Developing Loop Mediated Isothermal Amplification (LAMP) assays for rapid, presumptive DNA detection of an invasive reptile (Boa constrictor)
Nathan Deliveyne A B * , Jeremy J. Austin
A B * , Jeremy J. Austin  B and Phillip Cassey
B and Phillip Cassey  A B
A B
A
B
Abstract
Wildlife trade is a prominent pathway for invasive species introductions into novel environments. Deliberate or accidental release of exotic pets can result in the establishment of alien populations, with damaging impacts for native species and environmental assets. This process is well documented for reptiles globally and is of considerable biosecurity concern in Australia. Boa constrictor is one species at high risk of establishment in Australia, and has insufficient biosecurity detection and post-border control capacity.
We aimed to develop rapid DNA-based presumptive testing capacity for detecting B. constrictor, with appropriate sensitivity and specificity to operate in a trace DNA biosecurity context.
Loop Mediated Isothermal Amplification (LAMP) is an emerging biosecurity tool that provides highly specific, sensitive, low-resource methods for detection of trace DNA in the absence of physical evidence. We developed colourimetric and fluorescent LAMP assays targeting the mitochondrial DNA control region of B. constrictor. We tested and validated these assays against synthetic DNA fragments, as well as DNA extracted from: (1) vouchered museum B. constrictor tissue; (2) shed B. constrictor skin samples; (3) a range of non-target species to test specificity; and (4) trace DNA recovered from glass tanks post B. constrictor presence.
We successfully detected synthetic target DNA down to 1 fg and genomic B. constrictor DNA from tissue and shed skins down to <10 pg in under 30 minutes with our fluorescence-based LAMP assay. Additionally, we were able to detect B. constrictor trace DNA following 24 h of presence utilising a traditional laboratory-based DNA extraction method (approximately 180 min) and a rapid lysis step (approximately 8 min).
Both colourimetric and fluorescent assays show promise for the specific detection of B. constrictor in biosecurity contexts, including post-border enforcement and compliance checks in the domestic illicit wildlife trade.
Our findings greatly strengthen the ongoing development of biosecurity tools for trace DNA detection of commonly traded and trafficked species (i.e. reptiles) in wildlife enforcement contexts, advancing both preparedness and surveillance.
Keywords: Biosecurity, Boa constrictor, DNA detection, invasive species, Loop Mediated Isothermal Amplification (LAMP), reptiles, wildlife forensics, wildlife trade.
Introduction
Wildlife trade is a prominent driver of extinction (Scheffers et al. 2019; Hinsley et al. 2023) that affects species from across the tree of life (Fukushima et al. 2020), including terrestrial (Morton et al. 2021) and aquatic vertebrates (Ripple et al. 2019), invertebrates (Purcell et al. 2014; Lassaline et al. 2023), plants (Margulies et al. 2019), and fungi (Sills et al. 2021). The scale of wildlife trade is increasing in the face of ongoing globalisation, with vertebrate trade involving over a quarter of all terrestrial species (Scheffers et al. 2019). One group that is particularly prominent in the global illegal wildlife trade is reptiles (Class: Reptilia) (Marshall et al. 2020). The reason for this popularity is often attributed to their uniqueness, with certain species harbouring patterns and colourations that are valuable in international markets (Heinrich et al. 2022). The importation of alien reptile species as pets has been identified as a major source of invasive species (García-Díaz et al. 2017), which is a key extinction driver when pets are released or escape into the wild (Lockwood et al. 2019). In addition to intentional trade, herpetofauna also present serious biosecurity concerns due to incursions as stowaways (García-Díaz and Cassey 2014; Pili et al. 2023).
Several biosecurity measures have been recommended or implemented to detect and monitor live reptile trade. These include DNA based methods for species identification and detection (e.g. Short Tandem Repeat (STR) assays for the detection of carpet python trafficking (Ciavaglia and Linacre 2018)), the use of stable isotope analysis to determine whether animals are released pets or wild individuals (e.g. Trachemys scripta elegans (red eared slider) as either pet release or wild caught individuals (Hill et al. 2020)), and recent developments in 3D X-ray technologies for imaging and detecting cases of reptile trafficking (Pirotta et al. 2022). Predictive modelling of future incursions (Stringham et al. 2021a) and web scraping of existing trade have also advanced biosecurity preparedness (Stringham et al. 2021b). Most recently, a forensically validated genetic toolkit has been developed for Tiliqua rugosa (shingleback lizards), which shows great promise for detection of illegal wildlife trafficking (Brown et al. 2023).
One component of the biosecurity toolkit that currently requires greater adoption is targeted invasive species presumptive detection from trace DNA samples. This is particularly true for situations in which reptiles have been hidden, moved, or destroyed, and only trace evidence remains, as commonly encountered in elusive illegal trade pathways. In addition, there is growing interest in detecting reptiles from environmental DNA (eDNA) in a variety of contexts, including biodiversity surveys (Kyle et al. 2022), invasive species detection (Hunter et al. 2015), and threatened species detection (Katz et al. 2021). Limited attention has been directed towards rapid DNA-based detection of traded reptiles, with some previous work conducted on trace DNA visualisation (Deliveyne et al. 2022). Loop Mediated Isothermal Amplification (LAMP) has recently emerged as a promising biosecurity tool (Deliveyne et al. 2023). LAMP provides robust DNA or RNA detection with limited processing time and apparatus required, as well as simple visualisation of results (Notomi et al. 2015). This contrasts with conventional Polymerase Chain Reaction (PCR), which often requires significant time, resources, and external expertise (Lee 2017). Current applications of LAMP have primarily addressed the detection of invertebrate species as stowaways crossing transnational boundaries by air and sea (Blaser et al. 2018; Rako et al. 2021), detection of adulterated meat products (Nikunj and Vivek 2019), and illegitimate fur products (Yu et al. 2019). Currently, LAMP has not been applied to the detection of live illegal wildlife trade (Deliveyne et al. 2023); however, the method has been proposed as a rapid field-based detection system in studies addressing interrelated issues (Wimbles et al. 2021), and for biosecurity screening, including the detection of one reptile species, the Asian House Gecko, which is of key biosecurity concern in Australia (Marks 2022).
Several alien reptile species warrant development of advanced DNA detection methods in Australia, including those that are frequently detected in Australian border-level seizure and post border seizure records. One such species is Boa constrictor, with 89 incidents and 176 animals recovered in post-border seizures between 1999 and 2016 (Toomes et al. 2020). Climate matching indicates high suitability for establishment across the northern half of Australia for B. constrictor, with sufficient introductions (Henderson et al. 2011). Consequently, B. constrictor is a biosecurity risk and is included on the Australian Department of Agriculture, Fisheries and Forestry (DAFF) national vertebrate pest priority list as a species of highest concern (DAFF 2023). Additionally, B. constrictor is a host of Inclusion Body Disease (IBD) (Schumacher et al. 1994), a disease mainly of the family Boidae – but cases have also been diagnosed in captive Australian pythons (Carlisle-Nowak et al. 1998). There are concerns this disease could become established in native Australian species, which presents substantial risks due to very high mortality rates (Chang and Jacobson 2010).
Our aim was to develop LAMP assays for the rapid biosecurity detection of B. constrictor from trace DNA sources. We focused our efforts on development and optimisation of LAMP methods validating the specificity, sensitivity, and efficacy of these methods for detecting trace DNA. This included: (1) primer design validated in silico; (2) in vitro optimisation using synthetic DNA, tissue samples, and shed skin as trace DNA proxies; and (3) experimental conditions designed to mimic reptile holdings used to hold B. constrictor. We successfully detected B. constrictor DNA from a range of samples using the developed LAMP assays without considerable cross-reactivity for other reptiles common in the domestic Australian pet trade. In this article we discuss further development, necessary steps, and current obstacles to the implementation of the described B. constrictor assay, to deliver a rapid, presumptive, and cost-effective detection tool for an emerging biosecurity threat in Australia.
Materials and methods
Primer design and in silico validation
LAMP assay development followed a systematic approach (Fig. 1), with mitochondrial DNA genome sequences for B. constrictor obtained from GenBank, (accession numbers, NC_007398, and AB177354) and the tRNA-lle, control region (accession number D84260). The mitochondrial DNA control region was selected, and the New England Biolabs (NEB) LAMP Primer Design Tool (https://lamp.neb.com/#!/) was used to develop LAMP primer sets. Additionally, primer binding sites were investigated for mismatches at primer target sites for Australian snakes common in the legal pet trade, based on previously published python mtDNA sequences (Supplementary Appendix A) (GenBank accession numbers EF545015–107) (Rawlings et al. 2008). Primers were designed using the normal default parameters and selected based on the largest delta G for dimerisation and with end stability of less than −4 kcal/mol at the 3′ end of the F2 region, 5′ end of the F1c region, 3′ end of the B2 region, and 5′ end of the B1c region. Additionally, all primers were designed to have GC content between 40 and 60%. Species-specific LAMP primer sequences were imported into Geneious Prime 2023.1 (Biomatters, New Zealand) for visual inspection against B. constrictor mtDNA sequences, and Megablast searches of the nucleotide collection (nr/nt) database were conducted for the target regions. As an assessment of primer pair specificity, the FastPCR software was used to test cross-species amplification in silico. FastPCR was used because this tool allows for linked searches of multiple primer pairs (Kalendar et al. 2017). In FastPCR the LAMP primers were uploaded, and the in silico PCR function was used to assess potential amplification for the top 20 Megablast hit table results. Based on these primer design conditions, a 207-bp fragment of the mitochondrial D-loop control region was ultimately selected for in vitro testing. LAMP primers were ordered from Sigma Aldrich, USA, with the synthesis scale of 0.025 μmole, desalt purification, at a concentration of 100 μM in TRIS solution.
Schematic outlining the methodological approach to B. constrictor Loop Mediated Isothermal Amplification (LAMP) assay development. (a) B. constrictor sequences were collected from NCBI nucleotide, with the New England Biolabs LAMP Primer Design Tool used for primer design in tandem with Geneious. (b) DNA was extracted from tissue and a range of reptile shed skins and (c) serially diluted 1:10 in tandem to synthetic DNA matching the target region, to assess the sensitivity and specificity. (d) Colourimetric reactions were assessed quantitatively as the hue component of colour and (e) LAMP target validation was conducted by PCR amplification using outer most LAMP primers followed by Sanger sequencing. (f) The applicability to trace DNA was assessed for two extraction methods, a rapid approach and a more traditional laboratory method, with detection facilitated by B. constrictor fluorescence LAMP.
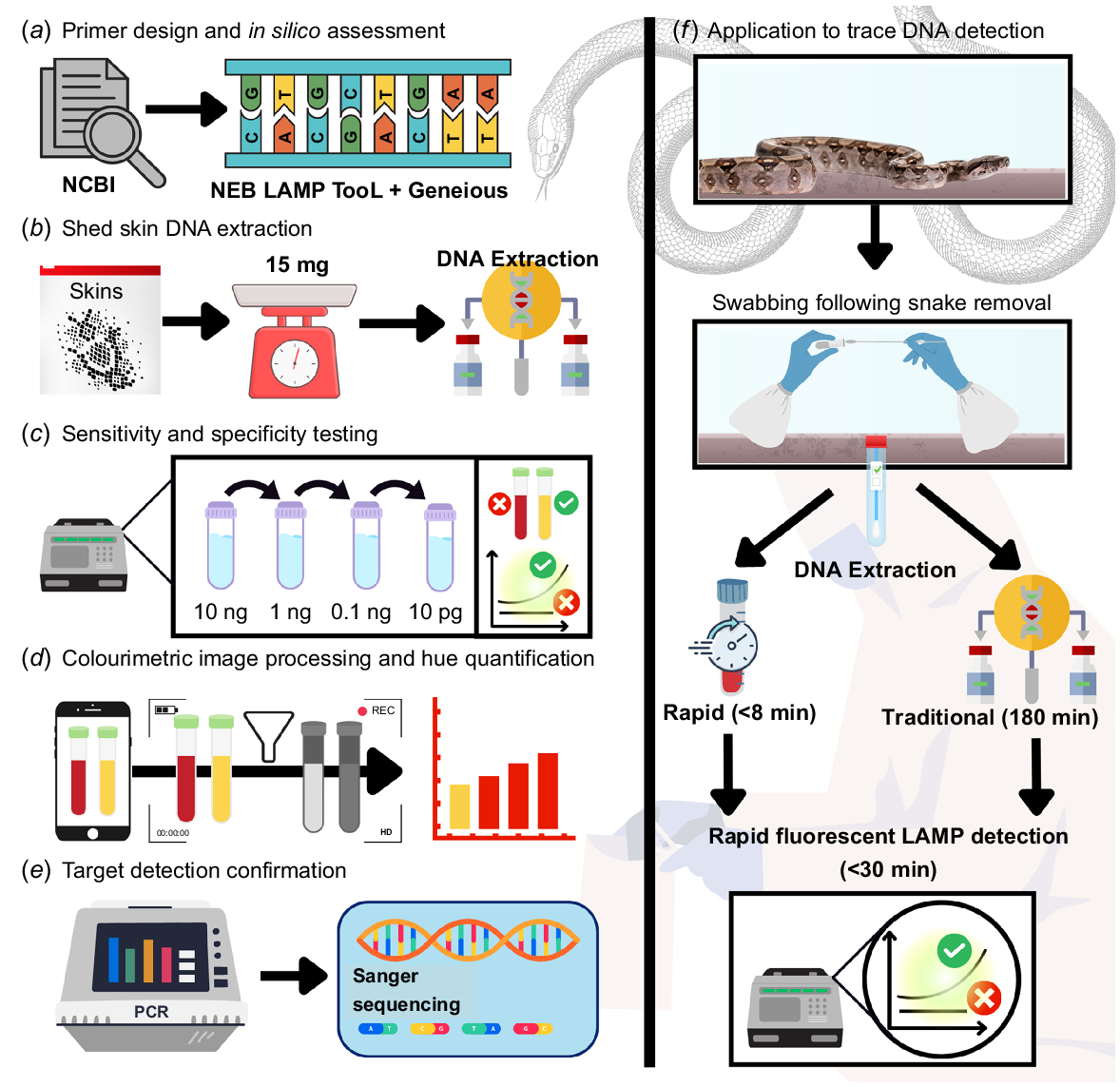
Synthetic gBlock development and use in validation
The LAMP target region for B. constrictor mitochondrial control region was used to design synthetic DNA ordered as gBlocks™ Gene Fragments (IDT, USA) to provide positive controls of known concentration, and for assessing reaction sensitivity (Agarwal et al. 2020; Rako et al. 2021; Starkie et al. 2022). Developing gBlocks™ with slight differences in annealing derivative temperature can facilitate differentiation between sample and synthetic positive control when separating priming regions by triplicate GGG or CCC (Agarwal et al. 2022). Thus, we designed two gBlocks™, BC.MCR that matched the target region for which the primers were designed and BC.CR.P that includes each primer binding site separated by CCC.
BC.MCR: 5 – AT TGT GTC CCT TAA TTC TGC CCT TCC CGT GAA ATC CTC TAT CCT TTC ATA CAT GCT AAC AGT CCT GCT TTT CAC GTC CAT ATA ATG TAA CCC CTCCCT ACT GTA CTT TCC AAG ACC ACT GGT TAC ACC TTC AAG TCC ATT TCA ACG GCC CGG AAC CAT CCC TCC CTA CTT GCT CTT TCC AAG ACC TTT GGT CGCACC CTT TAT TTA AGT AC – 3
BC.CR.P: 5 – CC TGT GTC CCT TAA TTC TGC CCC CAT CCT CTA TCC TTT CAT ACA TGC CCC TAA CAG TCC TGC TTT TCA CGT CCC ATA ATG TAA CCC CTC CCT ACTGTA CCC AGA CCA CTG GTT ACA CCT TCA CCC TCC ATT TCA ACG GCC CGG CCC ACT TGC TCT TTC CAA GAC CCC CGT CGC ACC CTT TAT TTA AGT CCC – 3
BC.MCR was used as a positive control and serially diluted from 1 ng/μL to 1 fg/μL as standards used for assessing the limit of detection. BC.CR.P was developed and utilised to test primer binding affinity and as an additional synthetic control.
In vitro validation of LAMP primers
Positive control DNA was extracted from a B. constrictor liver sample (10 mg), collected on 13 June 1997 (ex-Adelaide Zoo) and obtained from the Australian Biological Tissue Collection (ABTC accession #74730, South Australian Museum). Samples used for in vitro validation included shed skins from B. constrictor and non-target reptiles common in the legal Australian reptile trade covering several genera (Morelia carinata, Morelia spilota, Antaresia childreni, Antaresia maculosa, Liasis olivaceus, Liasis fuscus, Aspidites ramsayi, Aspidites melanocephalus, Simalia kinghorni), and those identified as key incursion species (Pantherophis guttatus and Python brongersmai (Stringham et al. 2021a). Shed skins were selected because they are morphologically indistinguishable without expert training but provide a reliable source of DNA (Bricker et al. 1996). These samples were sourced from the Gorge Wildlife Park, South Australia, and some shed skins were received from members of the public. DNA extraction was conducted in duplicate using the QIAamp DNA Investigator Kit (Qiagen, USA), alongside extraction blank controls. Initial DNA yields were quantified using the Quantus™ Fluorometer and QuantiFluor dsDNA System (Promega, USA) and then serially diluted 1:10 to assess the limit of detection for LAMP assays.
The WarmStart® Colourimetric LAMP 2x Master Mix (New England Biolabs, USA) was used to amplify synthetic gBlock DNA, ABTC reference DNA, and DNA from shed skin extracts. Reactions were conducted for B. constrictor and all non-target species following the manufacturer's protocol. LAMP primers were combined in a 10x primer mix at two different concentrations: ‘standard’ – containing 16 μM FIP/BIP, 2 μM F3/B3, and 4 μM LF/LB; and ‘high’ – 20 μM/2 μM/10 μM. LAMP reactions were conducted using 12.5 μL 2x WarmStart MasterMix, 2.5 μL 10x primer mix (standard or high), 1 μL of DNA, and dH2O to a total volume of 25 μL, using an Eppendorf Mastercycler X50 at 65°C for 30–75 min.
Sensitivity was assessed by performing serial 1:10 dilutions of the synthetic gBlock DNA ranging from 1 ng/μL to 0.1 fg/μL, and for DNA extracted from the ABTC B. constrictor positive control ranging from 63 ng/μL to 0.63 pg/μL. We included 12 technical replicates at each concentration (eight in 12.5 μL, and four in 25 μL reaction volumes). Samples were photographed following 30, 45, and 60 min of incubation at 65°C, to assess colour transition. We defined the limit of detection (LOD) using a discrete threshold of 100% positive amplification for all replicates at the lowest concentration (Klymus et al. 2020), without detectable primer-dimer (NTC transitioned to yellow).
Specificity assessments were conducted for all non-target Australian snakes with four technical replicates of each DNA extract, followed by an additional four technical replicates for 1:10 dilutions of DNA extracts that transitioned away from pink towards yellow.
Colour component analysis was conducted on all colourimetric LAMP reactions to determine the earliest point of colour change, colour change sensitivity, and conditions for colour change, including initial template concentration and primer concentrations. This analysis was conducted in the Fiji version of ImageJ (Schindelin et al. 2012). Images were collected under standardised conditions by use of a lightbox placed underneath samples after incubation for 30, 45, and 60 min, halting at the earliest point of non-specific amplification. At each time interval an image was captured using a Xiaomi Pocophone F1 using the manual mode, with the shutter speed locked at 1/500 and the ISO set to 400.
The images were imported into ImageJ and the image type was changed to HSB stack. A circular region of interest was placed under the meniscus of each sample with a 40-unit height and width encapsulating 1264 pixels. For each region of interest, the mean hue values were recorded (Fig. 2). Hue is an established indicator for colourimetric LAMP transition, exemplified in studies concerning human body fluid identification (Scott et al. 2020; Layne et al. 2021). The hue values for each of the reactions were plotted using ggplot2 (Wickham 2011) package in the R software environment (v 1.3.1073) for statistical and graphical computing (R Core Team 2023) to provide an objective and quantitative assessment of colourimetric transition success. This approach was used to establish upper and lower bounds for positive LAMP reactions.
Image capture, transformation, and hue quantification of LAMP colourimetric detection of B. constrictor DNA. (a) Reaction series captured under standardised conditions using a light pad placed underneath the samples, captured using a Pocophone F1 with a shutter speed of 1/500 s and ISO locked to 400. (b) The same image is then converted to a HSB stack. (c) A circular selection of 1264 pixels is made under the meniscus of the reaction, and the hue mean is quantified.

In parallel, we tested fluorescence-based real-time LAMP detection. We followed the LAMP user guide published by Optigene to develop our assay and optimise primer concentrations (Optigene 2017). We firstly tested fluorescence LAMP reactions in 25 μL volumes containing 15 μL of the ISO-004 GspSSD2.0 Isothermal Master Mix (Optigene, UK), 2.5 μL of the ‘high’ primer mix, 1 μL sample, and 6.5 μL water. We also tested ‘low’ primer concentrations (FIP/BIP = 0.8 μM, F3/B3 = 0.2 μM, LF/LB = 0.4 μM), in 12.5 μL final volumes containing 7.5 μL of the ISO-004 GspSSD2.0 Isothermal Master Mix (Optigene, UK), 1.25 μL primer mix, 1 μL sample, and 2.75 μL dH2O. All real-time isothermal LAMP incubations were conducted at 65°C for 30 min, with measurements of fluorescence recorded at 90 intervals of 20 s on the QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, USA). Additionally, melt curve analysis was conducted starting at 98°C for 15 s, 70°C for 60 s, 0.05°C/S ramping to 98°C for 15 s.
Sensitivity of the fluorescent LAMP approach was assessed by including 12 technical replicates of synthetic gBlock DNA diluted at concentrations ranging from 1 ng/μL to 0.1 fg/μL. Similarly, DNA extracted from the ABTC B. constrictor sample was diluted in series 1:10 ranging from 63 ng/μL to 0.63 pg/μL, with 12 technical replicates at each concentration. We used a discrete threshold approach to determine the LOD, defined here as 100% positive replicates (Klymus et al. 2020). Specificity was assessed for all non-target species in at least triplicate, and additional testing was conducted for genera including Morelia, Antaresia, Liasis, and Aspidites. This included 10 technical replicates per species belonging to each genus, because these species were identified as common native Australian snakes kept as pets.
Following LAMP testing, B. constrictor DNA extracts from shed skin underwent PCR amplification using the species specific F3/B3 LAMP primers. PCRs were conducted containing PowerUp™ SYBR™ Green Master Mix (Applied Biosystems, USA), 0.4 μM of forward and reverse Primer, 1 μL of DNA extract, and dH2O to a volume of 20 μL. Thermocycling conditions on the QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems, USA) were as follows. Initial denaturation at 95°C for 2 min, 40 cycles of denaturation at 95°C for 15 s, annealing at 54°C for 20 s, extension at 72°C for 60 s. This also included a melt curve following amplification with the following conditions: 95°C for 15 s, 60°C for 60 s, 0.05°C/S ramping to 95°C for 15 s.
Subsequent bi-directional Sanger sequencing of PCR amplicons was conducted by the Australian Genome Research Facility (AGRF, Australia) to confirm correct target amplification. Sequences were analysed in Geneious Prime (Biomatters, New Zealand), with ends trimmed at the 3′ and 5′ ends and an error probability limit of 0.05. De novo assemblies were created using the reference sequence NC_007398, F3 and B3 primers and the bi-directional Sanger sequencing data. The consensus sequence was subject to a BLAST search.
Following optimisation of LAMP assays using synthetic DNA and DNA extracted from tissue and shed skins, we tested the detection efficacy of the colourimetric and fluorescence assays on detection of remanent trace DNA left behind post B. constrictor presence in glass tanks (following The University of Adelaide animal ethics approval ID S-2020–024). Prior to reptile exposure, 65-L glass tanks were sterilised using 10% bleach followed by a wipe down with absolute ethanol. Tanks were placed in a constant-temperature room (25°C) next to one another for the duration of each experimental trial. For each of five replicates and one negative control (B. constrictor absence), one B. constrictor individual was placed in a 65-L glass tank for 24 h, the snake was then removed, and the inside surface of the base was swabbed with five different swabs, one targeting each corner and one targeting the centre of the tank. Copan FLOQSwabs pre-wetted with 40 μL of 0.1% Triton X-100 (Bio-Rad, USA) in DNA-free water were used for sample collection. DNA was extracted from the tank swabs using QIAamp DNA Investigator Kit (Qiagen, USA) and quantified using the Quantus™ Fluorometer and QuantiFluor dsDNA System (Promega, USA).
We also tested a rapid lysis DNA extraction method QuickExtract (Lucigen, USA) as an alternative to the more time-consuming QIAamp DNA Investigator Kit, facilitating rapid detection. An additional six replicate tanks, including a negative control, were set up and swabbed as previously described. Each swab was placed in 250 μL of the QuickExtract buffer and incubated following the manufacturer’s instructions. DNA was quantified using the Quantus™ Fluorometer and QuantiFluor dsDNA System (Promega, USA).
Results
LAMP primer development
LAMP primers were successfully developed targeting a 207-bp portion of the B. constrictor mitochondrial control region (Table 1). BLAST searches of this 207-bp sequence showed the highest non-target percentage identity was 91.09%, corresponding to Eunectes notaeus (yellow anaconda, JN967236.1). A linked search conducted in FastPCR (Kalendar et al. 2017) indicated no cross amplification concerns for the LAMP primers, returning only B. constrictor (D84260, and AB177354) sites when queried against the top 20 Megablast hits.
| Species | Regions | Primers | |
|---|---|---|---|
| B. constrictor | Mitochondrial DNA control region | F3: TGTGTCCCTTAATTCTGCC | |
| B3: ACTTAAATAAAGGGTGCGAC | |||
| FIP: TACAGTAGGGAGGGGTTACATTAT ATCCTCTATCCTTTCATACATGC | |||
| BIP: AGACCACTGGTTACACCTTCA GGTCTTGGAAAGAGCAAGT | |||
| LF: ACGTGAAAAGCAGGACTGTTA | |||
| LB: TCCATTTCAACGGCCCGG |
Primers target a 207-bp fragment of the mitochondrial control region with each primer in the 5′ to 3′ direction.
Primer and reaction condition optimisation for B. constrictor colourimetric LAMP assay
Greater specificity, and shorter reaction time to positive detection (45 min), was achieved with ‘high’ primer concentrations (Fig. 3a). No cross amplification of P. guttatus DNA was observed. B. constrictor template transitioned to yellow (positive) with input DNA ranging from 83 ng to 83 pg after 75-min incubation, when using the ‘standard’ primer concentrations (Fig. 3b). Cross amplification (mild colour change) was detected for P. guttatus containing 16 ng of input DNA after 75-min of incubation (Fig. 3b). Maintaining the higher primer concentrations, LAMP reactions containing synthetic DNA diluted down to 10 fg (100% of replicates) transitioned to yellow after 30 min (Fig. 3c, d), including the primer gBlock (BC.CR.P). Colour change for the ABTC B. constrictor liver DNA extract (i.e. down to 63 pg) was achieved after 45 min (Fig. 3e, f).
Colourimetric B. constrictor LAMP assay with varied primer concentrations, incubation periods, and constant 65°C incubation temperature. (a) Results of higher primer concentrations post 45 min of incubation, with B. constrictor and P. guttatus DNA included in the reactions. (b) Reaction series incubated for 75 min including standard primer concentration, all reactions containing B. constrictor template (83 ng–83 pg) transitioning to yellow. (c, d) Sensitivity testing, for high primer concentrations, incubated for 45 min, including 1:10 dilution series of synthetic DNA (BC.MCR) with four replicates ranging from 1 pg–0.1 fg, shown as the raw image and the HSB transformed image. (e, f) Sensitivity testing, with high primer concentrations for a 1:10 dilution series including genomic B. constrictor DNA (ABTC Boa) ranging from 63 ng–0.63 pg, incubated for 45 min.
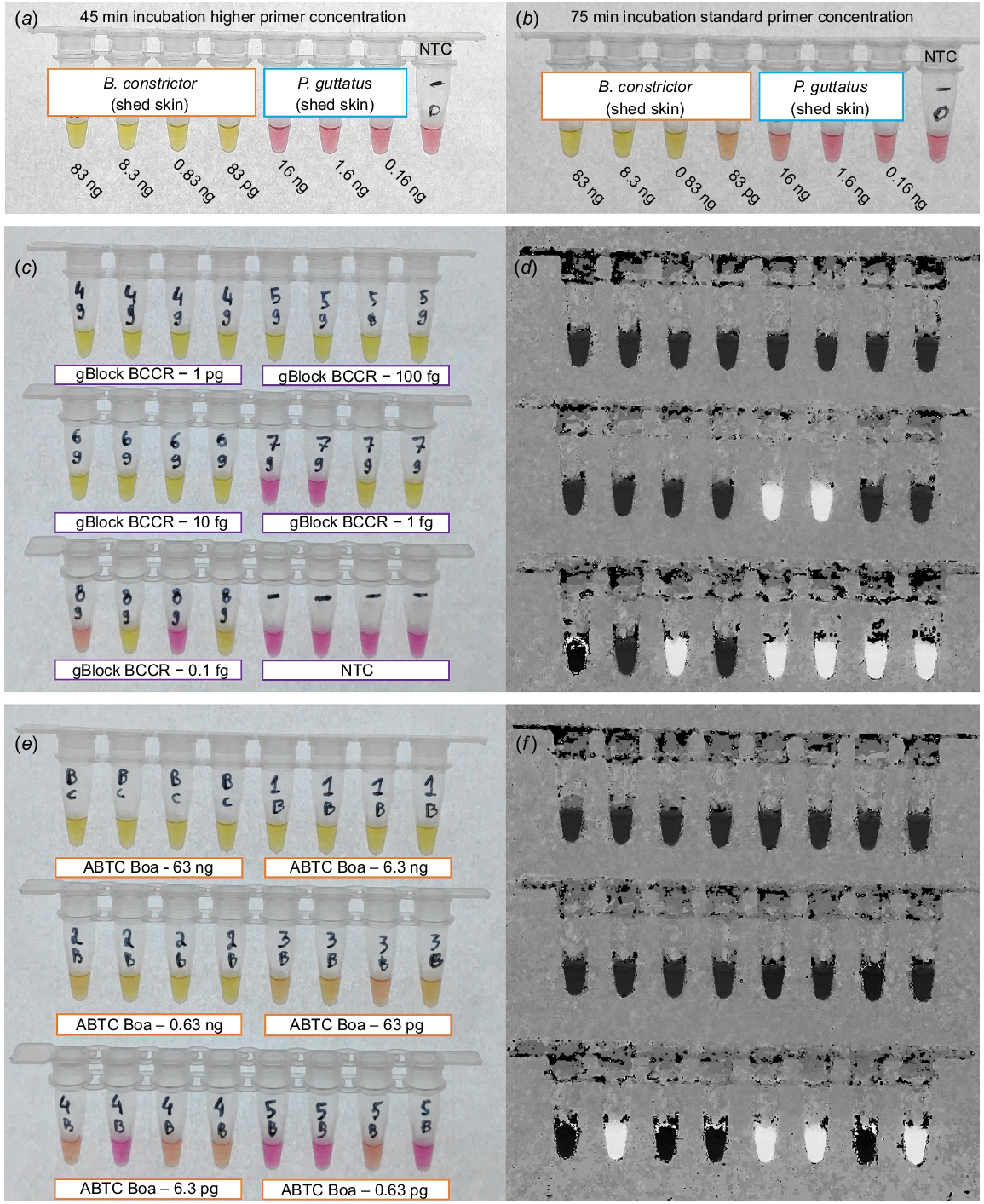
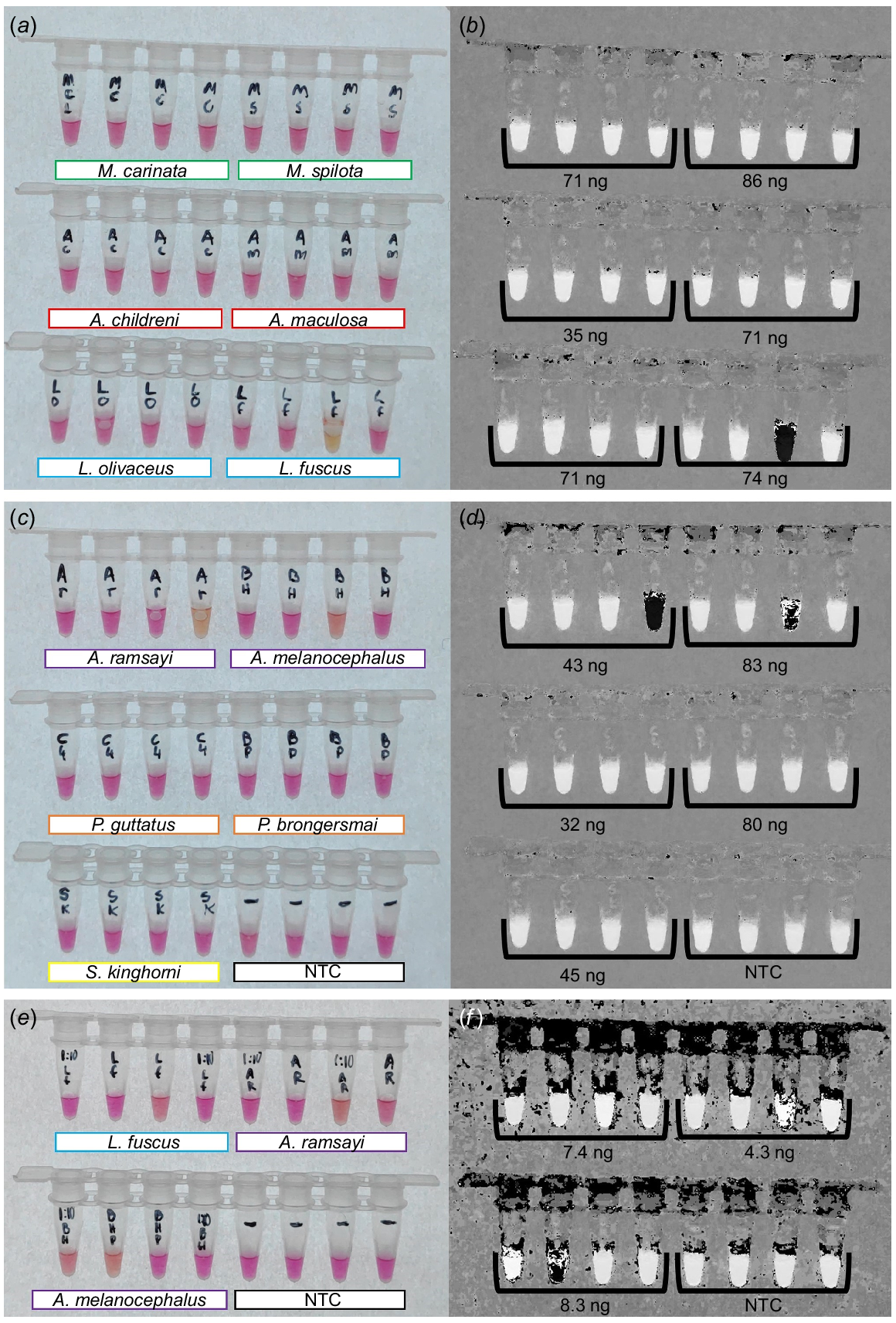
Maintaining the higher primer concentration while testing specificity led to promising results for native Australian snake species and new incursion species in the pet trade (Fig. 4). We did not detect cross amplification for M. carinata, M. spilota, A. childreni, A. maculosa, L. olivaceus, P. guttatus, P. brongersmai, or S. kinghorni (Fig. 4a–d). Mild colour transitions were observed for L. fuscus, A. ramsayi, and A. melanocephalus with DNA inputs >10 ng (Fig. 4a–d). Reducing the template to <10 ng following 1:10 dilutions reduced the severity of cross amplifications, as clearly indicated on the hue-transformed image, with none of the reactions transitioning to solid black (Fig. 4f). Incubating reactions for 60 min resulted in far greater cross amplification, visible as stronger transitions to yellow, and is thus strongly recommended against.
Cross amplification testing for colourimetric LAMP assay targeting a 207-bp section of the mitochondrial D-loop of B. constrictor. All reactions were incubated at 65°C for 45 min with F3/B3, FIP/BIP, and LF/LB primers included at 2.0 μM/0.2 μM/1.0 μM respectively. (a, b) Colourimetric transitions for M. carinata, M. spilota, A. childreni, A. maculosa, L. olivaceus, and L. fuscus, shown as a raw image and HSB transformed image, including amount of added DNA. (c, d) Colourimetric transition for A. ramsayi, A. melanocephalus, P. guttatus, P. brongersmai, S. kinghorni, and the no template control (NTC), shown as a raw image and the HSB transformed image, including DNA input amounts. (e, f) Colourimetric transitions of 1:10 diluted L. fuscus, A. ramsayi, and A. melanocephalus, and NTC reactions to re-assess the colour transition at lower concentrations.
Colour component analysis
Quantifying colourimetric reactions as the hue component of colour provided an objective quantitative delimitation of positive and negative reactions, establishing the optimal primer concentrations, incubation times, and limits of detection. The resulting optimal reaction conditions utilised the ‘high’ primer concentration with 45-min incubation at 65°C. Assessing the selected reaction conditions as the hue component of colour for gBlock fragments BC.MCR and BC.CR.P, the ABTC B. constrictor DNA extract and several snakeskin extracts (B1, B2, B3, B5, B6) serially diluted 1:10 ranging from 83 ng to 35 pg led to positive detection for all LAMP reactions containing B. constrictor target DNA after 45 min of incubation at 65°C (Fig. 5). The positive detection range (orange dashed lines) spanned mean hue values of 20–60 units (Fig. 5) and defined the quantitative range for target detection. Incubating the reactions for 60 min resulted in occasional colour change in the NTC’s (primer-dimer) and is thus strongly recommended against.
Hue component of colour assessed at different time points during the progress of the LAMP reaction for several samples with F3/B3, FIP/BIP, and LF/LB primers included at 2.0 μM/0.2 μM/1.0 μM respectively. Points indicate different individual sample hue measurements, with bars indicating the average hue for each category. Category B0 includes ABTC liver DNA sample at 63 ng/μL and B. constrictor shed skin DNA extracts B1, B2, B3, B5, B6 ranging from 89 to 35 ng/μL of input DNA. Categories B0.1–B0.3 showcase a serial 1:10 dilution of the B0 samples. Synthetic gBlock DNA fragments BC.MCR, BC.CR.P, and a no-template control (NTC) are included for reference. Orange dashed lines indicates the range of positive detection.

Fluorescent LAMP detection
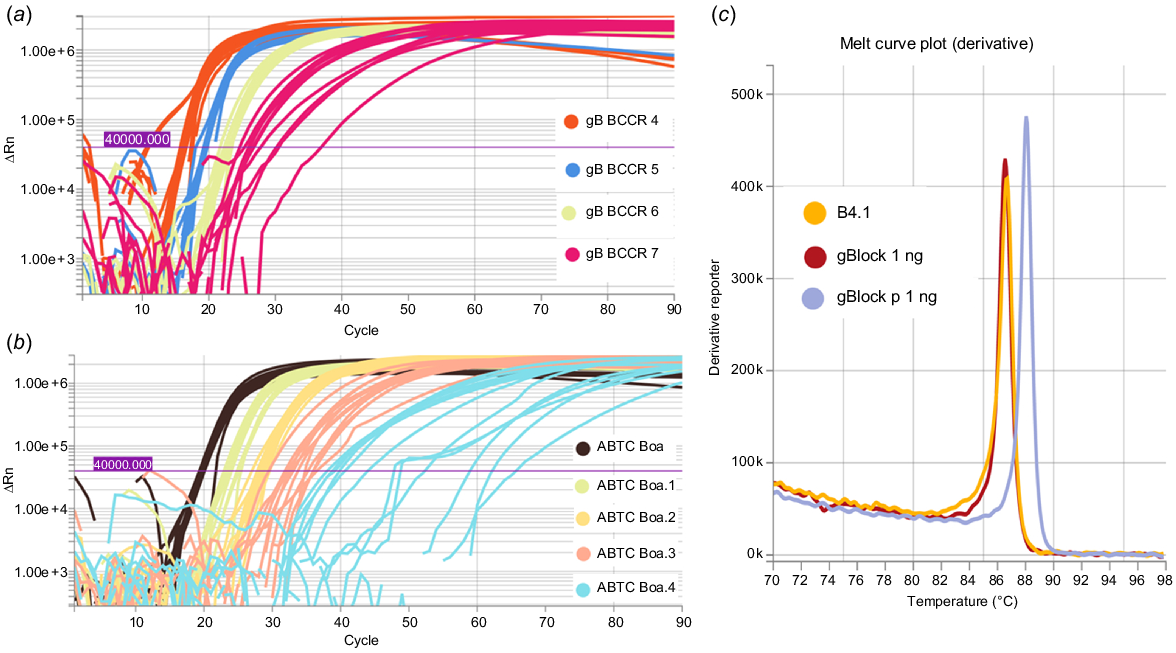
Synthetic B. constrictor gBlock DNA and genomic DNA from the ABTC reference sample and shed skin samples were detected using the fluorescent LAMP assay, at two different primer concentrations (0.8 μM/0.2 μM/0.4 μM and 2.0 μM/0.2 μM/1.0 μM for FIP/BIP, F3/B3, and LF/LB respectively). At higher primer concentrations we detected primer dimer using the DNA 1000 kit (Agilent, USA) on the 2100 Bioanalyser system, so we opted for lower primer concentrations. Serial 1:10 dilutions of BC.MCR, B. constrictor ABTC reference DNA, and shed skin samples led to the positive detection of synthetic DNA down to 1 fg and genomic B. constrictor DNA extracts down to <10 pg (Fig. 6a, b). The LOD for synthetic DNA was 1 fg as at 0.1 fg only 5/12 replicates amplified, and c. 1 pg for genomic B. constrictor DNA as at 0.63 pg only 2/12 replicates amplified. Additionally, the melt curves confirmed the target LAMP amplicons were generated for synthetic DNA (BC.MCR, BC.CR.P), the B. constrictor ABTC reference sample, and skin extracts (Fig. 6c).
Amplification curves for LAMP reactions conducted for 30 min at an incubation temperature of 65°C. (a) Synthetic gBlock designed to match priming regions diluted 1:10 (1 pg/μL–1 fg/μL) and (b) template DNA extracted from B. constrictor liver sample diluted 1:10 (63 ng/μL–6.3 pg/μL). (c) Melt curve plot indicating the LAMP amplicons for synthetic (gBlock BC.MCR) and B. constrictor shed skin DNA (B4.1) extracts match, with a different anneal derivative temperature for the synthetic primer gBlock (BC.CR.P).
Based on the amplification curves and melt curve temperatures, no cross amplification was detected for most of the native Australian snakes common in the pet trade. However, cross amplification was detected for one native snake species, L. olivaceus (99 ng), and one invasive species, P. guttatus (16 ng), at high template DNA inputs. Each cross amplification event was limited to only one out of each triplicate reaction. Thus, only results with two or more positive amplification curves crossing the fluorescence threshold were further considered positive detections for B. constrictor.
Additional dilution series to assess cross amplification sensitivity for species where cross amplification was detected initially resulted in no amplification for two shed skin extracts of L. olivaceus (L.oli1 99 ng–99 pg and L.oli2 at 78 ng–78 pg). Additional cross amplification assessed for P. guttatus (C3 and C4 at 32 ng–32 pg) led to cross amplification for one reaction within a technical triplicate with 32 ng of input P. guttatus DNA. Reactions including less than 10 ng of non-target DNA did not lead to conclusive amplification.
We identified high initial DNA template (>10 ng) as a potential source of undesirable cross amplification. Further cross amplification testing conducted for two species of each genus of common Australian pet snake (M. carinata, M. spilota, A. childreni, A. maculosa, L. olivaceus, L. fuscus, A. ramsayi, and A. melanocephalus) at concentrations of input DNA between 10 and 4 ng did not lead to any cross amplification for 10 technical replicates of each species.
PCR and bi-directional Sanger sequencing
Synthetic gBlocks and B. constrictor skin extracts were successfully amplified using the F3 and B3 primers. Bi-directional Sanger sequencing confirmed the intended target PCR amplicons, with the gBlock BC.MCR amplicon matching the NC_007398 reference with 100% query cover and percentage identity. Similarly, DNA extracts derived from shed snakeskin of B. constrictor (B4) also matched reference samples AB177354 and D84260, with PCR reactions including 37 ng of template returning 100% Query cover and 98.08% identity and 37 pg of template returning 100% query cover and 97.60% identity.
DNA detection from swabbed tanks
The colourimetric LAMP assay successfully detected B. constrictor DNA from swab samples retrieved from empty tanks (following 24-h B. constrictor presence). However, repeatability and colourimetric transition were low and inconsistent, so we abandoned this approach for the specific application of trace DNA detection from contact samples. The fluorescent assay resulted in less time to detection (<30 min in contrast to <45 min for colourimetric LAMP), greater repeatability (consistent amplification in triplicate), and an order of magnitude greater sensitivity. As such, the remainder of B. constrictor touch samples from glass tanks were amplified using the fluorescent assay.
Initial testing conducted for sample swabs extracted using QIAamp investigator kit retrieved post removal after 24-h of B. constrictor presence yielded positive results, with at least one swab extract amplifying in duplicate for each tank replicate (Table 2a). Swabs retrieved from Tank 1 had a minimum of four swabs with at least duplicate amplification. Swabs retrieved from Tank 2 led to positive triplicate amplification for a minimum of three swabs. Tank 3 led to positive amplification for two swabs in at least duplicate. Tank 4 led to positive amplification of a single swab in duplicate. Tank 5 led to positive amplification for one swab in triplicate. No amplification was detected in the negative control tank.
Testing conducted for tank swabs subject to QuickExtract DNA extractions led to positive detections for a minimum of two swabs in duplicate for each tank (Table 2b). Swabs retrieved from Tank 1 led to at least duplicate amplification for two swabs. Tank 2 led to the recovery and amplification of two swabs in triplicate. Swabs recovered from Tank 3 led to triplicate amplification for two swabs. Tank 4 included four swabs with at least duplicate amplification. Tank 5 included two swabs that amplified in at least duplicate. We did not observe any amplification from the negative control tank.
Discussion
Colourimetric and fluorescent LAMP assays are a promising novel tool for detecting reptile species of highest biosecurity concern in Australia. Both assays detected the presence of target DNA with high specificity under optimal primer concentrations and showed appropriate sensitivity to operate in a trace DNA environment, detecting as little as 1 fg of synthetic template and less than 10 pg of genomic B. constrictor DNA extracted from reference tissue and shed skins. Additionally, LAMP detection for both assays was possible within 45 min. We present promising solutions to field testing of trace DNA samples due to the low expertise, equipment, and time required to carry out the set-up and incubation of LAMP reactions. As such, we have provided an important first step to developing field-based detection capacity for biosecurity risk reptile trafficking.
Several additional steps and considerations could be taken to improve and build on our work presented here. These improvements would primarily involve further increasing the specificity, sensitivity, and robustness of the outlined detection protocol, including considerations such as comparisons with alternative amplification methods and field-ready apparatus.
Our fluorescence LAMP assay, developed using the Optigene GspSSD2.0 Isothermal Master Mix (ISO-004), provided the best diagnostic capacity. We confidently detected described gBlock fragments, DNA extracts from tissue, shed snakeskin, and trace DNA recovered from glass tank surfaces. The fluorescent assay also provided a faster time to detection, with incubation limited to 30 min. The difference in assay performance could be due to the polymerase involved in the reactions with the colourimetric approach utilising the bst 2.0 polymerase, as opposed to the GspSSD2.0 used in the fluorescent assay. Differences in polymerase performance have been observed in the presence of inhibitors (Jevtuševskaja et al. 2017). Additionally, this could be due to fundamental differences in the chemistries of the two approaches, with the colourimetric approach relying on a change in pH due to the by-product magnesium pyrophosphate, resulting in a colour change from pink to yellow (Tanner et al. 2015). The fluorescent approach utilises an intercalating dye that binds to amplified DNA and thus increases over time as amplification progresses (Hardinge and Murray 2019). The sensitivity of the QuantStudio may also play a role due to its capacity for detecting small changes in fluorescence, as opposed to naked-eye or hue component analysis at the end point of the reaction. This gap could potentially be bridged by use of a spectrophotometer, with additional potential for quantification (Nguyen et al. 2019). However, such specialised equipment is commonly confined to a dedicated lab so is not well suited to rapid in situ biosecurity detection. Fluorescent real-time detection also allowed us to assess the amplification curves and corresponding melt curves. This was a useful tool in determining whether amplification followed the expected sigmoidal pattern and melt temperature corresponded to the synthetic and positive ABTC control sample. End point detection using the colorimetric approach does not offer this benefit, leading to limited capacity to assess and exclude false positives due to non-target amplification.
Suboptimal results encountered when applying the colourimetric LAMP to tank samples may stem from the quality of DNA retrieved from swabs. This template DNA could have been degraded or low quality because it was indirectly retrieved from surfaces using swabs. DNA extracted directly from shed skins may be of higher quality because it is from a direct source (Bricker et al. 1996; Fetzner 1999; Horreo et al. 2015). As such, we can only recommend the application of our colourimetric assay to the detection of B. constrictor DNA from high-quality sources such as tissue, saliva swabs, and shed skins. These key sample types still present areas that require rapid biosecurity detection due to disease concerns, including IBD and scenarios in which the only remaining evidence of trafficking are morphologically indistinguishable shed skins. A similar issue was encountered when extracting DNA from snakeskin using the QuickExtract (Lucigen, USA) solution; this drastically impacted colour transition and resulted in inconclusive results, further demonstrating the requirement for high-quality DNA.
The primary advantage of the colourimetric approach is the limited resources required to conduct the experiment and the ease of interpretation for results. Fluorescent real-time detection requires a fluorometer, which is perhaps the largest barrier to biosecurity integration because these are traditionally benchtop qPCR machines confined to a dedicated laboratory space. This limitation can be overcome with the integration of field-ready LAMP apparatus, such as the Genie III developed by Optigene. The integration of in-field amplification and detection of biosecurity risk species has been demonstrated on several occasions (Rako et al. 2021; Agarwal et al. 2022), illustrating the suitability of a fluorescence LAMP assay in situations without overbearing financial concerns.
In addition to the greater sensitivity, repeatability, and capacity for field integration, specificity of our fluorescent LAMP assay could be increased with the integration of molecular probes. We encountered some non-specific amplification, which was limited to one technical replicate within each non-target triplicate. All non-target amplification was observed when relatively high DNA (>10 ng) was added into the reaction, which is much greater than what we would expect to encounter from trace DNA samples. Nonetheless, this presents a source of potential false positives. Consequently, we recommend diluting DNA and limiting input to less than 10 ng to avoid false positive detection. Alternatively, integrating molecular probes or beacons has previously led to increased specificity, reducing the risk of false positives due to non-specific amplification eliminating this issue altogether (Liu et al. 2017; Hardinge and Murray 2019). The molecular probe-based methods could additionally facilitate multiplexing, allowing for the detection of multiple biosecurity risk reptiles within one reaction (Tanner et al. 2012). The advantages for field integration using emerging in-field thermocyclers and the capacity for molecular probe integration means fluorescent LAMP methods are well poised for biosecurity screening, despite the higher financial costs in contrast to colourimetric LAMP.
The lack of amplification observed for some swabs retrieved from tanks subject to B. constrictor presence could stem from factors external to assay robustness, such as the shedding cycle of the individual present within the tank, the activity level of the individual while in the tank, or the lack of appropriate contact with the tank surfaces. Touch DNA is not distributed evenly or consistently, and deposition can vary due to a range of factors. DNA deposition dynamics is an understudied topic for reptiles, with a recent review highlighting the limited literature regarding terrestrial reptiles in contrast to other taxa (Nordstrom et al. 2022). A previous study has explored trace environmental DNA deposition, accumulation, and degradation for snakes in controlled lab and field environments (Kucherenko et al. 2018). In contrast to previous work, our study looks at recovery directly from surface contact samples as opposed to sand or soil. Additionally, our accumulation time (24 h) is far shorter compared with the 7-day accumulation and degradation assessed for P. guttatus by Kucherenko et al. (2018). This reflects the context to which we envision our work best applies – short term transportation and rapid compliance checks of existing collections. In the context of our study, temperature, activity level, position in shedding cycle, and time of last meal (and their interactions) are likely to drastically alter the movement of individuals within their enclosure and thus impact DNA deposition and downstream recovery and amplification. These factors will almost always remain unknown in a realistic biosecurity scenario; however, we predict they will play a large role in detectability.
Confirming LAMP amplification by quantitative PCR led to successful amplification of gBlock fragments and extracts from snake skins. This approach illustrates the utility of LAMP for presumptive testing because the results can easily be confirmed by use of the F3 and B3 primers for PCR amplification followed by Bi-directional Sanger sequencing for target confirmation. Presumptive positive LAMP detection provides the end-user with a rapid test (LAMP) that will reduce the number of samples sent off for more exhaustive laboratory confirmation testing (PCR followed by sequencing). The qPCR amplification protocol used throughout had an overall thermocycling period of 1 h and 40 min, indicating this approach would not be well suited for in-field detection of B. constrictor because it is time consuming and requires specialist equipment. It would, however, be beneficial to conduct a comparative assessment of qPCR and related methods such as TaqMan PCR against our developed LAMP approach to ensure the most appropriate method for any given biosecurity scenario can be recommended. This has been conducted for Trogoderma granarium (Khapra beetle) in biosecurity incursion scenarios, highlighting important concerns about LAMP and the lack of suitability for highly degraded sources of trace DNA (Trujillo-González et al. 2022). Assessing factors such as speed of detection, sensitivity, specificity, cost, resource requirements, and ease of integration in a comparative study across the most likely sample types to be encountered would be highly beneficial when contrasting the suitability of LAMP against PCR, qPCR, TaqMan PCR, and alternative forms of isothermal amplification, including recombinase polymerase amplification (Hsu et al. 2021). We have focused our efforts on developing methods for trace DNA detection in a biosecurity and compliance context, but our described assays could also be applied to field environmental DNA biomonitoring, if or when B. constrictor establishes in Australia. We addressed this somewhat by including S. kinghorni in our cross amplification testing – a species with a range that overlaps the climate matching conducted for B. constrictor (Freeman and Freeman 2009; Henderson and Bomford 2011). Implementation following the best-practice guidelines for Australian and New Zealand eDNA researchers and end-users will aid preparedness given the possibility and risk of establishment (De Brauwer et al. 2023). This includes accounting for B. constrictor sub-species (Card et al. 2016) and population-level differences in the mitochondrial control region, which will impact primer binding, LAMP efficiency, and broad applicability. This also extends to the detection of sub-species and populations for which trafficking has been identified as a high risk at the source. Having high-quality vouchered specimens as positive controls for individuals from these populations will be necessary to assess LAMP applicability, particularly if mismatches are present in primer-binding regions. International applications of our assays will also require the careful considerations of non-target native species common in local pet-trades, to minimise potential cross amplification.
In summary, we have developed two LAMP assays, one that relies on colourimetric chemistry and another that is based on fluorescence detection. We critically and rigorously tested both the described assays for key sample types, including synthetic DNA, DNA extracts from tissue, shed skins, and trace DNA recovered from glass surfaces. Both assays could be used to detect B. constrictor trace DNA in biosecurity contexts, with our fluorescence-based approach most broadly suited to all sample types with acceptable sensitivity, specificity, and robustness. We recommend further optimisation of sample recovery, DNA extraction, specificity, and comparison with alternative methods to best address current unknowns and weaknesses.
Data availability
The datasets related to this study are available on Figshare, doi:10.6084/m9.figshare.24136695.
Declaration of funding
This project was funded by the Australian Government Department of Agriculture, Fisheries and Forestry under the Environmental Biosecurity Project Fund (Reference ID: C08757).
Author contributions
ND, JJA, PC were involved in conceptualisation; ND, PC contributed to funding acquisition; ND conducted the experiments and laboratory analyses; JJA provided specialist technical advice; ND drafted the manuscript; JA, and PC edited the manuscript.
Acknowledgements
We acknowledge the traditional owners of the land on which our work was conducted, the Kaurna people. We wish to extend our gratitude to Gorge Wildlife Park for their continued support and assistance with the experimental component of the project, namely Daniel Markos. We also wish to thank Tamika Nash-Hahn for providing shed reptile skins and Jennifer M. Young for her previous support and assistance regarding conceptualisation. Funding was provided by the Australian Government Department for Agriculture, Fisheries and Forestry Environmental Biosecurity Project Fund (Project code: C08757). ND is a recipient of the Australian Government Research Training Program Stipend Scholarship. PC is a recipient of an ARC Discovery Project Grant (DP210103050) ‘Drivers of the live pet trade in Australian reptiles’ and an Australian Research Council Industry Laureate Fellow (IL230100175; ‘Combatting wildlife crime and preventing environmental harm’). All laboratory work was conducted in the Advanced Forensic identification Facility (ADIFF) at the University of Adelaide, and we wish to thank the lab manager Arif Malik for his support.
References
Agarwal A, Cunningham JP, Valenzuela I, Blacket MJ (2020) A diagnostic LAMP assay for the destructive grapevine insect pest, phylloxera (Daktulosphaira vitifoliae). Scientific Reports 10, 21229.
| Crossref | Google Scholar | PubMed |
Agarwal A, Rako L, Schutze MK, Starkie ML, Tay WT, Rodoni BC, Blacket MJ (2022) A diagnostic LAMP assay for rapid identification of an invasive plant pest, fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae). Scientific Reports 12, 1116.
| Crossref | Google Scholar | PubMed |
Blaser S, Diem H, von Felten A, Gueuning M, Andreou M, Boonham N, Tomlinson J, Müller P, Utzinger J, Frey JE, Bühlmann A (2018) From laboratory to point of entry: development and implementation of a loop-mediated isothermal amplification (LAMP)-based genetic identification system to prevent introduction of quarantine insect species. Pest Management Science 74, 1504-1512.
| Crossref | Google Scholar | PubMed |
Bricker J, Bushar LM, Reinert HK, Gelbert L (1996) Purification of high quality DNA from shed skins. Herpetological Review 27, 133.
| Google Scholar |
Brown AO, Ueland M, Stuart BH, Frankham GJ (2023) A forensically validated genetic toolkit for the species and lineage identification of the highly trafficked shingleback lizard (Tiliqua rugosa). Forensic Science International: Genetics 62, 102784.
| Crossref | Google Scholar |
Card DC, Schield DR, Adams RH, Corbin AB, Perry BW, Andrew AL, Pasquesi GIM, Smith EN, Jezkova T, Boback SM, Booth W, Castoe TA (2016) Phylogeographic and population genetic analyses reveal multiple species of Boa and independent origins of insular dwarfism. Molecular Phylogenetics and Evolution 102, 104-116.
| Crossref | Google Scholar | PubMed |
Carlisle-Nowak M, Sullivan N, Carrigan M, Knight C, Ryan C, Jacobson E (1998) Inclusion body disease in two captive Australian pythons (Morelia spilota variegata and Morelia spilota spilota). Australian Veterinary Journal 76, 98-100.
| Crossref | Google Scholar | PubMed |
Chang L-W, Jacobson ER (2010) Inclusion body disease, a worldwide infectious disease of boid snakes: a review. Journal of Exotic Pet Medicine 19, 216-225.
| Crossref | Google Scholar |
Ciavaglia S, Linacre A (2018) OzPythonPlex: an optimised forensic STR multiplex assay set for the Australasian carpet python (Morelia spilota). Forensic Science International: Genetics 34, 231-248.
| Crossref | Google Scholar | PubMed |
DAFF (2023) ‘The national priority list of exotic environmental pests, weeds and diseases’. (Australian Government Department of Agriculture, Fisheries and Forestry). Available at: https://www.agriculture.gov.au/biosecurity-trade/policy/environmental/priority-list
De Brauwer M, Clarke LJ, Chariton A, Cooper MK, de Bruyn M, Furlan E, MacDonald AJ, Rourke ML, Sherman CDH, Suter L, Villacorta-Rath C, Zaiko A, Trujillo-González A (2023) Best practice guidelines for environmental DNA biomonitoring in Australia and New Zealand. Environmental DNA 5, 417-423.
| Crossref | Google Scholar |
Deliveyne N, Cassey P, Linacre A, Delean S, Austin JJ, Young JM (2022) Recovering trace reptile DNA from the illegal wildlife trade. Forensic Science International: Animals and Environments 2, 100040.
| Crossref | Google Scholar |
Deliveyne N, Young JM, Austin JJ, Cassey P (2023) Shining a LAMP on the applications of isothermal amplification for monitoring environmental biosecurity. NeoBiota 82, 119-144.
| Crossref | Google Scholar |
Fetzner JW, Jr. (1999) Extracting high-quality DNA from shed reptile skins: a simplified method. BioTechniques 26, 1052-1054.
| Crossref | Google Scholar | PubMed |
Freeman A, Freeman A (2009) Habitat use in a large rainforest python (Morelia kinghorni) in the wet tropics of North Queensland, Australia. Herpetological Conservation and Biology 4, 252-260.
| Google Scholar |
Fukushima CS, Mammola S, Cardoso P (2020) Global wildlife trade permeates the Tree of Life. Biological Conservation 247, 108503.
| Crossref | Google Scholar | PubMed |
García-Díaz P, Cassey P (2014) Patterns of transport and introduction of exotic amphibians in Australia. Diversity and Distributions 20, 455-466.
| Crossref | Google Scholar |
García-Díaz P, Ross JV, Woolnough AP, Cassey P (2017) The illegal wildlife trade is a likely source of alien species. Conservation Letters 10, 690-698.
| Crossref | Google Scholar |
Hardinge P, Murray JAH (2019) Reduced false positives and improved reporting of loop-mediated isothermal amplification using quenched fluorescent primers. Scientific Reports 9, 7400.
| Crossref | Google Scholar | PubMed |
Heinrich S, Toomes A, Shepherd CR, Stringham OC, Swan M, Cassey P (2022) Strengthening protection of endemic wildlife threatened by the international pet trade: the case of the Australian shingleback lizard. Animal Conservation 25, 91-100.
| Crossref | Google Scholar |
Henderson W, Bomford M, Cassey P (2011) Managing the risk of exotic vertebrate incursions in Australia. Wildlife Research 38, 501-508.
| Crossref | Google Scholar |
Hill KGW, Nielson KE, Tyler JJ, McInerney FA, Doubleday ZA, Frankham GJ, Johnson RN, Gillanders BM, Delean S, Cassey P (2020) Pet or pest? Stable isotope methods for determining the provenance of an invasive alien species. NeoBiota 59, 21-37.
| Crossref | Google Scholar |
Hinsley A, Willis J, Dent AR, Oyanedel R, Kubo T, Challender DWS (2023) Trading species to extinction: evidence of extinction linked to the wildlife trade. Cambridge Prisms: Extinction 1, e10.
| Crossref | Google Scholar |
Horreo JL, Peláez ML, Fitze PS (2015) Skin sheds as a useful DNA source for lizard conservation. Phyllomedusa: Journal of Herpetology 14, 73-77.
| Crossref | Google Scholar |
Hsu Y-H, Yang W-C, Chan K-W (2021) Bushmeat species identification: recombinase polymerase amplification (RPA) combined with lateral flow (LF) strip for identification of Formosan Reeves’ muntjac (Muntiacus reevesi micrurus). Animals 11, 426.
| Crossref | Google Scholar | PubMed |
Hunter ME, Oyler-McCance SJ, Dorazio RM, Fike JA, Smith BJ, Hunter CT, Reed RN, Hart KM (2015) Environmental DNA (eDNA) sampling improves occurrence and detection estimates of invasive burmese pythons. PLoS ONE 10, e0121655.
| Crossref | Google Scholar | PubMed |
Jevtuševskaja J, Krõlov K, Tulp I, Langel Ü (2017) The effect of main urine inhibitors on the activity of different DNA polymerases in loop-mediated isothermal amplification. Expert Review of Molecular Diagnostics 17, 403-410.
| Crossref | Google Scholar | PubMed |
Kalendar R, Khassenov B, Ramankulov Y, Samuilova O, Ivanov KI (2017) FastPCR: an in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 109, 312-319.
| Crossref | Google Scholar | PubMed |
Katz AD, Harper LR, Sternhagen EC, Pearce SE, Melder CA, Sperry JH, Davis MA (2021) Environmental DNA is effective in detecting the federally threatened Louisiana Pinesnake (Pituophis ruthveni). Environmental DNA 3, 409-425.
| Crossref | Google Scholar |
Klymus KE, Merkes CM, Allison MJ, Goldberg CS, Helbing CC, Hunter ME, Jackson CA, Lance RF, Mangan AM, Monroe EM, Piaggio AJ, Stokdyk JP, Wilson CC, Richter CA (2020) Reporting the limits of detection and quantification for environmental DNA assays. Environmental DNA 2, 271-282.
| Crossref | Google Scholar |
Kucherenko A, Herman JE, III EME, Urakawa H (2018) Terrestrial snake environmental DNA accumulation and degradation dynamics and its environmental application. Herpetologica 74, 38-49.
| Crossref | Google Scholar |
Kyle KE, Allen MC, Dragon J, Bunnell JF, Reinert HK, Zappalorti R, Jaffe BD, Angle JC, Lockwood JL (2022) Combining surface and soil environmental DNA with artificial cover objects to improve terrestrial reptile survey detection. Conservation Biology 36, e13939.
| Crossref | Google Scholar |
Lassaline CR, Stringham OC, Moncayo S, Toomes A, Cassey P (2023) Untangling the web: dynamics of Australia’s online terrestrial invertebrate trade. Austral Entomology 62, 372-387.
| Crossref | Google Scholar |
Layne T, Jackson K, Scott A, Tanner NA, Piland A, Haverstick DM, Landers JP (2021) Optimization of novel loop-mediated isothermal amplification with colorimetric image analysis for forensic body fluid identification. Journal of Forensic Sciences 66, 1033-1041.
| Crossref | Google Scholar | PubMed |
Lee PLM (2017) DNA amplification in the field: move over PCR, here comes LAMP. Molecular Ecology Resources 17, 138-141.
| Crossref | Google Scholar | PubMed |
Liu W, Huang S, Liu N, Dong D, Yang Z, Tang Y, Ma W, He X, Ao D, Xu Y, Zou D, Huang L (2017) Establishment of an accurate and fast detection method using molecular beacons in loop-mediated isothermal amplification assay. Scientific Reports 7, 40125.
| Crossref | Google Scholar | PubMed |
Lockwood JL, Welbourne DJ, Romagosa CM, Cassey P, Mandrak NE, Strecker A, Leung B, Stringham OC, Udell B, Episcopio-Sturgeon DJ, Tlusty MF, Sinclair J, Springborn MR, Pienaar EF, Rhyne AL, Keller R (2019) When pets become pests: the role of the exotic pet trade in producing invasive vertebrate animals. Frontiers in Ecology and the Environment 17, 323-330.
| Crossref | Google Scholar |
Margulies JD, Bullough L-A, Hinsley A, Ingram DJ, Cowell C, Goettsch B, Klitgård BB, Lavorgna A, Sinovas P, Phelps J (2019) Illegal wildlife trade and the persistence of “plant blindness”. Plants, People, Planet 1, 173-182.
| Crossref | Google Scholar |
Marks BA (2022) Technological advances in biosecurity monitoring. The APPEA Journal 62, S306-S309.
| Crossref | Google Scholar |
Marshall BM, Strine C, Hughes AC (2020) Thousands of reptile species threatened by under-regulated global trade. Nature Communications 11, 4738.
| Crossref | Google Scholar | PubMed |
Morton O, Scheffers BR, Haugaasen T, Edwards DP (2021) Impacts of wildlife trade on terrestrial biodiversity. Nature Ecology & Evolution 5, 540-548.
| Crossref | Google Scholar | PubMed |
Nguyen DV, Nguyen VH, Seo TS (2019) Quantification of colorimetric loop-mediated isothermal amplification process. BioChip Journal 13, 158-164.
| Crossref | Google Scholar |
Nikunj B, Vivek U (2019) An application of loop mediated iso-thermal amplification technology in forensic science. International Journal of Research in Advent Technology 7, 1247-1252.
| Crossref | Google Scholar |
Nordstrom B, Mitchell N, Byrne M, Jarman S (2022) A review of applications of environmental DNA for reptile conservation and management. Ecology and Evolution 12, e8995.
| Crossref | Google Scholar | PubMed |
Notomi T, Mori Y, Tomita N, Kanda H (2015) Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. Journal of Microbiology 53, 1-5.
| Crossref | Google Scholar | PubMed |
Optigene (2017) LAMP user guide – assay design & primers. Available at www.optigene.co.uk
Pili AN, Tingley R, van Winkel D, Maria L, Chapple DG (2023) The escalating global problem of accidental human-mediated transport of alien species: a case study using alien herpetofauna interceptions in New Zealand. Biological Conservation 278, 109860.
| Crossref | Google Scholar |
Pirotta V, Shen K, Liu S, Phan HTH, O’Brien JK, Meagher P, Mitchell J, Willis J, Morton E (2022) Detecting illegal wildlife trafficking via real time tomography 3D X-ray imaging and automated algorithms. Frontiers in Conservation Science 3, 757950.
| Crossref | Google Scholar |
Purcell SW, Polidoro BA, Hamel J-F, Gamboa RU, Mercier A (2014) The cost of being valuable: predictors of extinction risk in marine invertebrates exploited as luxury seafood. Proceedings of the Royal Society B: Biological Sciences 281, 20133296.
| Crossref | Google Scholar |
Rako L, Agarwal A, Semeraro L, Broadley A, Rodoni BC, Blacket MJ (2021) A LAMP (Loop-mediated isothermal amplification) test for rapid identification of Khapra beetle (Trogoderma granarium). Pest Management Science 77, 5509-5521.
| Crossref | Google Scholar | PubMed |
Rawlings LH, Rabosky DL, Donnellan SC, Hutchinson MN (2008) Python phylogenetics: inference from morphology and mitochondrial DNA. Biological Journal of the Linnean Society 93, 603-619.
| Crossref | Google Scholar |
R Core Team (2023) ‘R: a language and environment for statistical computing.’ (R Foundation for Statistical Computing: Vienna, Austria). Available at https://www.R-project.org/
Ripple WJ, Wolf C, Newsome TM, Betts MG, Ceballos G, Courchamp F, Hayward MW, Van Valkenburgh B, Wallach AD, Worm B (2019) Are we eating the world’s megafauna to extinction? Conservation Letters 12, e12627.
| Crossref | Google Scholar |
Scheffers BR, Oliveira BF, Lamb I, Edwards DP (2019) Global wildlife trade across the tree of life. Science 366, 71-76.
| Crossref | Google Scholar | PubMed |
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676-682.
| Crossref | Google Scholar | PubMed |
Schumacher J, Jacobson ER, Homer BL, Gaskin JM (1994) Inclusion body disease in boid snakes. Journal of Zoo and Wildlife Medicine 25, 511-524.
| Google Scholar |
Scott AT, Layne TR, O’Connell KC, Tanner NA, Landers JP (2020) Comparative evaluation and quantitative analysis of loop-mediated isothermal amplification indicators. Analytical Chemistry 92, 13343-13353.
| Crossref | Google Scholar | PubMed |
Sills J, Gonçalves SC, Haelewaters D, Furci G, Mueller GM (2021) Include all fungi in biodiversity goals. Science 373, 403.
| Crossref | Google Scholar |
Starkie ML, Fowler EV, Zhu X, Agarwal A, Rako L, Schneider IC, Schutze MK, Royer JE, Gopurenko D, Gillespie P, Blacket MJ (2022) Loop-mediated isothermal amplification (LAMP) assays for detection of the New Guinea fruit fly Bactrocera trivialis (Drew) (Diptera: Tephritidae). Scientific Reports 12, 12602.
| Crossref | Google Scholar | PubMed |
Stringham OC, García-Díaz P, Toomes A, Mitchell L, Ross JV, Cassey P (2021a) Live reptile smuggling is predicted by trends in the legal exotic pet trade. Conservation Letters 14, e12833.
| Crossref | Google Scholar |
Stringham OC, Toomes A, Kanishka AM, Mitchell L, Heinrich S, Ross JV, Cassey P (2021b) A guide to using the internet to monitor and quantify the wildlife trade. Conservation Biology 35, 1130-1139.
| Crossref | Google Scholar | PubMed |
Tanner NA, Zhang Y, Evans TC, Jr. (2012) Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. BioTechniques 53, 81-89.
| Crossref | Google Scholar | PubMed |
Tanner NA, Zhang Y, Evans TC, Jr. (2015) Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. BioTechniques 58, 59-68.
| Crossref | Google Scholar | PubMed |
Toomes A, García-Díaz P, Wittmann TA, Virtue J, Cassey P (2020) New aliens in Australia: 18 years of vertebrate interceptions. Wildlife Research 47, 55-67.
| Crossref | Google Scholar |
Trujillo-González A, Thuo DN, Divi U, Sparks K, Wallenius T, Gleeson D (2022) Detection of khapra beetle environmental DNA using portable technologies in Australian biosecurity. Frontiers in Insect Science 2, 795379.
| Crossref | Google Scholar |
Wickham H (2011) ggplot2. WIREs Computational Statistics 3, 180-185.
| Crossref | Google Scholar |
Wimbles R, Melling LM, Cain B, Davies N, Doherty J, Johnson B, Shaw KJ (2021) On-site genetic analysis for species identification using lab-on-a-chip. Ecology and Evolution 11, 1535-1543.
| Crossref | Google Scholar | PubMed |
Yu S, Gu W, Yu Y, Qu Q, Zhang Y (2019) Species identification of felis and vulpes by a novel loop-mediated isothermal amplification assay in fur products. Journal of Engineered Fibers and Fabrics 14, 1558925018820720.
| Crossref | Google Scholar |



