sequences and protein identities were assigned using Sequest. All mass spectrometric data were manually inspected. Only proteins with at least two peptide matches were considered positively identified. Not DS-affinity-enriched HS-Sultan cell protein extracts were used as reference control samples.
COVID data comparison
DS-affinity proteins were compared with currently available COVID-19 multi-omic data compiled in the Coronascape database.[17–38] These data have been obtained with proteomics, phosphoproteomics, interactome, ubiquitome, and RNA-seq techniques. Up- and down-regulated proteins or genes were identified by comparing cells infected vs uninfected by SARS-CoV-2 or COVID-19 patients vs healthy controls. Similarity searches were conducted to identify DS-affinity proteins that are up- and/or down-regulated in viral infection at any omic level.
Protein network analysis
Protein–protein interactions were analyzed by STRING[217] based on both direct physical interaction and indirect functional associations derived from genomic context predictions, high-throughput lab experiments, co-expression, automated text mining, and previous knowledge in databases. Each interaction received a confidence score from 0 to 1, with 1 denoting the highest confidence, indicating the likelihood of an interaction to be true. Pathways and processes enrichment were analyzed with Metascape,[17] using KEGG Pathway, GO Biological Process, Reactome Gene Sets, Canonical Pathways, CORUM, TRRUST, and DiGenBase. All genes in the genome were used as the enrichment background. Terms with a P-value <0.01, a minimum count of three, and an enrichment factor (ratio between the observed counts and the counts expected by chance) >1.5 were collected and grouped into clusters based on their membership similarities. The most statistically significant term within a cluster was chosen to represent the cluster.
Autoantigen literature text mining
Every DS-affinity protein identified in this study was searched for specific autoAbs reported in the PubMed literature. Search keywords included the MeSH keyword ‘autoantibodies’, the protein name or its gene symbol, or alternative names and symbols. Only proteins for which specific autoAbs are reported in PubMed-listed journal articles were considered ‘confirmed’ autoAgs in this study.
Data availability
The data that support this study are available in the article and accompanying online Supplementary material. An earlier version of this manuscript is available as a preprint on bioRxiv (https://doi.org/10.1101/2021.04.05.438500).
Conflicts of interest
JYW is the founder and Chief Scientific Officer of Curandis. WZ was supported by the NIH and declares no competing interests. MWR and VBR are volunteers of Curandis. MHR is a member of the Scientific Advisory Boards of Trans-Hit (Azenta), Universal DX, and Acumino, but these companies have no relation to the study.
Declaration of funding
This work was partially supported by Curandis. MHR acknowledges grants from the NIH/NCI (R21 CA251992, R21 CA263262, and U01 CA263986), a Cycle for Survival Equinox Innovation Grant, an Investigator Grant from the Neuroendocrine Tumor Research Foundation (NETRF), and MSKCC Cancer Center Support Grant P30 CA008748. The funding bodies were not involved in the design of the study and the collection, analysis, and interpretation of data.
Acknowledgements
The authors dedicate this paper to Prof. Ed Nice as part of a special issue for the occasion of his 75th birthday. The authors thank Jung-hyun Rho for technical assistance with experiments. They thank Ross Tomaino and the Taplin Biological Mass Spectrometry facility of Harvard Medical School for expert service with protein sequencing.
References
1 Wang JY, Zhang W, Roehrl MW, Roehrl VB, Roehrl MH. An Autoantigen Atlas from Human Lung HFL1 Cells Offers Clues to Neurological and Diverse Autoimmune Manifestations of COVID-19. Front Immunol 2022; 13: 831849.
| Crossref | Google Scholar | PubMed |
2 Wang JY, Zhang W, Roehrl MW, Roehrl VB, Roehrl MH. An Autoantigen Profile of Human A549 Lung Cells Reveals Viral and Host Etiologic Molecular Attributes of Autoimmunity in COVID-19. J Autoimmun 2021; 120: 102644.
| Crossref | Google Scholar | PubMed |
3 Wang JY, Lee J, Yan M, Rho J-h, Roehrl MHA. Dermatan sulfate interacts with dead cells and regulates CD5+ B-cell fate: implications for a key role in autoimmunity. Am J Pathol 2011; 178(5): 2168-76.
| Crossref | Google Scholar |
4 Rho J-h, Zhang W, Murali M, Roehrl MHA, Wang JY. Human proteins with affinity for dermatan sulfate have the propensity to become autoantigens. Am J Pathol 2011; 178(5): 2177-90.
| Crossref | Google Scholar |
5 Lee J, Rho J-h, Roehrl MH, Wang JY. Dermatan Sulfate Is a Potential Master Regulator of IgH via Interactions with Pre-BCR, GTF2I, and BiP ER Complex in Pre-B Lymphoblasts. Front Immunol 2021; 12: 680212.
| Crossref | Google Scholar |
6 Zhang W, Rho J-h, Roehrl MH, Wang JY. A comprehensive autoantigen-ome of autoimmune liver diseases identified from dermatan sulfate affinity enrichment of liver tissue proteins. BMC Immunol 2019; 20(1): 21.
| Crossref | Google Scholar |
7 Zhang W, Rho J-h, Roehrl MW, Roehrl MH, Wang JY. A repertoire of 124 potential autoantigens for autoimmune kidney diseases identified by dermatan sulfate affinity enrichment of kidney tissue proteins. PLoS One 2019; 14(6): e0219018.
| Crossref | Google Scholar |
8 Wang JY, Zhang W, Rho J-h, Roehrl MW, Roehrl MH. A proteomic repertoire of autoantigens identified from the classic autoantibody clinical test substrate HEp-2 cells. Clin Proteomics 2020; 17: 35.
| Crossref | Google Scholar |
9 Chang SE, Feng A, Meng W, Apostolidis SA, Mack E, Artandi M, et al. New-Onset IgG Autoantibodies in Hospitalized Patients with COVID-19. Nat Commun 2021; 12(1): 5417.
| Crossref | Google Scholar |
10 Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science (New York, NY) 2020; 370(6515): eabd4585.
| Crossref | Google Scholar |
11 Consiglio CR, Cotugno N, Sardh F, Pou C, Amodio D, Rodriguez L, et al. The Immunology of Multisystem Inflammatory Syndrome in Children with COVID-19. Cell. 2020; 183(4): 968-81.e7.
| Crossref | Google Scholar | PubMed |
12 Gruber CN, Patel RS, Trachtman R, Lepow L, Amanat F, Krammer F, et al. Mapping Systemic Inflammation and Antibody Responses in Multisystem Inflammatory Syndrome in Children (MIS-C). Cell. 2020; 183(4): 982-95.e14.
| Crossref | Google Scholar |
13 Franke C, Ferse C, Kreye J, Reincke SM, Sanchez-Sendin E, Rocco A, et al. High frequency of cerebrospinal fluid autoantibodies in COVID-19 patients with neurological symptoms. Brain Behav Immun 2020; 93: 415-9.
| Crossref | Google Scholar |
14 Zhou Y, Han T, Chen J, Hou C, Hua L, He S, et al. Clinical and Autoimmune Characteristics of Severe and Critical Cases of COVID-19. Clin Transl Sci 2020; 13(6): 1077-86.
| Crossref | Google Scholar |
15 Sacchi MC, Tamiazzo S, Stobbione P, Agatea L, De Gaspari P, Stecca A, et al. SARS-CoV-2 Infection as a trigger of autoimmune response. Clin Transl Sci 2020; 14: 898-907.
| Crossref | Google Scholar |
16 Lerma LA, Chaudhary A, Bryan A, Morishima C, Wener MH, Fink SL. Prevalence of autoantibody responses in acute coronavirus disease 2019 (COVID-19). J Transl Autoimmun 2020; 3: 100073.
| Crossref | Google Scholar |
17 Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019; 10(1): 1523.
| Crossref | Google Scholar |
18 Zhang JY, Wang XM, Xing X, Xu Z, Zhang C, Song JW, et al. Single-cell landscape of immunological responses in patients with COVID-19. Nat Immunol 2020; 21(9): 1107-18.
| Crossref | Google Scholar |
19 Davies JP, Almasy KM, McDonald EF, Plate L. Comparative Multiplexed Interactomics of SARS-CoV-2 and Homologous Coronavirus Nonstructural Proteins Identifies Unique and Shared Host-Cell Dependencies. ACS Infect Dis 2020; 6(12): 3174-89.
| Crossref | Google Scholar |
20 Klann K, Bojkova D, Tascher G, Ciesek S, Münch C, Cinatl J. Growth Factor Receptor Signaling Inhibition Prevents SARS-CoV-2 Replication. Mol Cell 2020; 80(1): 164-74.e4.
| Crossref | Google Scholar |
21 Sun J, Ye F, Wu A, Yang R, Pan M, Sheng J, et al. Comparative Transcriptome Analysis Reveals the Intensive Early Stage Responses of Host Cells to SARS-CoV-2 Infection. Front Microbiol 2020; 11: 593857.
| Crossref | Google Scholar |
22 Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 2020; 583(7816): 469-72.
| Crossref | Google Scholar |
23 Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med 2020; 26(7): 1070-6.
| Crossref | Google Scholar |
24 Lieberman NAP, Peddu V, Xie H, Shrestha L, Huang ML, Mears MC, et al. In vivo antiviral host transcriptional response to SARS-CoV-2 by viral load, sex, and age. PLoS Biol 2020; 18(9): e3000849.
| Crossref | Google Scholar |
25 Riva L, Yuan S, Yin X, Martin-Sancho L, Matsunaga N, Pache L, et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature 2020; 586(7827): 113-9.
| Crossref | Google Scholar |
26 Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M, et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020; 182(3): 685-712.e19.
| Crossref | Google Scholar |
27 Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Møller R, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020; 181(5): 1036-45.e9.
| Crossref | Google Scholar |
28 Shen B, Yi X, Sun Y, Bi X, Du J, Zhang C, et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell. 2020; 182(1): 59-72.e15.
| Crossref | Google Scholar |
29 Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 productively infects human gut enterocytes. Science (New York, NY) 2020; 369(6499): 50-4.
| Crossref | Google Scholar |
30 Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020; 583(7816): 459-68.
| Crossref | Google Scholar |
31 Xiong Y, Liu Y, Cao L, Wang D, Guo M, Jiang A, et al. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients. Emerg Microbes Infect 2020; 9(1): 761-70.
| Crossref | Google Scholar |
32 Vanderheiden A, Ralfs P, Chirkova T, Upadhyay AA, Zimmerman MG, Bedoya S, et al. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J Virol 2020; 94(19): e00985-20.
| Crossref | Google Scholar | PubMed |
33 Appelberg S, Gupta S, Svensson Akusjärvi S, Ambikan AT, Mikaeloff F, Saccon E, et al. Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg Microbes Infect 2020; 9(1): 1748-60.
| Crossref | Google Scholar |
34 Stukalov A, Girault V, Grass V, Bergant V, Karayel O, Urban C, et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2020; 594(7862): 246-252.
| Crossref | Google Scholar |
35 Wyler E, Mösbauer K, Franke V, Diag A, Gottula LT, Arsiè R, et al. Transcriptomic profile of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. iScience 2020; 24(3): 102151.
| Crossref | Google Scholar |
36 Li Y, Wang Y, Liu H, Sun W, Ding B, Zhao Y, et al. Urine Proteome of COVID-19 Patients. Urine 2020; 2: 1-8.
| Crossref | Google Scholar |
37 Liao M, Liu Y, Yuan J, Wen Y, Xu G, Zhao J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med 2020; 26(6): 842-4.
| Crossref | Google Scholar |
38 Laurent EMN, Sofianatos Y, Komarova A, Gimeno J-P, Tehrani PS, Kim D-K, et al. Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms [Preprint]. bioRxiv 2020; 2020.08.28.272955.
| Crossref | Google Scholar |
39 Saunders RD, Nakajima ST, Rai SN, Pan J, Gercel-Taylor C, Taylor DD. Alterations in antibody subclass immune reactivity to trophoblast-derived fetal fibronectin and α2-macroglobulin in women with recurrent pregnancy loss. Am J Reprod Immunol 2012; 68(5): 438-49.
| Crossref | Google Scholar |
40 Petrohai A, Nagy G, Bosze S, Hudecz F, Zsiros E, Paragh G, et al. Detection of citrate synthase-reacting autoantibodies after heart transplantation: an epitope mapping study. Transpl Int 2005; 17(12): 834-40.
| Crossref | Google Scholar |
41 Mande PV, Parikh FR, Hinduja I, Zaveri K, Vaidya R, Gajbhiye R, et al. Identification and validation of candidate biomarkers involved in human ovarian autoimmunity. Reprod Biomed Online 2011; 23(4): 471-83.
| Crossref | Google Scholar |
42 van Beers JJBC, Schwarte CM, Stammen-Vogelzangs J, Oosterink E, Božič B, Pruijn GJM. The rheumatoid arthritis synovial fluid citrullinome reveals novel citrullinated epitopes in apolipoprotein E, myeloid nuclear differentiation antigen, and β-actin. Arthritis Rheum 2013; 65(1): 69-80.
| Crossref | Google Scholar |
43 Zhao Z, Deocharan B, Scherer PE, Ozelius LJ, Putterman C. Differential binding of cross-reactive anti-DNA antibodies to mesangial cells: the role of α-actinin. J Immunol 2006; 176(12): 7704-14.
| Crossref | Google Scholar |
44 Moritz CP, Tholance Y, Stoevesandt O, Ferraud K, Camdessanché JP, Antoine JC. CIDP Antibodies Target Junction Proteins and Identify Patient Subgroups: An Autoantigenomic Approach. Neurol Neuroimmunol Neuroinflamm 2021; 8(2): e944.
| Crossref | Google Scholar |
45 Rucksaken R, Haonon O, Pinlaor P, Pairojkul C, Roytrakul S, Yongvanit P, et al. Plasma IgG autoantibody against actin-related protein 3 in liver fluke Opisthorchis viverrini infection. Parasite Immunol 2015; 37(7): 340-8.
| Crossref | Google Scholar |
46 Wang T, Liu M, Zheng SJ, Bian DD, Zhang JY, Yao J, et al. Tumor-associated autoantibodies are useful biomarkers in immunodiagnosis of α-fetoprotein-negative hepatocellular carcinoma. World J Gastroenterol 2017; 23(19): 3496-504.
| Crossref | Google Scholar |
47 Privitera D, Corti V, Alessio M, Volontè A, Lampasona V, Comi G, et al. Proteomic identification of aldolase A as an autoantibody target in patients with atypical movement disorders. Neurol Sci 2013; 34(3): 313-20.
| Crossref | Google Scholar |
48 Morohoshi K, Ohbayashi M, Patel N, Chong V, Bird AC, Ono SJ. Identification of anti-retinal antibodies in patients with age-related macular degeneration. Exp Mol Pathol 2012; 93(2): 193-9.
| Crossref | Google Scholar |
49 Pott MC, Frede N, Wanders J, Hammarström L, Glocker EO, Glocker C, et al. Autoantibodies against BAFF, APRIL or IL21 - an alternative pathogenesis for antibody-deficiencies? BMC Immunol 2017; 18(1): 34.
| Crossref | Google Scholar |
50 Hrycek A, Cieślik P. Annexin A5 and anti-annexin antibodies in patients with systemic lupus erythematosus. Rheumatol Int 2012; 32(5): 1335-42.
| Crossref | Google Scholar |
51 Seko Y, Matsumoto A, Fukuda T, Imai Y, Fujimura T, Taka H, et al. A case of neonatal lupus erythematosus presenting delayed dilated cardiomyopathy with circulating autoantibody to annexin A6. Int Heart J 2007; 48(3): 407-15.
| Crossref | Google Scholar |
52 Katsumata Y, Kawaguchi Y, Baba S, Hattori S, Tahara K, Ito K, et al. Identification of three new autoantibodies associated with systemic lupus erythematosus using two proteomic approaches. Mol Cell Proteomics 2011; 10(6): M110.005330.
| Crossref | Google Scholar |
53 Kamburova EG, Gruijters ML, Kardol-Hoefnagel T, Wisse BW, Joosten I, Allebes WA, et al. Antibodies against ARHGDIB are associated with long-term kidney graft loss. Am J Transplant 2019; 19(12): 3335-44.
| Crossref | Google Scholar |
54 Heo CK, Hwang HM, Lim WH, Lee HJ, Yoo JS, Lim KJ, et al. Cyclic Peptide Mimotopes for the Detection of Serum Anti-ATIC Autoantibody Biomarker in Hepato-Cellular Carcinoma. Int J Mol Sci 2020; 21(24): 9718.
| Crossref | Google Scholar |
55 Creaney J, Dick IM, Yeoman D, Wong S, Robinson BWS. Auto-antibodies to β-F1-ATPase and vimentin in malignant mesothelioma. PLoS One 2011; 6(10): e26515.
| Crossref | Google Scholar |
56 Beutgen VM, Schmelter C, Pfeiffer N, Grus FH. Autoantigens in the trabecular meshwork and glaucoma-specific alterations in the natural autoantibody repertoire. Clin Transl Immunol 2020; 9(3): e01101.
| Crossref | Google Scholar |
57 Boehm J, Orth T, Van Nguyen P, Söling H-D. Systemic lupus erythematosus is associated with increased auto-antibody titers against calreticulin and Grp94, but calreticulin is not the Ro/SS-A antigen. Eur J Clin Invest 1994; 24(4): 248-57.
| Crossref | Google Scholar |
58 Matsuo K, Xiang Y, Nakamura H, Masuko K, Yudoh K, Noyori K, et al. Identification of novel citrullinated autoantigens of synovium in rheumatoid arthritis using a proteomic approach. Arthritis Res Ther 2006; 8(6): R175.
| Crossref | Google Scholar |
59 Li WH, Zhao J, Li HY, Liu H, Li AL, Wang HX, et al. Proteomics-based identification of autoantibodies in the sera of healthy Chinese individuals from Beijing. Proteomics 2006; 6(17): 4781-9.
| Crossref | Google Scholar |
60 Yokota SI, Hirata D, Minota S, Higashiyama T, Kurimoto M, Yanagi H, et al. Autoantibodies against chaperonin CCT in human sera with rheumatic autoimmune diseases: comparison with antibodies against other Hsp60 family proteins. Cell Stress Chaperones 2000; 5(4): 337-46.
| Crossref | Google Scholar |
61 Hirai K, Maeda H, Omori K, Yamamoto T, Kokeguchi S, Takashiba S. Serum antibody response to group II chaperonin from Methanobrevibacter oralis and human chaperonin CCT. Pathog Dis 2013; 68(1): 12-9.
| Crossref | Google Scholar |
62 Goto M, Kuribayashi K, Takahashi Y, Kondoh T, Tanaka M, Kobayashi D, et al. Identification of autoantibodies expressed in acquired aplastic anaemia. Br J Haematol 2013; 160(3): 359-62.
| Crossref | Google Scholar |
63 Rouwette M, Noben JP, Van Horssen J, Van Wijmeersch B, Hupperts R, Jongen PJ, et al. Identification of coronin-1a as a novel antibody target for clinically isolated syndrome and multiple sclerosis. J Neurochem 2013; 126(4): 483-92.
| Crossref | Google Scholar |
64 Bénistant C, Bourgaux JF, Chapuis H, Mottet N, Roche S, Bali JP. The COOH-terminal Src kinase Csk is a tumor antigen in human carcinoma. Cancer Res 2001; 61(4): 1415-20.
| Google Scholar |
65 Schneider F, Aggarwal R, Bi D, Gibson K, Oddis C, Yousem SA. The pulmonary histopathology of anti-KS transfer RNA synthetase syndrome. Arch Pathol Lab Med 2015; 139(1): 122-5.
| Crossref | Google Scholar |
66 Netter HJ, Will H, Szostecki C, Guldner HH. Repetitive P68-autoantigen specific epitopes recognized by human anti-(U1) small nuclear ribonucleoprotein autoantibodies. J Autoimmun 1991; 4(4): 651-63.
| Crossref | Google Scholar |
67 Yamasaki Y, Narain S, Yoshida H, Hernandez L, Barker T, Hahn PC, et al. Autoantibodies to RNA helicase A: a new serologic marker of early lupus. Arthritis Rheum 2007; 56(2): 596-604.
| Crossref | Google Scholar |
68 Yoneyama K, Shibata R, Igarashi A, Kojima S, Kodani Y, Nagata K, et al. Proteomic identification of dihydrolipoamide dehydrogenase as a target of autoantibodies in patients with endometrial cancer. Anticancer Res 2014; 34(9): 5021-7.
| Google Scholar |
69 Kim EG, Kwak SH, Hwang D, Yi EC, Park KS, Koo BK, et al. The Level of Autoantibodies Targeting Eukaryote Translation Elongation Factor 1 α1 and Ubiquitin-Conjugating Enzyme 2L3 in Nondiabetic Young Adults. Diabetes Metab J 2016; 40(2): 154-60.
| Crossref | Google Scholar |
70 Mooney CJ, Dunphy EJ, Stone B, McNeel DG. Identification of autoantibodies elicited in a patient with prostate cancer presenting as dermatomyositis. Int J Urol 2006; 13(3): 211-7.
| Crossref | Google Scholar |
71 Ohkouchi K, Mizutani H, Tanaka M, Takahashi M, Nakashima K, Shimizu M. Anti-elongation factor-1α autoantibody in adult atopic dermatitis patients. Int Immunol 1999; 11(10): 1635-40.
| Crossref | Google Scholar |
72 Fernández-Madrid F, Tang N, Alansari H, Granda JL, Tait L, Amirikia KC, et al. Autoantibodies to Annexin XI-A and Other Autoantigens in the Diagnosis of Breast Cancer. Cancer Res 2004; 64(15): 5089-96.
| Crossref | Google Scholar |
73 Bach M, Winkelmann G, Lührmann R. 20S small nuclear ribonucleoprotein U5 shows a surprisingly complex protein composition. Proc Natl Acad Sci U S A 1989; 86(16): 6038-42.
| Crossref | Google Scholar |
74 Heo CK, Hwang HM, Lee HJ, Kwak SS, Yoo JS, Yu DY, et al. Serum anti-EIF3A autoantibody as a potential diagnostic marker for hepatocellular carcinoma. Sci Rep 2019; 9(1): 11059.
| Crossref | Google Scholar |
75 Chen X, Fu S, Chen F, Chen H, Chen Z. Identification of tumor-associated antigens in human hepatocellular carcinoma by autoantibodies. Oncol Rep 2008; 20(4): 979-85.
| Crossref | Google Scholar |
76 Betteridge Z, Chinoy H, Vencovsky J, Winer J, Putchakayala K, Ho P, et al. Identification of a novel autoantigen eukaryotic initiation factor 3 associated with polymyositis. Rheumatology (Oxford, England) 2020; 59(5): 1026-30.
| Crossref | Google Scholar |
77 Suwarnalata G, Tan AH, Isa H, Gudimella R, Anwar A, Loke MF, et al. Augmentation of Autoantibodies by Helicobacter pylori in Parkinson's Disease Patients May Be Linked to Greater Severity. PLoS One 2016; 11(4): e0153725.
| Crossref | Google Scholar |
78 Pagaza-Straffon C, Marchat LA, Herrera L, Díaz-Chávez J, Avante MG, Rodríguez YP, et al. Evaluation of a panel of tumor-associated antigens in breast cancer. Cancer Biomark 2020; 27(2): 207-11.
| Crossref | Google Scholar |
79 Nabors LB, Furneaux HM, King PH. HuR, a novel target of anti-Hu antibodies, is expressed in non-neural tissues. J Neuroimmunol 1998; 92(1-2): 152-9.
| Crossref | Google Scholar |
80 O'Dwyer DT, Clifton V, Hall A, Smith R, Robinson PJ, Crock PA. Pituitary autoantibodies in lymphocytic hypophysitis target both γ- and a-Enolase – a link with pregnancy? Arch Physiol Biochem 2002; 110(1–2): 94-8.
| Crossref | Google Scholar |
81 Targoff IN, Trieu EP, Miller FW. Reaction of anti-OJ autoantibodies with components of the multi-enzyme complex of aminoacyl-tRNA synthetases in addition to isoleucyl-tRNA synthetase. J Clin Invest 1993; 91(6): 2556-64.
| Crossref | Google Scholar |
82 Capello M, Cappello P, Linty FC, Chiarle R, Sperduti I, Novarino A, et al. Autoantibodies to Ezrin are an early sign of pancreatic cancer in humans and in genetically engineered mouse models. J Hematol Oncol 2013; 6: 67.
| Crossref | Google Scholar |
83 Betteridge Z, Gunawardena H, North J, Slinn J, McHugh N. Anti-synthetase syndrome: a new autoantibody to phenylalanyl transfer RNA synthetase (anti-Zo) associated with polymyositis and interstitial pneumonia. Rheumatology (Oxford, England) 2007; 46(6): 1005-8.
| Crossref | Google Scholar |
84 Heo CK, Woo MK, Yu DY, Lee JY, Yoo JS, Yoo HS, et al. Identification of autoantibody against fatty acid synthase in hepatocellular carcinoma mouse model and its application to diagnosis of HCC. Int J Oncol 2010; 36(6): 1453-9.
| Crossref | Google Scholar |
85 Desmetz C, Bascoul-Mollevi C, Rochaix P, Lamy PJ, Kramar A, Rouanet P, et al. Identification of a new panel of serum autoantibodies associated with the presence of in situ carcinoma of the breast in younger women. Clin Cancer Res 2009; 15(14): 4733-41.
| Crossref | Google Scholar |
86 Terrier B, Tamby MC, Camoin L, Guilpain P, Broussard C, Bussone G, et al. Identification of target antigens of antifibroblast antibodies in pulmonary arterial hypertension. Am J Respir Crit Care Med 2008; 177(10): 1128-34.
| Crossref | Google Scholar |
87 Kit Y, Starykovych M, Vajrychova M, Lenco J, Zastavna D, Stoika R. Detection of novel auto-antigens in patients with recurrent miscarriage: description of an approach and preliminary findings. Croat Med J 2014; 55(3): 259-64.
| Crossref | Google Scholar |
88 Takasaki Y, Kaneda K, Matsushita M, Yamada H, Nawata M, Matsudaira R, et al. Glyceraldehyde 3-phosphate dehydrogenase is a novel autoantigen leading autoimmune responses to proliferating cell nuclear antigen multiprotein complexes in lupus patients. Int Immunol 2004; 16(9): 1295-304.
| Crossref | Google Scholar |
89 Schneider F, Yousem SA, Bi D, Gibson KF, Oddis CV, Aggarwal R. Pulmonary pathologic manifestations of anti-glycyl-tRNA synthetase (anti-EJ)-related inflammatory myopathy. J Clin Pathol 2014; 67(8): 678-83.
| Crossref | Google Scholar |
90 Kiyota A, Iwama S, Sugimura Y, Takeuchi S, Takagi H, Iwata N, et al. Identification of the novel autoantigen candidate Rab GDP dissociation inhibitor alpha in isolated adrenocorticotropin deficiency. Endocr J 2015; 62(2): 153-60.
| Crossref | Google Scholar |
91 Massa O, Alessio M, Russo L, Nardo G, Bonetto V, Bertuzzi F, et al. Serological Proteome Analysis (SERPA) as a tool for the identification of new candidate autoantigens in type 1 diabetes. J Proteomics 2013; 82: 263-73.
| Crossref | Google Scholar |
92 Carlsson L, Ronquist G, Nilsson BO, Larsson A. Dominant prostasome immunogens for sperm-agglutinating autoantibodies of infertile men. J Androl 2004; 25(5): 699-705.
| Crossref | Google Scholar |
93 Belousov PV, Afanasyeva MA, Gubernatorova EO, Bogolyubova AV, Uvarova AN, Putlyaeva LV, et al. Multi-dimensional immunoproteomics coupled with in vitro recapitulation of oncogenic NRASQ61R identifies diagnostically relevant autoantibody biomarkers in thyroid neoplasia. Cancer Lett 2019; 467: 96-106.
| Crossref | Google Scholar |
94 Rubin RL, Bell SA, Burlingame RW. Autoantibodies associated with lupus induced by diverse drugs target a similar epitope in the (H2A-H2B)-DNA complex. J Clin Invest 1992; 90(1): 165-73.
| Crossref | Google Scholar |
95 Burlingame RW, Boey ML, Starkebaum G, Rubin RL. The central role of chromatin in autoimmune responses to histones and DNA in systemic lupus erythematosus. J Clin Invest 1994; 94(1): 184-92.
| Crossref | Google Scholar |
96 Rosenberg AM, Cordeiro DM. Relationship between sex and antibodies to high mobility group proteins 1 and 2 in juvenile idiopathic arthritis. J Rheumatol 2000; 27(10): 2489-93.
| Google Scholar |
97 Wesierska-Gadek J, Penner E, Lindner H, Hitchman E, Sauermann G. Autoantibodies against different histone H1 subtypes in systemic lupus erythematosus sera. Arthritis Rheum 1990; 33(8): 1273-8.
| Crossref | Google Scholar |
98 Vordenbäumen S, Böhmer P, Brinks R, Fischer-Betz R, Richter J, Bleck E, et al. High diagnostic accuracy of histone H4-IgG autoantibodies in systemic lupus erythematosus. Rheumatology (Oxford, England) 2018; 57(3): 533-7.
| Crossref | Google Scholar |
99 Baranova SV, Dmitrienok PS, Ivanisenko NV, Buneva VN, Nevinsky GA. Antibodies to H2a and H2b histones from the sera of HIV-infected patients catalyze site-specific degradation of these histones. Mol Biosyst 2017; 13(6): 1090-101.
| Crossref | Google Scholar |
100 Bruschi M, Galetti M, Sinico RA, Moroni G, Bonanni A, Radice A, et al. Glomerular Autoimmune Multicomponents of Human Lupus Nephritis In Vivo (2): Planted Antigens. J Am Soc Nephrol 2015; 26(8): 1905-24.
| Crossref | Google Scholar |
101 van Bavel CC, Dieker J, Muller S, Briand JP, Monestier M, Berden JH, et al. Apoptosis-associated acetylation on histone H2B is an epitope for lupus autoantibodies. Mol Immunol 2009; 47(2-3): 511-6.
| Crossref | Google Scholar |
102 Baranova SV, Dmitrenok PS, Zubkova AD, Ivanisenko NV, Odintsova ES, Buneva VN, et al. Antibodies against H3 and H4 histones from the sera of HIV-infected patients catalyze site-specific degradation of these histones. J Mol Recognit 2018; 31(7): e2703.
| Crossref | Google Scholar |
103 Astaldi Ricotti GCB, Bestagno M, Cerino A, Negri C, Caporali R, Cobianchi F, et al. Antibodies to hnRNP core protein A1 in connective tissue diseases. J Cell Biochem 1989; 40(1): 43-7.
| Crossref | Google Scholar |
104 Konig MF, Giles JT, Nigrovic PA, Andrade F. Antibodies to native and citrullinated RA33 (hnRNP A2/B1) challenge citrullination as the inciting principle underlying loss of tolerance in rheumatoid arthritis. Ann Rheum Dis 2016; 75(11): 2022-8.
| Crossref | Google Scholar |
105 Siapka S, Patrinou-Georgoula M, Vlachoyiannopoulos PG, Guialis A. Multiple specificities of autoantibodies against hnRNP A/B proteins in systemic rheumatic diseases and hnRNP L as an associated novel autoantigen. Autoimmunity 2007; 40(3): 223-33.
| Crossref | Google Scholar |
106 Vencovský J, Kafková J, Staněk D, Raška I. Heterogenous nuclear RNP C1 and C2 core proteins are targets for an autoantibody found in the serum of a patient with systemic sclerosis and psoriatic arthritis. Arthritis Rheum 1997; 40(12): 2172-7.
| Crossref | Google Scholar |
107 Skriner K, Hueber W, Süleymanoglu E, Höfler E, Krenn V, Smolen J, et al. AUF1, the regulator of tumor necrosis factor α messenger RNA decay, is targeted by autoantibodies of patients with systemic rheumatic diseases. Arthritis Rheum 2008; 58(2): 511-20.
| Crossref | Google Scholar |
108 Zhang Y, Zhao H, Liu B, Li L, Zhang L, Bao M, et al. Low Level Antibodies Against Alpha-Tropomyosin Are Associated With Increased Risk of Coronary Heart Disease. Front Pharmacol 2020; 11: 195.
| Crossref | Google Scholar |
109 Op De Beéck K, Maes L, Van den Bergh K, Derua R, Waelkens E, Van Steen K, et al. Heterogeneous nuclear RNPs as targets of autoantibodies in systemic rheumatic diseases. Arthritis Rheum 2012; 64(1): 213-21.
| Crossref | Google Scholar |
110 Rauch J, Ahlemann M, Schaffrik M, Mack B, Ertongur S, Andratschke M, et al. Allogenic antibody-mediated identification of head and neck cancer antigens. Biochem Biophys Res Commun 2004; 323(1): 156-62.
| Crossref | Google Scholar |
111 Zhang JY, Chan EKL, Peng XX, Tan EM. A novel cytoplasmic protein with RNA-binding motifs is an autoantigen in human hepatocellular carcinoma. J Exp Med 1999; 189(7): 1101-10.
| Crossref | Google Scholar |
112 Hassfeld W, Chan EKL, Mathison DA, Portman D, Dreyfuss G, Steiner G, et al. Molecular definition of heterogeneous nuclear ribonucleoprotein R (hnRNP R) using autoimmune antibody: immunological relationship with hnRNP P. Nucleic Acids Res 1998; 26(2): 439-45.
| Crossref | Google Scholar |
113 Britton S, Froment C, Frit P, Monsarrat B, Salles B, Calsou P. Cell nonhomologous end joining capacity controls SAF-A phosphorylation by DNA-PK in response to DNA double-strand breaks inducers. Cell Cycle 2009; 8(22): 3717-22.
| Crossref | Google Scholar |
114 Harlow L, Rosas IO, Gochuico BR, Mikuls TR, Dellaripa PF, Oddis CV, et al. Identification of citrullinated hsp90 isoforms as novel autoantigens in rheumatoid arthritis-associated interstitial lung disease. Arthritis Rheum 2013; 65(4): 869-79.
| Crossref | Google Scholar |
115 Qin HY, Mahon JL, Atkinson MA, Chaturvedi P, Lee-Chan E, Singh B. Type 1 diabetes alters anti-hsp90 autoantibody isotype. J Autoimmun 2003; 20(3): 237-45.
| Crossref | Google Scholar |
116 Pashov A, Kenderov A, Kyurkchiev S, Kehayov I, Hristova S, Lacroix‐Desmazes S, et al. Autoantibodies to heat shock protein 90 in the human natural antibody repertoire. Int Immunol 2002; 14(5): 453-61.
| Crossref | Google Scholar |
117 Tishler M, Shoenfeld Y. Anti-heat-shock protein antibodies in rheumatic and autoimmune diseases. Semin Arthritis Rheum 1996; 26(2): 558-63.
| Crossref | Google Scholar |
118 Bläß S, Union A, Raymackers J, Schumann F, Ungethüm U, Müller-Steinbach S, et al. The stress protein BiP is overexpressed and is a major B and T cell target in rheumatoid arthritis. Arthritis Rheum 2001; 44(4): 761-71.
| Crossref | Google Scholar |
119 Munakata S, Chen M, Aosai F, Kawaguchi N, Nemoto Y, Norose K, et al. The clinical significance of anti-heat shock cognate protein 71 antibody in myasthenia gravis. J Clin Neurosci 2008; 15(2): 158-65.
| Crossref | Google Scholar |
120 Alard JE, Dueymes M, Mageed RA, Saraux A, Youinou P, Jamin C. Mitochondrial heat shock protein (HSP) 70 synergizes with HSP60 in transducing endothelial cell apoptosis induced by anti-HSP60 autoantibody. FASEB J 2009; 23(8): 2772-9.
| Crossref | Google Scholar |
121 Horváth L, Cervenak L, Oroszlán M, Prohászka Z, Uray K, Hudecz F, et al. Antibodies against different epitopes of heat-shock protein 60 in children with type 1 diabetes mellitus. Immunol Lett 2002; 80(3): 155-62.
| Crossref | Google Scholar |
122 Minohara M. [Heat shock protein 105 in multiple sclerosis]. Nihon Rinsho 2003; 61(8): 1317-22.
| Google Scholar |
123 Bremer HD, Landegren N, Sjöberg R, Hallgren Å, Renneker S, Lattwein E, et al. ILF2 and ILF3 are autoantigens in canine systemic autoimmune disease. Sci Rep 2018; 8(1): 4852.
| Crossref | Google Scholar |
124 Ola TO, Biro PA, Hawa MI, Ludvigsson J, Locatelli M, Puglisi MA, et al. Importin beta: a novel autoantigen in human autoimmunity identified by screening random peptide libraries on phage. J Autoimmun 2006; 26(3): 197-207.
| Crossref | Google Scholar |
125 Luo LY, Herrera I, Soosaipillai A, Diamandis EP. Identification of heat shock protein 90 and other proteins as tumour antigens by serological screening of an ovarian carcinoma expression library. Br J Cancer 2002; 87(3): 339-43.
| Crossref | Google Scholar |
126 Presslauer S, Hinterhuber G, Cauza K, Wolff K, Foedinger D, Horvat R, et al. RasGAP-like protein IQGAP1 is expressed by human keratinocytes and recognized by autoantibodies in association with bullous skin disease. J Invest Dermatol 2003; 120(3): 365-71.
| Crossref | Google Scholar |
127 Ueda K, Nakanishi T, Shimizu A, Takubo T, Matsuura N. Identification of L-plastin autoantibody in plasma of patients with non-Hodgkin's lymphoma using a proteomics-based analysis. Ann Clin Biochem 2008; 45(1): 65-9.
| Crossref | Google Scholar |
128 Forti S, Scanlan MJ, Invernizzi A, Castiglioni F, Pupa S, Agresti R, et al. Identification of breast cancer-restricted antigens by antibody screening of SKBR3 cDNA library using a preselected patient's serum. Breast Cancer Res Treat 2002; 73(3): 245-56.
| Crossref | Google Scholar |
129 Gemma R, Suzuki Y, Tanaka I, Taminato T, Yoshimi T, Kanno T. Lactate dehydrogenase (LDH)-linked immunoglobulin in a patient with Graves' disease treated with methimazole. Intern Med 1992; 31(3): 377-9.
| Crossref | Google Scholar |
130 Tanaka M, Kishimura M, Ozaki S, Osakada F, Hashimoto H, Okubo M, et al. Cloning of novel soluble gp130 and detection of its neutralizing autoantibodies in rheumatoid arthritis. J Clin Invest 2000; 106(1): 137-44.
| Crossref | Google Scholar |
131 Frampton G, Moriya S, Pearson JD, Isenberg DA, Ward FJ, Smith TA, et al. Identification of candidate endothelial cell autoantigens in systemic lupus erythematosus using a molecular cloning strategy: a role for ribosomal P protein P0 as an endothelial cell autoantigen. Rheumatology (Oxford, England) 2000; 39(10): 1114-20.
| Crossref | Google Scholar |
132 Suzuki K, Nagao T, Itabashi M, Hamano Y, Sugamata R, Yamazaki Y, et al. A novel autoantibody against moesin in the serum of patients with MPO-ANCA-associated vasculitis. Nephrol Dial Transplant 2014; 29(6): 1168-77.
| Crossref | Google Scholar |
133 Mossabeb R, Seiberler S, Mittermann I, Natter S, Kraft D, Valenta R, et al. Characterization of a novel isoform of α-nascent polypeptide-associated complex as IgE-defined autoantigen. J Invest Dermatol 2002; 119(4): 820-9.
| Crossref | Google Scholar |
134 Beaulande M, Kron M, Härtlein M. Human anti-asparaginyl-tRNA synthetase autoantibodies (anti-KS) increase the affinity of the enzyme for its tRNA substrate. FEBS Lett 2001; 494(3): 170-4.
| Crossref | Google Scholar |
135 Batova IN, Richardson RT, Widgren EE, O'Rand MG. Analysis of the autoimmune epitopes on human testicular NASP using recombinant and synthetic peptides. Clin Exp Immunol 2000; 121(2): 201-9.
| Crossref | Google Scholar |
136 Qin Z, Lavingia B, Zou Y, Stastny P. Antibodies against nucleolin in recipients of organ transplants. Transplantation 2011; 92(7): 829-35.
| Crossref | Google Scholar |
137 Le Naour F, Brichory F, Misek DE, Bréchot C, Hanash SM, Beretta L. A distinct repertoire of autoantibodies in hepatocellular carcinoma identified by proteomic analysis. Mol Cell Proteomics 2002; 1(3): 197-203.
| Crossref | Google Scholar |
138 Ulanet DB, Torbenson M, Dang CV, Casciola-Rosen L, Rosen A. Unique conformation of cancer autoantigen B23 in hepatoma: a mechanism for specificity in the autoimmune response. Proc Natl Acad Sci U S A 2003; 100(21): 12361-6.
| Crossref | Google Scholar |
139 Nagayama S, Yokoi T, Tanaka H, Kawaguchi Y, Shirasaka T, Kamataki T. Occurrence of autoantibody to protein disulfide isomerase in patients with hepatic disorder. J Toxicol Sci 1994; 19(3): 163-9.
| Crossref | Google Scholar |
140 Becker A, Ludwig N, Keller A, Tackenberg B, Eienbröker C, Oertel WH, et al. Myasthenia gravis: analysis of serum autoantibody reactivities to 1827 potential human autoantigens by protein macroarrays. PLoS One 2013; 8(3): e58095.
| Crossref | Google Scholar |
141 Caorsi C, Niccolai E, Capello M, Vallone R, Chattaragada MS, Alushi B, et al. Protein disulfide isomerase A3-specific Th1 effector cells infiltrate colon cancer tissue of patients with circulating anti-protein disulfide isomerase A3 autoantibodies. Transl Res 2016; 171: 17-28.e1-2.
| Crossref | Google Scholar |
142 Gut J, Christen U, Frey N, Koch V, Stoffler D. Molecular mimicry in halothane hepatitis: biochemical and structural characterization of lipoylated autoantigens. Toxicology 1995; 97(1–3): 199-224.
| Crossref | Google Scholar |
143 Chang DC, Piaggi P, Hanson RL, Knowler WC, Bogardus C, Krakoff J. Autoantibodies against PFDN2 are associated with an increased risk of type 2 diabetes: a case-control study. Diabetes Metab Res Rev 2017; 33(8): e2922.
| Crossref | Google Scholar |
144 Kimura A, Sakurai T, Koumura A, Yamada M, Hayashi Y, Tanaka Y, et al. High prevalence of autoantibodies against phosphoglycerate mutase 1 in patients with autoimmune central nervous system diseases. J Neuroimmunol 2010; 219(1–2): 105-8.
| Crossref | Google Scholar |
145 Xiang D, Xia Q, Chen D, Feng X, Zhao Y, Liu Y, et al. Detection of D-3-phosphoglycerate dehydrogenase autoantibodies in patients with autoimmune hepatitis: clinical significance evaluation. Hepatol Res 2011; 41(9): 867-76.
| Crossref | Google Scholar |
146 Loshaj-Shala A, Colzani M, Brezovska K, Poceva Panovska A, Suturkova L, Beretta G. Immunoproteomic identification of antigenic candidate Campylobacter jejuni and human peripheral nerve proteins involved in Guillain-Barré syndrome. J Neuroimmunol 2018; 317: 77-83.
| Crossref | Google Scholar |
147 Kratz A, Harding MW, Craft J, Mackworth-Young CG, Handschumacher RE. Autoantibodies against cyclophilin in systemic lupus erythematosus and Lyme disease. Clin Exp Immunol 1992; 90(3): 422-7.
| Crossref | Google Scholar |
148 Karasawa R, Ozaki S, Nishioka K, Kato T. Autoantibodies to peroxiredoxin I and IV in patients with systemic autoimmune diseases. Microbiol Immunol 2005; 49(1): 57-65.
| Crossref | Google Scholar |
149 Lin LH, Xu YW, Huang LS, Hong CQ, Zhai TT, Liao LD, et al. Serum proteomic-based analysis identifying autoantibodies against PRDX2 and PRDX3 as potential diagnostic biomarkers in nasopharyngeal carcinoma. Clin Proteomics 2017; 14: 6.
| Crossref | Google Scholar |
150 Sun KH, Tang SJ, Wang YS, Lin WJ, You RI. Autoantibodies to dsDNA cross-react with the arginine–glycine-rich domain of heterogeneous nuclear ribonucleoprotein A2 (hnRNP A2) and promote methylation of hnRNP A2. Rheumatology (Oxford) 2003; 42(1): 154-61.
| Crossref | Google Scholar |
151 Mayo I, Arribas J, Villoslada P, Alvarez DoForno R, Rodriguez-Vilarino S, Montalban X, et al. The proteasome is a major autoantigen in multiple sclerosis. Brain 2002; 125(12): 2658-67.
| Crossref | Google Scholar |
152 Feist E, Kuckelkorn U, Dörner T, Dönitz H, Scheffler S, Hiepe F, et al. Autoantibodies in primary Sjögren's syndrome are directed against proteasomal subunits of the α and β type. Arthritis Rheum 1999; 42(4): 697-702.
| Crossref | Google Scholar |
153 Feist E, Dörner T, Kuckelkorn U, Schmidtke G, Micheel B, Hiepe F, et al. Proteasome alpha-type subunit C9 is a primary target of autoantibodies in sera of patients with myositis and systemic lupus erythematosus. J Exp Med 1996; 184(4): 1313-8.
| Crossref | Google Scholar |
154 Bohring C, Krause W. Characterization of spermatozoa surface antigens by antisperm antibodies and its influence on acrosomal exocytosis. Am J Reprod Immunol 2003; 50(5): 411-9.
| Crossref | Google Scholar |
155 Sugimoto K, Hiwasa T, Shibuya K, Hirano S, Beppu M, Isose S, et al. Novel autoantibodies against the proteasome subunit PSMA7 in amyotrophic lateral sclerosis. J Neuroimmunol 2018; 325: 54-60.
| Crossref | Google Scholar |
156 Scheffler S, Kuckelkorn U, Egerer K, Dörner T, Reiter K, Soza A, et al. Autoimmune reactivity against the 20S-proteasome includes immunosubunits LMP2 (β1i), MECL1 (β2i) and LMP7 (β5i). Rheumatology (Oxford, England) 2008; 47(5): 622-6.
| Crossref | Google Scholar |
157 Luna Coronell JA, Sergelen K, Hofer P, Gyurján I, Brezina S, Hettegger P, et al. The Immunome of Colon Cancer: Functional In Silico Analysis of Antigenic Proteins Deduced from IgG Microarray Profiling. Genom Proteom Bioinform 2018; 16(1): 73-84.
| Crossref | Google Scholar |
158 Roessler M, Rollinger W, Mantovani-Endl L, Hagmann ML, Palme S, Berndt P, et al. Identification of PSME3 as a novel serum tumor marker for colorectal cancer by combining two-dimensional polyacrylamide gel electrophoresis with a strictly mass spectrometry-based approach for data analysis. Mol Cell Proteomics 2006; 5(11): 2092-101.
| Crossref | Google Scholar |
159 Montecucco C, Caporali R, Cobianchi F, Biamonti G. Identification of autoantibodies to the I protein of the heterogeneous nuclear ribonucleoprotein complex in patients with systemic sclerosis. Arthritis Rheum 1996; 39(10): 1669-76.
| Crossref | Google Scholar |
160 Thébault S, Gilbert D, Hubert M, Drouot L, Machour N, Lange C, et al. Orderly pattern of development of the autoantibody response in (New Zealand White × BXSB)F1 lupus mice: characterization of target antigens and antigen spreading by two-dimensional gel electrophoresis and mass spectrometry. J Immunol (Baltimore, MD: 1950) 2002; 169(7): 4046-53.
| Crossref | Google Scholar |
161 Wagatsuma M, Kimura M, Suzuki R, Takeuchi F, Matsuta K, Watanabe H. Ezrin, radixin and moesin are possible auto-immune antigens in rheumatoid arthritis. Mol Immunol 1996; 33(15): 1171-6.
| Crossref | Google Scholar |
162 Gerli R, Caponi L. Anti-ribosomal P protein antibodies. Autoimmunity 2005; 38(1): 85-92.
| Crossref | Google Scholar |
163 Tycowski KT, Shu MD, Steitz JA. A small nucleolar RNA is processed from an intron of the human gene encoding ribosomal protein S3. Genes Dev 1993; 7(7a): 1176-90.
| Crossref | Google Scholar |
164 Kaji K, Fertig N, Medsger Jr TA, Satoh T, Hoshino K, Hamaguchi Y, et al. Autoantibodies to RuvBL1 and RuvBL2: a novel systemic sclerosis-related antibody associated with diffuse cutaneous and skeletal muscle involvement. Arthritis Care Res 2014; 66(4): 575-84.
| Crossref | Google Scholar |
165 Pauling JD, Salazar G, Lu H, Betteridge ZE, Assassi S, Mayes MD, et al. Presence of anti-eukaryotic initiation factor-2B, anti-RuvBL1/2 and anti-synthetase antibodies in patients with anti-nuclear antibody negative systemic sclerosis. Rheumatology (Oxford, England) 2018; 57(4): 712-7.
| Crossref | Google Scholar |
166 Margutti P, Sorice M, Conti F, Delunardo F, Racaniello M, Alessandri C, et al. Screening of an endothelial cDNA library identifies the C-terminal region of Nedd5 as a novel autoantigen in systemic lupus erythematosus with psychiatric manifestations. Arthritis Res Ther 2005; 7(4): R896-903.
| Crossref | Google Scholar |
167 Lefranc D, Launay D, Dubucquoi S, de Seze J, Dussart P, Vermersch M, et al. Characterization of discriminant human brain antigenic targets in neuropsychiatric systemic lupus erythematosus using an immunoproteomic approach. Arthritis Rheum 2007; 56(10): 3420-32.
| Crossref | Google Scholar |
168 Chai Z, Sarcevic B, Mawson A, Toh BH. SET-related cell division autoantigen-1 (CDA1) arrests cell growth. J Biol Chem 2001; 276(36): 33665-74.
| Crossref | Google Scholar |
169 Hwang HM, Heo CK, Lee HJ, Kwak SS, Lim WH, Yoo JS, et al. Identification of anti-SF3B1 autoantibody as a diagnostic marker in patients with hepatocellular carcinoma. J Transl Med 2018; 16(1): 177.
| Crossref | Google Scholar |
170 Hosono Y, Nakashima R, Serada S, Murakami K, Imura Y, Yoshifuji H, et al. Splicing factor proline/glutamine-rich is a novel autoantigen of dermatomyositis and associated with anti-melanoma differentiation-associated gene 5 antibody. J Autoimmun 2017; 77: 116-22.
| Crossref | Google Scholar |
171 Hof D, Cheung K, de Rooij DJ, van den Hoogen FH, Pruijn GJ, van Venrooij WJ, et al. Autoantibodies specific for apoptotic U1-70K are superior serological markers for mixed connective tissue disease. Arthritis Res Ther 2005; 7(2): R302-9.
| Crossref | Google Scholar |
172 Yamamoto AM, Amoura Z, Johannet C, Jeronimo ALC, Campos H, Koutouzov S, et al. Quantitative radioligand assays using de novo–synthesized recombinant autoantigens in connective tissue diseases: new tools to approach the pathogenic significance of anti-RNP antibodies in rheumatic diseases. Arthritis Rheum 2000; 43(3): 689-98.
| Crossref | Google Scholar |
173 Mimori T, Hinterberger M, Pettersson I, Steitz JA. Autoantibodies to the U2 small nuclear ribonucleoprotein in a patient with scleroderma-polymyositis overlap syndrome. J Biol Chem 1984; 259(1): 560-5.
| Crossref | Google Scholar |
174 Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Lührmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem 2000; 275(22): 17122-9.
| Crossref | Google Scholar |
175 McClain MT, Ramsland PA, Kaufman KM, James JA. Anti-sm autoantibodies in systemic lupus target highly basic surface structures of complexed spliceosomal autoantigens. J Immunol (Baltimore, MD: 1950) 2002; 168(4): 2054-62.
| Crossref | Google Scholar |
176 Satoh M, Chan JYF, Ross SJ, Ceribelli A, Cavazzana I, Franceschini F, et al. Autoantibodies to survival of motor neuron complex in patients with polymyositis: immunoprecipitation of D, E, F, and G proteins without other components of small nuclear ribonucleoproteins. Arthritis Rheum 2011; 63(7): 1972-8.
| Crossref | Google Scholar |
177 Garbarz M, Dhermy D, Bournier O, Bezeaud A, Boivin P. Anti-spectrin in sera containing smooth muscle autoantibodies from patients with chronic active hepatitis. Clin Exp Immunol 1981; 43(1): 87-93.
| Google Scholar |
178 Imai H, Chan EK, Kiyosawa K, Fu XD, Tan EM. Novel nuclear autoantigen with splicing factor motifs identified with antibody from hepatocellular carcinoma. J Clin Invest 1993; 92(5): 2419-26.
| Crossref | Google Scholar |
179 Yang Q, Qin J, Sun G, Qiu C, Jiang D, Ye H, et al. Discovery and Validation of Serum Autoantibodies Against Tumor-Associated Antigens as Biomarkers in Gastric Adenocarcinoma Based on the Focused Protein Arrays. Clin Transl Gastroenterol 2020; 12(1): e00284.
| Crossref | Google Scholar |
180 Gordon P, Khamashta MA, Rosenthal E, Simpson JM, Sharland G, Brucato A, et al. Anti-52 kDa Ro, anti-60 kDa Ro, and anti-La antibody profiles in neonatal lupus. J Rheumatol 2004; 31(12): 2480-7.
| Google Scholar |
181 Cortini A, Bembich S, Marson L, Cocco E, Edomi P. Identification of novel non-myelin biomarkers in multiple sclerosis using an improved phage-display approach. PLoS One 2019; 14(12): e0226162.
| Crossref | Google Scholar |
182 Niland B, Miklossy G, Banki K, Biddison WE, Casciola-Rosen L, Rosen A, et al. Cleavage of transaldolase by granzyme B causes the loss of enzymatic activity with retention of antigenicity for multiple sclerosis patients. J Immunol (Baltimore, MD: 1950) 2010; 184(7): 4025-32.
| Crossref | Google Scholar |
183 Muto M, Mori M, Hiwasa T, Takiguchi M, Iwadate Y, Uzawa A, et al. Novel serum autoantibodies against talin1 in multiple sclerosis: Possible pathogenetic roles of the antibodies. J Neuroimmunol 2015; 284: 30-6.
| Crossref | Google Scholar |
184 Mariampillai K, Granger B, Amelin D, Guiguet M, Hachulla E, Maurier F, et al. Development of a New Classification System for Idiopathic Inflammatory Myopathies Based on Clinical Manifestations and Myositis-Specific Autoantibodies. JAMA Neurol 2018; 75(12): 1528-37.
| Crossref | Google Scholar |
185 Geng X, Biancone L, Dai HH, Lin JJ-C, Yoshizaki N, Dasgupta A, et al. Tropomyosin isoforms in intestinal mucosa: production of autoantibodies to tropomyosin isoforms in ulcerative colitis. Gastroenterology 1998; 114(5): 912-22.
| Crossref | Google Scholar |
186 Mahesh SP, Li Z, Buggage R, Mor F, Cohen IR, Chew EY, et al. Alpha tropomyosin as a self-antigen in patients with Behçet’s disease. Clin Exp Immunol 2005; 140(2): 368-75.
| Crossref | Google Scholar |
187 Kimura A, Sakurai T, Yamada M, Koumura A, Hayashi Y, Tanaka Y, et al. Anti-endothelial cell antibodies in patients with cerebral small vessel disease. Curr Neurovasc Res 2012; 9(4): 296-301.
| Crossref | Google Scholar |
188 Lorenz K, Beck S, Keilani MM, Wasielica-Poslednik J, Pfeiffer N, Grus FH. Course of serum autoantibodies in patients after acute angle-closure glaucoma attack. Clin Exp Ophthalmol 2017; 45(3): 280-7.
| Crossref | Google Scholar |
189 Zhao X, Cheng Y, Gan Y, Jia R, Zhu L, Sun X. Anti-tubulin-α-1C autoantibody in systemic lupus erythematosus: a novel indicator of disease activity and vasculitis manifestations. Clin Rheumatol 2018; 37(5): 1229-37.
| Crossref | Google Scholar |
190 Matthes T, Wolff A, Soubiran P, Gros F, Dighiero G. Antitubulin antibodies. II. Natural autoantibodies and induced antibodies recognize different epitopes on the tubulin molecule. J Immunol (Baltimore, MD: 1950) 1988; 141(9): 3135-41.
| Crossref | Google Scholar |
191 Kimura A, Yoshikura N, Koumura A, Hayashi Y, Kobayashi Y, Kobayashi I, et al. Identification of target antigens of naturally occurring autoantibodies in cerebrospinal fluid. J Proteomics 2015; 128: 450-7.
| Crossref | Google Scholar |
192 Prasannan L, Misek DE, Hinderer R, Michon J, Geiger JD, Hanash SM. Identification of beta-tubulin isoforms as tumor antigens in neuroblastoma. Clin Cancer Res 2000; 6(10): 3949-56.
| Google Scholar |
193 Stubbs Jr EB, Fisher MA, Siegel GJ. Anti-tubulin antibodies in a sensorimotor neuropathy patient alter tubulin polymerization. Acta Neuropathol 1998; 95(3): 302-5.
| Crossref | Google Scholar |
194 Muro Y, Ogawa Y, Kato Y, Hagiwara M. Autoantibody to thioredoxin reductase in an ovarian cancer patient. Biochem Biophys Res Commun 1998; 242(2): 267-71.
| Crossref | Google Scholar |
195 Betteridge ZE, Gunawardena H, Chinoy H, North J, Ollier WER, Cooper RG, et al. Clinical and human leucocyte antigen class II haplotype associations of autoantibodies to small ubiquitin-like modifier enzyme, a dermatomyositis-specific autoantigen target, in UK Caucasian adult-onset myositis. Ann Rheum Dis 2009; 68(10): 1621-5.
| Crossref | Google Scholar |
196 Miyachi K, Hosaka H, Nakamura N, Miyakawa H, Mimori T, Shibata M, et al. Anti-p97/VCP antibodies: an autoantibody marker for a subset of primary biliary cirrhosis patients with milder disease? Scand J Immunol 2006; 63(5): 376-82.
| Crossref | Google Scholar |
197 Bang H, Egerer K, Gauliard A, Lüthke K, Rudolph PE, Fredenhagen G, et al. Mutation and citrullination modifies vimentin to a novel autoantigen for rheumatoid arthritis. Arthritis Rheum 2007; 56(8): 2503-11.
| Crossref | Google Scholar |
198 Paley EL, Alexandrova N, Smelansky L. Tryptophanyl-tRNA synthetase as a human autoantigen. Immunol Lett 1995; 48(3): 201-7.
| Crossref | Google Scholar |
199 Wen J, Yaneva M. Mapping of epitopes on the 86 kDa subunit of the Ku autoantigen. Mol Immunol 1990; 27(10): 973-80.
| Crossref | Google Scholar |
200 Hoa S, Hudson M, Troyanov Y, Proudman S, Walker J, Stevens W, et al. Single-specificity anti-Ku antibodies in an international cohort of 2140 systemic sclerosis subjects: clinical associations. Medicine (Baltimore) 2016; 95(35): e4713.
| Crossref | Google Scholar |
201 Mahler M, Miller FW, Fritzler MJ. Idiopathic inflammatory myopathies and the anti-synthetase syndrome: a comprehensive review. Autoimmun Rev 2014; 13(4–5): 367-71.
| Crossref | Google Scholar |
202 Kistner A, Bigler MB, Glatz K, Egli SB, Baldin FS, Marquardsen FA, et al. Characteristics of autoantibodies targeting 14-3-3 proteins and their association with clinical features in newly diagnosed giant cell arteritis. Rheumatology (Oxford, England) 2017; 56(5): 829-34.
| Crossref | Google Scholar |
203 van Beers-Tas MH, Marotta A, Boers M, Maksymowych WP, van Schaardenburg D. A prospective cohort study of 14-3-3η in ACPA and/or RF-positive patients with arthralgia. Arthritis Res Ther 2016; 18: 76.
| Crossref | Google Scholar |
204 Qiu J, Choi G, Li L, Wang H, Pitteri SJ, Pereira-Faca SR, et al. Occurrence of autoantibodies to annexin I, 14-3-3 theta and LAMR1 in prediagnostic lung cancer sera. J Clin Oncol 2008; 26(31): 5060-6.
| Crossref | Google Scholar |
205 Chakravarti R, Gupta K, Swain M, Willard B, Scholtz J, Svensson LG, et al. 14-3-3 in Thoracic Aortic Aneurysms: Identification of a Novel Autoantigen in Large Vessel Vasculitis. Arthritis Rheumatol 2015; 67(7): 1913-21.
| Crossref | Google Scholar |
206 Teitsma XM, Devenport J, Jacobs JWG, Pethö-Schramm A, Borm MEA, Budde P, et al. Comprehensive exploratory autoantibody profiling in patients with early rheumatoid arthritis treated with methotrexate or tocilizumab. PLoS One 2020; 15(12): e0241189.
| Crossref | Google Scholar |
207 Alfraji N, Mazahir U, Chaudhri M, Miskoff J. Anti-synthetase syndrome: a rare and challenging diagnosis for bilateral ground-glass opacities-a case report with literature review. BMC Pulm Med 2021; 21(1): 11.
| Crossref | Google Scholar |
208 Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science (New York, NY) 2020; 369(6508): 1249-55.
| Crossref | Google Scholar |
209 Li Q, Zhang YY, Chiu S, Hu Z, Lan KH, Cha H, et al. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog 2014; 10(5): e1004163.
| Crossref | Google Scholar |
210 Hahn F, Schmalen A, Setz C, Friedrich M, Schlößer S, Kölle J, et al. Proteolysis of mature HIV-1 p6 Gag protein by the insulin-degrading enzyme (IDE) regulates virus replication in an Env-dependent manner. PLoS One 2017; 12(4): e0174254.
| Crossref | Google Scholar |
211 Carpenter JE, Jackson W, de Souza GA, Haarr L, Grose C. Insulin-degrading enzyme binds to the nonglycosylated precursor of varicella-zoster virus gE protein found in the endoplasmic reticulum. J Virol 2010; 84(2): 847-55.
| Crossref | Google Scholar |
212 Flach H, Rosenbaum M, Duchniewicz M, Kim S, Zhang SL, Cahalan MD, et al. Mzb1 protein regulates calcium homeostasis, antibody secretion, and integrin activation in innate-like B cells. Immunity 2010; 33(5): 723-35.
| Crossref | Google Scholar |
213 Miyagawa-Hayashino A, Yoshifuji H, Kitagori K, Ito S, Oku T, Hirayama Y, et al. Increase of MZB1 in B cells in systemic lupus erythematosus: proteomic analysis of biopsied lymph nodes. Arthritis Res Ther 2018; 20(1): 13.
| Crossref | Google Scholar |
214 Herold T, Mulaw MA, Jurinovic V, Seiler T, Metzeler KH, Dufour A, et al. High expression of MZB1 predicts adverse prognosis in chronic lymphocytic leukemia, follicular lymphoma and diffuse large B-cell lymphoma and is associated with a unique gene expression signature. Leuk Lymphoma 2013; 54(8): 1652-7.
| Crossref | Google Scholar |
215 Schiller HB, Mayr CH, Leuschner G, Strunz M, Staab-Weijnitz C, Preisendörfer S, et al. Deep Proteome Profiling Reveals Common Prevalence of MZB1-Positive Plasma B Cells in Human Lung and Skin Fibrosis. Am J Respir Crit Care Med 2017; 196(10): 1298-310.
| Crossref | Google Scholar |
216 Rosenbaum M, Andreani V, Kapoor T, Herp S, Flach H, Duchniewicz M, et al. MZB1 is a GRP94 cochaperone that enables proper immunoglobulin heavy chain biosynthesis upon ER stress. Genes Dev 2014; 28(11): 1165-78.
| Crossref | Google Scholar |
217 Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019; 47(D1): D607-13.
| Crossref | Google Scholar |