Kinship analysis reveals low dispersal in a hog deer (Axis porcinus) population in Wilsons Promontory National Park, Australia
Erin Hill A B * , Nicholas Murphy A C , Adrian Linacre D , Simon Toop E and Jan M. Strugnell A F
A B * , Nicholas Murphy A C , Adrian Linacre D , Simon Toop E and Jan M. Strugnell A F
A La Trobe University, Department of Environment and Genetics, Melbourne, Vic., Australia.
B CSIRO Health and Biosecurity, Canberra, ACT, Australia.
C La Trobe University, Research Centre for Future Landscapes, Melbourne, Vic., Australia.
D Flinders University, College of Science and Engineering, Adelaide, SA, Australia.
E Game Management Authority, Melbourne, Vic., Australia.
F James Cook University, Centre for Sustainable Tropical Fisheries and Aquaculture, Townsville, Qld, Australia.
Wildlife Research 50(9) 746-756 https://doi.org/10.1071/WR22098
Submitted: 6 June 2022 Accepted: 29 May 2023 Published: 21 June 2023
© 2023 The Author(s) (or their employer(s)). Published by CSIRO Publishing. This is an open access article distributed under the Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License (CC BY-NC-ND)
Abstract
Context: A wild population of non-native hog deer has established in the Gippsland region of Victoria, Australia, and there is particular concern about its impact on native vegetation in Wilsons Promontory National Park (WPNP). Since 2015, there has been annual culling of hog deer at WPNP to reduce deer abundances and impacts.
Aims: The aims of this study were to use a kinship approach based on genotyping to assess contemporary dispersal of hog deer across WPNP, by identifying close kin, to determine whether dispersal of deer into culled sites from unculled sites may affect the long-term success of management there. Differences in the dispersal of male and female hog deer were also investigated.
Methods: In total, 91 hog deer tissue samples were collected across WPNP and surrounding sites. Single nucleotide polymorphism (SNP) markers were sequenced, and a final dataset comprising 8275 SNPs was used for analysis. First-order, second-order, and intermediate relative pairs were identified, and the geographic distance between these pairs was assessed to determine inter-pair distances to infer dispersal. Spatial autocorrelation between male and female samples was evaluated to measure the effects of sex-biased dispersal.
Key results: Only seven second-order relative pairs were found across different sites, with a 30 km distance between the furthest pair observed. However, most inter-pair distances across sites were ~5–10 km. Analyses of sex-biased dispersal showed that movement by deer was not strongly influenced by one sex.
Conclusions: Although hog deer in WPNP are genetically similar, most relatives that were sampled were not widely dispersed. This suggests that there is limited dispersal of hog deer across this park.
Implications: Recolonisation of hog deer at culled sites via dispersal is likely to be infrequent in WPNP. Kinship analysis provides an effective method of assessing contemporary dispersal and could be applied to other species to assess fine-scale movement across landscapes.
Keywords: Axis, Cervidae, dispersal, hog deer, introduced species, kinship, population control, sex-biased dispersal.
Introduction
Many deer species have been deliberately released outside their native ranges to establish novel populations for game hunting (Long 2003; Dolman and Wäber 2008). Following introductions of deer, management of these new populations often involves encouraging increases in abundance and distribution through hunting restrictions and maintaining suitable habitat. These new populations can have undesirable impacts in their new environments, including increasing the biomass of unpalatable plant species through preferential grazing/browsing of palatable species, reducing understorey biomass, and browsing of seedlings that can lead to a reduction in tree growth and a lower density of mature stands (Côté et al. 2004; Davis et al. 2016; Ramsey et al. 2018). Overabundant deer have also been implicated in the declining abundance of songbirds in forest communities (Jirinec et al. 2017) and have been shown to positively affect the abundance of invasive plant species (Kalisz et al. 2014; Bourg et al. 2017).
In Australia, six deer species have established self-sustaining wild populations following their introduction by European settlers in the 19th and 20th centuries (Moriarty 2004): fallow deer (Dama dama); red deer (Cervus elaphus); sambar deer (Cervus unicolor); rusa deer (Cervus timorensis); chital (Axis axis); and hog deer (Axis porcinus), with at least one of these species found in every State or Territory. Hog deer were first released into Australia in the 1860s, where they have hybridised with the closely related chital (Hill et al. 2019), and they are now common along the coastal zone of the Gippsland region of Victoria (Mayze and Moore 1990). The species is present in Wilsons Promontory National Park (WPNP), a 505 km2 national park in southwest Gippsland. This park is considered significant because it contains several threatened flora and fauna species. It was recently estimated that there are approximately 2000 hog deer in WPNP (3.9 per km2; Ramsey et al. 2019). Hog deer and native swamp wallabies (Wallabia bicolor) in WPNP have highly similar diets and may compete for food (Davis et al. 2008). Also, hog deer ingest and disperse the seeds of native and exotic plant species in WPNP (Davis et al. 2010). A desire to reduce the negative impacts of hog deer in WPNP led to the implementation of annual culling there in 2015. There is now a proposal to eradicate hog deer from WPNP (Parks Victoria n.d.). Given the interest in controlling and potentially eradicating hog deer in WPNP, it is important to understand dispersal of this species there.
There is no well-defined breeding season for hog deer in Australia, but most births occur in spring (Toop, pers. obs.), with females able to conceive again soon after giving birth (Mayze and Moore 1990). Sexual maturity can be attained at 12 months of age (Taylor 1971), but most females give birth to their first offspring at about 2 years (Mayze and Moore 1990; Dhungel and O’Gara 1991). Dispersal in hog deer appears to be variable. The mean home ranges of female and male hog deer in Nepal were 60 ha and 80 ha, respectively, in one study (Dhungel and O’Gara 1991), and 40 ha and 50 ha in another study (Odden and Wegge 2007). Conversely, an average home range of only 20 ha was reported for hog deer in Australia, with individuals having overlapping home ranges (Taylor 1971). These home ranges are small in comparison with those of other deer species (Lesage et al. 2000; Borkowski and Pudełko 2007; Reinecke et al. 2014), and suggest that hog deer are sedentary (Dhungel and O’Gara 1991). However, extensive movement of hog deer following translocations has been observed, with collared deer being sighted up to 70 km from their initial release point (Mayze and Moore 1990). These findings suggest that male deer move much further than females, which is common amongst mammalian species because males are typically the dispersing sex (Dobson 1982). Hog deer are also adept swimmers and have been observed swimming off the coast of Victoria (Mayze and Moore 1990). These life-history traits, which have contributed to hog deer becoming established across the south-eastern coast of Victoria, could be important for the recolonisation of sites within WPNP that are subject to culling.
To help understand if hog deer are likely to recolonise culled sites at WPNP following culling, estimates of fine-scale dispersal are needed. Genetic analysis using kinship methods are a viable approach for estimating dispersal distances via measurement of inter-pair distances among identified kin (Lepais et al. 2010; Vangestel et al. 2011; Schunter et al. 2014; Escoda et al. 2017). If family groups are only detected within a localised region, this would suggest low dispersal, whereas family groups that are spread across the landscape are the result of a higher dispersal capability. Genetic data can also be used to measure the dispersal differences between the sexes (Banks and Peakall 2012) to determine if one sex is the predominant disperser. By utilising samples from deer culled at WPNP in 2015 and 2016, and using single nucleotide polymorphism (SNP) genotyping, this study aims to investigate fine-scale dispersal of hog deer across WPNP using kinship analyses, and to identify potential sources of reinvasion both within the park and at nearby mainland and island sites.
Methods
Sampling, DNA extraction and next generation sequencing
Fifty-five liver and tongue samples were collected by recreational hunters from wild, free-ranging hog deer at Yanakie, Snake Island, Sunday Island, and Boole Poole during the annual March–April hunting seasons in 2015–2017. Thirty-nine liver samples were collected during the culls conducted in WPNP in August 2015 and 2016 (Fig. 1, Table 1). Animal ethics approval was not required for the collection of samples in this study because animals were killed as part of established population management and recreational harvesting programs. Culls were conducted at four sites: Tidal River campgrounds; Oberon Bay; Darby River airstrip; and Darby River at Cotters South, with the latter two sites combined into a single site (Kangaroo Valley) in the present study (Game Management Authority 2017). Of the samples used for genetic analysis, approximate distances between the furthest samples at individual sites across WPNP were 1.57 km at Oberon Bay, 1.03 km at Tidal River, and 5.15 km at Kangaroo Valley, with estimated radiuses of 0.9 km, 0.58 km and 3.02 km, respectively. The sex of each deer was recorded and the age of each deer was estimated by experienced personnel through inspection of the jawbone, by examining the wear on the teeth and the number of teeth that had erupted. This method is considered accurate in aging hog deer up to 3 years in age (Scroggie et al. 2012), although within the present study, hog deer were aged up to 6 years using this method. Boole Poole samples were included to confirm the robustness of the SNP data returned for analysis. This site is towards the easternmost area of the hog deer distribution in Victoria and has previously been shown to be genetically distinct from the western sites of interest using short tandem repeat (STR) analysis (Hill et al. 2022).
Location of hog deer sampling sites (green circles) in eastern Victoria, Australia. Records of hog deer in the Atlas of Living Australia (2019) are shown in blue circles.

| Site | Land tenure | n | Male | Female |
|---|---|---|---|---|
| Boole Poole | Coastal Park | 13 | 10 | 3 |
| Sunday Island | Private Land | 12 | 6 | 6 |
| Snake Island | Coastal Park | 14 | 9 | 5 |
| Yanakie | Public Land | 13 | 8 | 5 |
| Kangaroo Valley* | National Park | 14 | 7 | 7 |
| Tidal River* | National Park | 14 | 7 | 7 |
| Oberon Bay* | National Park | 11 | 7 | 4 |
| TOTAL | – | 91 | 54 | 37 |
Sites with an asterisk represent areas within WPNP.
We extracted DNA using a DNeasy Blood and Tissue Kit (QIAGEN) following the manufacturer’s instructions, with negative controls run throughout, and DNA was quantified using a Qubit dsDNA broad range quantitation kit with a Qubit 2.0 Fluorometer (Invitrogen). Samples were then either diluted or concentrated to obtain a final concentration between 15 and 30 ng/μL. To concentrate samples an RVC 2-18 Rotational Vacuum Concentrator (John Morris Scientific) was used. Samples were sent to Diversity Arrays Technology (DArT) in Canberra, Australia, for genotyping by sequencing. Briefly, DArT utilises a reduced representation sequencing method, using restriction-enzyme (RE) digestion, adapter ligation, and amplification of adapter-ligated fragments (Kilian et al. 2012). Typically, 50 000 DNA fragments are assayed for polymorphism using this method. Sequencing was conducted on an Illumina Hiseq2500 yielding 2.5 million reads per sample, and reads were aligned back to a red deer (Cervus elaphus) genome available on GenBank (BioProject PRJNA324173). Technical replicates for 10% of samples were included to determine genotyping accuracy. One sample from Yanakie and two samples from Sunday Island did not produce sufficient data after genotyping, so a total of 91 samples were retained for further analysis.
SNP filtering
DArT sequencing returned 12 881 SNPs, of which 9159 were successfully aligned to the red deer genome. 295 SNPs were mapped to the red deer X chromosome, and two SNPs were mapped to the red deer Y chromosome, and so to avoid any potential sex linkage, all SNPs aligned to an X or Y chromosome were removed from further analysis. The remaining markers were then additionally filtered using the dartR 2.1.4 package implemented in R 4.2.0 (Gruber et al. 2018; R Core Team 2020). SNPs were filtered based on average reproducibility across technical replicates, using a threshold of 1, which removed 2763 loci. Loci with >10% missing data were removed, resulting in the loss of 684 loci. SNPs that appeared on the same sequencing fragment and were therefore likely affected by linkage were filtered, using the ‘best’ method to retain one SNP per sequencing fragment, and removing 178 loci. 593 SNPs with minor allele frequencies at or below a threshold of 0.05 were removed. SNPs identified as being out of Hardy Weinberg Equilibrium (HWE) with a P-value of 0.05 in more than two populations (where all WPNP sites were considered one population) were removed, filtering 38 loci. Filtering based on read depth was then undertaken, removing loci with read depths below 5 and above 75, resulting in the loss of 33 SNPs. To assess heterozygote excess amongst SNP loci, observed heterozygosity was calculated for each locus on the entire dataset, as well as individual populations (where all WPNP sites were combined into one population). Observed heterozygosity ranged from 0.03 to 0.53 across the entire dataset; however, if two or more populations comprised an observed heterozygosity >0.7 at a locus, then this SNP was removed from the dataset. In total, 20 SNPs were removed using this method. SNPs were also filtered for monomorphic and sex-linked loci; however, these tests did not result in the removal of any loci. Filtering of individuals with >5% missing data and overall heterozygosity >0.7 was also conducted, with no individuals removed during either of these steps. The final dataset consisted of 91 individuals and 8275 loci, with an average read depth per locus of 18.92. A summary of the filtering steps can be found in Supplementary material Table S1.
Data analysis
Principal components analysis (PCA) was performed using the ‘tab’ and ‘dudi.pca’ functions implemented in the R package adegenet 2.1.1 (Jombart 2008; Jombart and Ahmed 2011), with data transformed into relative frequencies, the NA.method set to ‘mean’, and seven axes retained. Analyses of genetic diversity and inbreeding were calculated for each population using a combination of the R packages hierfstat 0.5, dartR and inbreedR 0.3.3 (Goudet 2005; Stoffel et al. 2016). DartR and hierfstat were used to calculate observed heterozygosity and gene diversity (HO and Hs) and inbreeding coefficient FIS, and inbreedR was used to calculate identity disequilibrium (g2) and standardised multi-locus heterozygosity (sMLH). Results of sMLH were averaged per population and the standard error reported. These calculations were conducted for each site separately, as well as combining all WPNP sites into a single population and combining all WPNP and Yanakie sites into a single population.
The program Colony 2.0.6.4 was used to assess kinship among individuals within each genetic cluster identified from the PCA (Jones and Wang 2010). This program uses a maximum-likelihood approach, with likelihood estimated over the entire pedigree arrangement rather than between pairs of individuals (Jones and Wang 2010). Assumptions included inbreeding, polygamy in both males and females and no sibship prior. The pairwise full likelihood combined method for analysis was chosen with high precision, and a 0.05 allele dropout rate and false allele rate per locus was assumed. Colony was then run three times to ensure convergence of results. Colony assigns related pairs as either full siblings or half siblings, but these categories can be misleading when including samples of different ages because other relationships are possible within the dataset. For example, full siblings and parent–offspring would be expected to comprise a similar related value of 0.5, whereas half sibling relationships may also represent grandparent–grandchild, or aunt/uncle and niece/nephew relationships with related values of 0.25. Given the data presented here comprises hog deer with a range of ages (0.5–6+ years), it is not possible to accurately confirm if such pairs are true siblings. Therefore, hereafter full sibling pairs identified from Colony are referred to as first-order relatives, and half sibling pairs as second-order relatives. Relatives identified in Colony with a probability of 0.99 or higher were retained for final analysis, resulting in the removal of three related pairs.
Pairwise relatedness between individuals was calculated using the related 1.0 package in R (Pew et al. 2015), using modified ‘compareestimators’ and ‘familysim’ functions, which allowed more than 100 SNP loci to be used (https://github.com/James-Odwyer/related_extended_function). The ‘compareestimators’ function identified the dyadml estimator as the best fit for the data, which was subsequently used to calculate pairwise relatedness estimates. This estimator uses a dyadic maximum-likelihood method, which applies likelihood estimations over pairs of individuals. The ‘familysim’ function was used to generate relatedness values for simulated pairs of known relatedness, with 100 simulated pairs being created for each familial relationship and a density plot generated showing the overlap between parent–offspring, full sibling, half sibling, and unrelated pairwise relatedness (Fig. S1). Overlap among these relationship classes from the simulated dataset was only observed between parent–offspring and full siblings, but pairwise relatedness values within the hog deer dataset fell between the ranges of each simulated relationship class. For example, overlap between half siblings and full sibling/parent–offspring was not observed in the simulated dataset; however, related individuals comprising pairwise values that fell between these relationship categories were observed within the hog deer dataset and are likely to represent true related pairs. In order to include these related individuals in the dataset, three categories were assigned to the pairs identified in related: first-order relatives that correspond to the full sibling/parent–offspring categories; second-order relatives that correspond to the half sibling category; and intermediate relatives that fall between the pairwise values of the preceding two relative classes. First-order relatives comprised pairwise values of 0.476 and above, second-order relatives between 0.226 and 0.279, and intermediate relatives between 0.280 and 0.475, as identified in the simulated dataset.
Genetic networks were created from all pairwise relationships identified in Colony and related using edgebundleR 0.1.4 to visualise the connectivity of relatives among sites and the differences between relative pairs in the two datasets (Bostock et al. 2016). Geographic distance between each relative pair identified in Colony and related was calculated using the ‘distm’ function in the R package geosphere 1.5 (Hijmans et al. 2017). Only approximate geographic coordinates were available for samples collected from Boole Poole, Sunday Island, and Yanakie; therefore, genetic distances between relatives within these populations were not calculated. A histogram of the distances between each relative pair was then plotted using ggplot2 3.2.1 (Wickham 2016).
To assess sex-biased dispersal within hog deer, samples from WPNP and Yanakie were split into male and female groups and spatial autocorrelation of genetic and geographic distances following Smouse and Peakall (1999) was calculated in dartR. Correlograms of the results were then generated using ggplot2. This analysis was then repeated with kin identified in Colony removed from each group. The function ‘sexbias.test’ was used in hierfstat to additionally assess sex-biased dispersal using the methods proposed by Goudet et al. (2002). All four test methods were calculated (F-statistics (FIS and FST), mean assignment index (mAIc), and variance of assignment index (vAIc)), with 1000 permutations used for each test.
Results
PCA analyses identified four distinct genetic clusters (Fig. 2). All WPNP sites, together with Yanakie, clustered into a single group, but Sunday Island, Snake Island, and Boole Poole samples each formed distinct, separate genetic groups. Sunday Island and Snake Island clusters were closely related but genetically distinct, and Boole Poole samples were separate from all other plotted clusters. Planes 1 and 2 explained 8.12% and 5.07% of the variance, respectively, in the PCA plot. Genetic diversity and inbreeding indices were similar across all populations, with inbreeding indices FIS and g2 only deviating slightly from 0 (FIS 0.0244–0.075; g2 0.0018–0.0166), indicating minor levels of inbreeding (FIS) and variance of inbreeding (g2); however, all g2 estimates were statistically significant (P (g2 > 0) = 0.001). A summary of the diversity and inbreeding indices can be found in Table S2.
Principal component analysis plot for all hog deer samples using 8275 SNP loci. BP, Boole Poole; SU, Sunday Island; SN, Snake Island; YA, Yanakie; KV, Kangaroo Valley; OB, Oberon Bay; TR, Tidal River.

Colony detected five first-order relative pairs and 50 second-order relative pairs with a probability >0.99, and related identified five first-order relative pairs, 51 second-order relative pairs and 29 intermediate relative pairs (Table 2). Across all kin pairs identified, 54 were unique to related, 24 were unique to Colony, and 31 were identified using both programs. Although the overall number of related pairs were similar for both programs, Colony detected many more relatives within Tidal River than related (25 and 10 total kin pairs, respectively), and related detected more relatives at Sunday Island and Boole Poole (14 and 44 total kin pairs, respectively) than Colony (4 and 1 total kin pairs, respectively) (Fig. 3). These differences were primarily from the detection of second-order relatives.
| Colony | Related | ||||||
|---|---|---|---|---|---|---|---|
| Site | First-order pairs | Second-order pairs | Unrelated individuals | First-order pairs | Intermediate pairs | Second-order pairs | Unrelated individuals |
| Yanakie | 0 | 3 | 7 | 0 | 2 | 0 | 9 |
| Kangaroo Valley | 2 | 5 | 4 | 2 | 0 | 2 | 7 |
| Tidal River | 1 | 24 | 1 | 1 | 4 | 5 | 2 |
| Oberon Bay | 2 | 5 | 1 | 1 | 4 | 2 | 2 |
| Snake Island | 0 | 1 | 12 | 0 | 0 | 1 | 12 |
| Sunday Island | 0 | 4 | 4 | 1 | 7 | 6 | 0 |
| Boole Poole | 0 | 1 | 11 | 0 | 12 | 32 | 0 |
| Mixed Sites | 0 | 7 | – | 0 | 0 | 3 | – |
| TOTAL | 5 | 50 | 40 | 5 | 29 | 51 | 32 |
Genetic networks of all hog deer relative pairs detected by Colony and related. Circles represent individual samples, with lines between samples indicating relatives. Sample names for each individual are displayed and samples coloured by site.

All first-order pairs were sampled from the same sites; in Colony this included two pairs each at Kangaroo Valley and Oberon Bay, and one pair from Tidal River (Table 3). First-order pairs were relatively similar in the related dataset, but one pair from Oberon Bay identified from Colony was classed as an intermediate relative pair in related, and a pair from Sunday Island was identified as first-order relatives. Of the second-order kin pairs identified through Colony, 24 pairs were detected from Tidal River, and five second-order pairs each were present in Oberon Bay and Kangaroo Valley (Table 2). From related, a majority of the second-order kin pairs were detected in Boole Poole, comprising 32 pairs (Table 2). All second-order pairs from WPNP sites identified by related were also found to be second-order relatives in the Colony dataset; this included two pairs each detected at Kangaroo Valley and Oberon Bay, and five pairs present in Tidal River. Similar patterns were seen in the intermediate relative pairs detected by related, with the majority from Boole Poole and Sunday Island.
| Relationship | Program identified | Site relative 1 | Sample | Sex | Estimated birth year | Site relative 2 | Sample | Sex | Estimated birth year | Distance between relatives (km) |
|---|---|---|---|---|---|---|---|---|---|---|
| First-order | Colony | Oberon Bay | OB140 | M | 2011† | Oberon Bay | OB643 | M | 2013 | 1.18 |
| First-order | Both | Oberon Bay | OB644 | M | 2010† | Oberon Bay | OB7 | F | 2010† | 0.11 |
| First-order | Both | Tidal River | TR640 | F | 2012† | Tidal River | TRZ52209 | M | 2010† | 0.18 |
| First-order | Both | Kangaroo Valley | KV148 | F | 2014 | Kangaroo Valley | KV150 | F | 2014 | 0.00 |
| First-order | Both | Kangaroo Valley | KVZ52241 | F | 2012 | Kangaroo Valley | KVZ52264 | F | 2013 | 0.67 |
| First-order | Related* | Sunday Island | SUI425 | M | – | Sunday Island | SUI437 | F | – | – |
| Second-order | Colony* | Oberon Bay | OB645 | M | 2015 | Tidal River | TR8 | M | 2015 | 4.44 |
| Second-order | Both | Oberon Bay | OB7 | F | 2010† | Tidal River | TR136 | F | 2010† | 5.16 |
| Second-order | Both | Oberon Bay | OB644 | M | 2010† | Tidal River | TR136 | F | 2010† | 5.18 |
| Second-order | Both | Kangaroo Valley | KVZ52247 | F | 2013 | Tidal River | TR637 | M | 2010† | 10.13 |
| Second-order | Colony* | Kangaroo Valley | KVZ52264 | F | 2013 | Tidal River | TR637 | M | 2010† | 10.51 |
| Second-order | Colony* | Kangaroo Valley | KVZ52241 | F | 2012 | Tidal River | TR637 | M | 2010† | 10.51 |
| Second-order | Colony* | Oberon Bay | OB137 | F | 2013 | Yanakie | YA619 | M | 2012† | 31.49 |
Relationship indicates the highest order relationship identified by either Colony or related, with an asterisk indicating a kin pair was only identified in that program. Birth year was calculated by subtracting the estimated age of an individual from the collection year. Samples for which the estimated age is >3 years are indicated by †.
A majority of all relative pairs detected by both Colony and related were present at the same sites. From Colony, 87.3% of all pairs were detected from the same sites, and 96.5% of pairs were from the same site in the related dataset. Inter-pair distances were calculated for 47 relative pairs across WPNP, and of these, 35 pairs were sampled <1 km apart (Fig. 4). Only seven relative pairs in total were sampled from different sites across both kin programs. From Colony, three of these were recorded each between Kangaroo Valley and Tidal River, and Oberon Bay and Tidal River, and a single pair was observed between Oberon Bay and Yanakie (Table 3). Only three of the seven second-order relatives detected between sites by Colony were also recognised as second-order relatives by related (Table 3). Related did not detect any additional relative pairs of any relationship class between sites. Inter-pair distances across sites ranged from 4.44 to 31.49 km, with the largest distance observed between deer sampled from Oberon Bay and Yanakie (Fig. 4, Table 3). These second-order relatives represent the only case of deer movement between WPNP and a site outside WPNP. This pair was detected by the Colony analysis only.
Dispersal distances between hog deer relative pairs detected by Colony, related, and both programs.

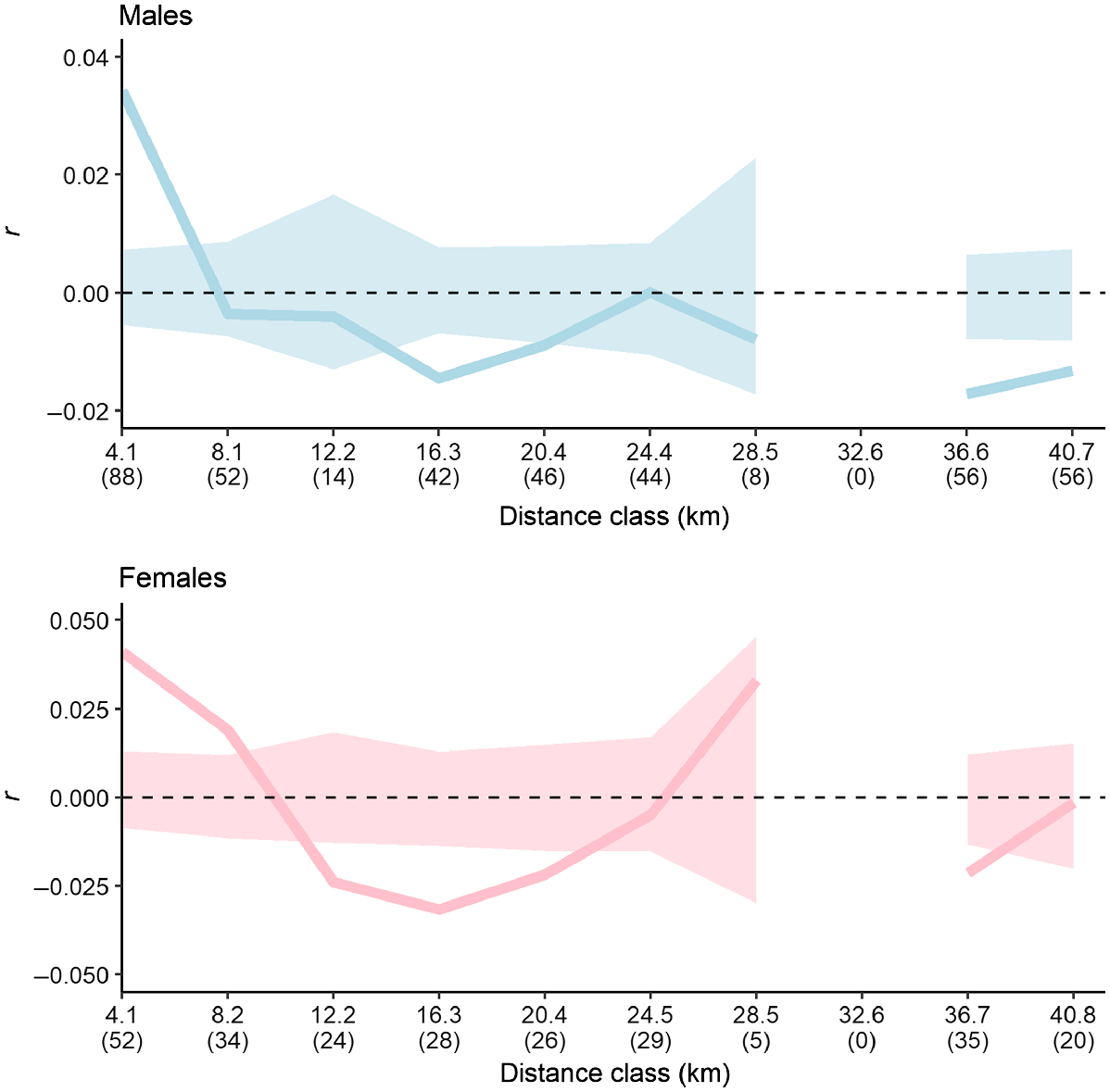
Spatial autocorrelation analyses across WPNP and Yanakie revealed similar patterns in males and females (Fig. 5). The correlations were positive and significant for the first 4 km, but beyond that distance the correlations became weaker. In males, this positive correlation was observed for the first distance class (4.1 km), followed by a significant negative correlation at 16.3 km. A similar trend was evident in females, with a significant positive correlation observed at the first two distance classes (4.1 km and 8.2 km), before rapidly decreasing to a significantly negative correlation at 12.2 km. Spatial autocorrelation results were similar when kin were removed from each dataset, but the frequency of significant negative correlations in both sexes was reduced (Fig. S2). None of the sex-bias tests conducted in hierfstat were statistically significant (Table 4).
Correlogram showing genetic similarity (r) at different distance classes for all male samples and all female samples. Solid lines show genetic similarity across distances, and shaded areas show 95% confidence intervals, with values inside these confidence intervals representing a random distribution of genotypes and therefore no correlation. Values outside these confidence intervals indicate a significant correlation. Numbers in parentheses are sample sizes.
| Test | Statistic | P-value |
|---|---|---|
| mAIc | −0.815 | 0.434 |
| vAIc | 0.790 | 0.922 |
| FIS | 0.014 | 0.422 |
| FST | 0.004 | 0.804 |
Further descriptions of each test are given in Goudet et al. (2002).
Discussion
Although genetic similarity was detected among sites at WPNP and Yanakie, family groups were not widely dispersed across this region, suggesting limited movement of ~5–10 km across the park. The largest inter-pair distance identified in this study (~30 km) was observed between a second-order relative pair identified from Yanakie and Oberon Bay; however, it is important to note that the results presented here are unable to elucidate whether this is a dispersal event by a single individual, or both individuals dispersing at a smaller scale with an accumulated distance of 30 km. Dispersal capability of terrestrial mammals appears to be strongly associated with home ranges, and to a lesser extent body size of the species (Santini et al. 2013), and given that hog deer are a small-bodied mammal with mature males reaching on average 50 kg and females 35 kg (Groves and Grubb 2011), and a mean home range size of 20 ha in Victoria (Taylor 1971), the low inter-pair distances observed between kin in the present study are unsurprising. Additionally, no evidence of dispersal was detected between WPNP and nearby Snake Island and Sunday Island, despite reports of hog deer being adept swimmers capable of moving between island and mainland sites (Mayze and Moore 1990). Dispersal of hog deer between island and mainland sites may be more common at closer distances than sampled in the present study and deserves further investigation.
Dispersal of hog deer across WPNP does not appear to be strongly influenced by one sex. A significant positive spatial autocorrelation was observed for two distance classes in the females compared with only one distance class in the males, but a significant negative autocorrelation was observed beginning at a lower distance class in females than in males. These negative spatial autocorrelation values likely represent dispersal limits beyond the capabilities of the species and suggest some genetic differentiation among sites at these distances, likely influenced by the close relatives sampled in this study (see Fig. S2). These results, coupled with the non-significant sex bias tests, suggest that dispersal rates may be relatively uniform between the sexes, and at a small scale (<10 km). Mayze and Moore (1990) present evidence that males move much further than females in Victoria, with females moving up to 39 km and males moving in excess of 70 km; a dispersal scale not observed in the present study. However, these large-scale dispersal distances of 39 km and 70 km were for deer that had recently been translocated, which could be a result of these deer attempting to return to their original location and therefore not representative of the typical movements of hog deer (Mayze and Moore 1990).
Kinship and spatial autocorrelation analyses showed that hog deer dispersal distances appear to be relatively small scale relative to some other deer species (Hjeljord 2001; Long et al. 2005). However, questions remain about how this species has colonised the coastal zone of the Gippsland region of Victoria. Given that male hog deer are not territorial and that there is not a well-defined breeding season (Taylor 1971; Mayze and Moore 1990), reproduction may not be an important driver of dispersal in this species, as reported in other deer species (Wahlström 1994; Jarnemo 2011; Debeffe et al. 2014). Translocations of hog deer across Gippsland, (both legally in government programs and illegally by hunters seeking to establish new populations to hunt; Mayze and Moore 1990; Scroggie et al. 2012; Hill et al. 2022), have likely assisted the colonisation of parts of Victoria by this species. However, other biological attributes of hog deer could have contributed to the dispersal of this species and therefore could be important for understanding recolonisation of culled areas. For example, it has been suggested that roe deer (Capreolus capreolus) dispersal occurs though natal movements and rearrangement of home ranges during the breeding season rather than sex-biased dispersal, because both sexes appear to be sedentary as adults (Coulon et al. 2005; Bonnot et al. 2010; Biosa et al. 2015). Tests for natal dispersal could not be performed for hog deer in our study due to a lack of juvenile samples, but observations of hog deer behaviour suggest that male offspring are driven away by mothers much sooner after birth than female offspring (Mayze and Moore 1990). Further investigation of sex-biased natal dispersal is warranted.
The inclusion of kinship analyses to inform culling strategies can provide much more fine-scale information than has been presented previously in genetic analyses of invasive species. Many genetic studies focusing on invasive species identify management units for targeted eradication through genetic similarity among sites (Abdelkrim et al. 2005; Fraser et al. 2013; Adams et al. 2014). However, the addition of kinship data provides further information regarding movement of individuals among sites that appear to be genetically similar and shows that dispersal is not always uniform across sites within one management unit. This can have important implications for how culling is conducted, particularly if the goal is to eradicate hog deer. Our study has shown that long-distance dispersal of hog deer occurs, and that hog deer are capable of dispersing between WPNP and sites outside the park. If long-distance dispersal into WPNP during an eradication program were to occur, then the likelihood of eradication would be reduced. The construction of deer-proof fencing along the northern boundary of the park could substantially reduce the probability of dispersing hog deer entering the park.
Data availability
Raw data and sample metadata are available on Figshare https://doi.org/10.6084/m9.figshare.22301251
Acknowledgements
We thank hunters who provided access to samples, and Victorian Hog Deer Checking Station operators, Parks Victoria, and Museums Victoria for assisting with the collection of hog deer samples. We would also like to thank Katherine Harrisson for her assistance with SNP filtering, and James O’Dwyer for his heroic efforts with related analyses.
References
Abdelkrim J, Pascal M, Samadi S (2005) Island colonization and founder effects: the invasion of the Guadeloupe islands by ship rats (Rattus rattus). Molecular Ecology 14, 2923-2931.
| Crossref | Google Scholar |
Adams A, van Heezik Y, Dickinson K, Robertson B (2014) Identifying eradication units in an invasive mammalian pest species. Biological Invasions 16, 1481-1496.
| Google Scholar |
Atlas of Living Australia (2019) Occurrence records. Available at https://biocache.ala.org.au/occurrences/search?q=lsid%3Aurn%3Alsid%3Abiodiversity.org.au%3Aafd.taxon%3A751e6627-f63b-4c1b-911e-f4ad688be569 [Accessed 4 September 2019]
Banks SC, Peakall R (2012) Genetic spatial autocorrelation can readily detect sex-biased dispersal. Molecular Ecology 21, 2092-2105.
| Crossref | Google Scholar |
Biosa D, Grignolio S, Sica N, Pagon N, Scandura M, Apollonio M (2015) Do relatives like to stay closer? Spatial organization and genetic relatedness in a mountain roe deer population. Journal of Zoology 296, 30-37.
| Crossref | Google Scholar |
Bonnot N, Gaillard J-M, Coulon A, Galan M, Cosson J-F, Delorme D, Klein F, Hewison AJM (2010) No difference between the sexes in fine-scale spatial genetic structure of roe deer. PLoS ONE 5, e14436.
| Crossref | Google Scholar |
Borkowski J, Pudełko M (2007) Forest habitat use and home-range size in radio-collared fallow deer. Annales Zoologici Fennici 44, 107-114.
| Google Scholar |
Bostock MP, Ellis RK, Tarr G (2016) Package “edgebundler.” Circle Plot with Bundled Edges. Available at https://cran.r-project.org/web/packages/edgebundleR/index.html
Bourg NA, McShea WJ, Herrmann V, Stewart CM (2017) Interactive effects of deer exclusion and exotic plant removal on deciduous forest understory communities. AoB PLANTS 9, plx046.
| Crossref | Google Scholar |
Côté SD, Rooney TP, Tremblay J-P, Dussault C, Waller DM (2004) Ecological impacts of deer overabundance. Annual Review of Ecology, Evolution, and Systematics 35, 113-147.
| Crossref | Google Scholar |
Coulon A, Cosson JF, Morellet N, Angibault JM, Cargnelutti B, Galan M, Aulagnier S, Hewison AJM (2005) Dispersal is not female biased in a resource defence mating ungulate, the European roe deer. Proceedings of the Royal Society B: Biological Sciences 273, 341-348.
| Google Scholar |
Davis NE, Coulson G, Forsyth DM (2008) Diets of native and introduced mammalian herbivores in shrub-encroached grassy woodland, south-eastern Australia. Wildlife Research 35, 684-694.
| Crossref | Google Scholar |
Davis NE, Forsyth DM, Coulson G (2010) Facilitative interactions between an exotic mammal and native and exotic plants: hog deer (Axis porcinus) as seed dispersers in south-eastern Australia. Biological Invasions 12, 1079-1092.
| Crossref | Google Scholar |
Davis NE, Bennett A, Forsyth DM, Bowman DMJS, Lefroy EC, Wood SW, Woolnough AP, West P, Hampton JO, Johnson CN (2016) A systematic review of the impacts and management of introduced deer (family Cervidae) in Australia. Wildlife Research 43, 515-532.
| Crossref | Google Scholar |
Debeffe L, Focardi S, Bonenfant C, Hewison AJM, Morellet N, Vanpé C, Heurich M, Kjellander P, Linnell JDC, Mysterud A, Pellerin M, Sustr P, Urbano F, Cagnacci F (2014) A one night stand? Reproductive excursions of female roe deer as a breeding dispersal tactic. Oecologia 176, 431-443.
| Crossref | Google Scholar |
Dobson FS (1982) Competition for mates and predominant juvenile male dispersal in mammals. Animal Behaviour 30, 1183-1192.
| Crossref | Google Scholar |
Dolman PM, Wäber K (2008) Ecosystem and competition impacts of introduced deer. Wildlife Research 35, 202-214.
| Crossref | Google Scholar |
Escoda L, González-Esteban J, Gómez A, Castresana J (2017) Using relatedness networks to infer contemporary dispersal: application to the endangered mammal Galemys pyrenaicus. Molecular Ecology 26, 3343-3357.
| Crossref | Google Scholar |
Fraser EJ, Macdonald DW, Oliver MK, Piertney S, Lambin X (2013) Using population genetic structure of an invasive mammal to target control efforts – an example of the American mink in Scotland. Biological Conservation 167, 35-42.
| Crossref | Google Scholar |
Goudet J (2005) HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Molecular Ecology Notes 5, 184-186.
| Crossref | Google Scholar |
Goudet J, Perrin N, Waser P (2002) Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Molecular Ecology 11, 1103-1114.
| Crossref | Google Scholar |
Gruber B, Unmack PJ, Berry OF, Georges A (2018) DARTR: an R package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Molecular Ecology Resources 18, 691-699.
| Crossref | Google Scholar |
Hijmans RJ, Williams E, Vennes C (2017) Package ‘geosphere’. Spherical Trigonometry 1(7), 1-45.
| Google Scholar |
Hill E, Linacre A, Toop S, Murphy N, Strugnell J (2019) Widespread hybridization in the introduced hog deer population of Victoria, Australia, and its implications for conservation. Ecology and Evolution 9, 10828-10842.
| Crossref | Google Scholar |
Hill E, Murphy N, Toop S, Linacre A, Strugnell JM (2022) Genetic analysis of hog deer (Axis porcinus) in Victoria, Australia, and its applications to invasive species and game management. European Journal of Wildlife Research 68, 45.
| Crossref | Google Scholar |
Hjeljord O (2001) Dispersal and migration in northern forest deer – are there unifying concepts? Alces 37, 353-370.
| Google Scholar |
Jarnemo A (2011) Male red deer (Cervus elaphus) dispersal during the breeding season. Journal of Ethology 29(2), 329-336.
| Crossref | Google Scholar |
Jirinec V, Cristol DA, Leu M (2017) Songbird community varies with deer use in a fragmented landscape. Landscape and Urban Planning 161, 1-9.
| Crossref | Google Scholar |
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403-1405.
| Crossref | Google Scholar |
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070-3071.
| Crossref | Google Scholar |
Jones OR, Wang J (2010) COLONY: a program for parentage and sibship inference from multilocus genotype data. Molecular Ecology Resources 10, 551-555.
| Crossref | Google Scholar |
Kalisz S, Spigler RB, Horvitz CC (2014) In a long-term experimental demography study, excluding ungulates reversed invader’s explosive population growth rate and restored natives. Proceedings of the National Academy of Sciences 111, 4501-4506.
| Crossref | Google Scholar |
Kilian A, Wenzl P, Huttner E, Carling J, Xia L, Blois H, Caig V, Heller-Uszynska K, Jaccoud D, Hopper C (2012) Diversity arrays technology: a generic genome profiling technology on open platforms. In ‘Data production and analysis in population genomics’. (Eds Pompanon F, Bonin A) pp. 67–89. (Springer)
Lepais O, Darvill B, O’Connor S, Osborne JL, Sanderson RA, Cussans J, Goffe L, Goulson D (2010) Estimation of bumblebee queen dispersal distances using sibship reconstruction method. Molecular Ecology 19, 819-831.
| Crossref | Google Scholar |
Lesage L, Crête M, Huot J, Dumont A, Ouellet J-P (2000) Seasonal home range size and philopatry in two northern white-tailed deer populations. Canadian Journal of Zoology 78, 1930-1940.
| Crossref | Google Scholar |
Long ES, Diefenbach DR, Rosenberry CS, Wallingford BD, Grund MD (2005) Forest cover influences dispersal distance of white-tailed deer. Journal of Mammalogy 86, 623-629.
| Crossref | Google Scholar |
Moriarty A (2004) The liberation, distribution, abundance and management of wild deer in Australia. Wildlife Research 31, 291-299.
| Crossref | Google Scholar |
Odden M, Wegge P (2007) Predicting spacing behavior and mating systems of solitary cervids: a study of hog deer and Indian muntjac. Zoology 110(4), 261-270.
| Crossref | Google Scholar |
Parks Victoria (n.d.) Wilsons Prom Sanctuary. Available at https://www.parks.vic.gov.au/get-into-nature/conservation-and-science/conserving-our-parks/wilsons-prom-sanctuary [Accessed 24 April 2023]
Pew J, Muir PH, Wang J, Frasier TR (2015) related: an R package for analysing pairwise relatedness from codominant molecular markers. Molecular Ecology Resources 15, 557-561.
| Crossref | Google Scholar |
Ramsey DSL, Forsyth DM, Veltman CJ, Richardson SJ, Allen RB, Allen WJ, Barker RJ, Bellingham PJ, Jacobson CL, Nicol SJ, Robertson AW, Todd CR (2018) A management experiment reveals the difficulty of altering seedling growth and palatable plant biomass by culling invasive deer. Wildlife Research 44, 623-636.
| Crossref | Google Scholar |
Reinecke H, Leinen L, Thißen I, Meißner M, Herzog S, Schütz S, Kiffner C (2014) Home range size estimates of red deer in Germany: environmental, individual and methodological correlates. European Journal of Wildlife Research 60, 237-247.
| Crossref | Google Scholar |
Santini L, Di Marco M, Visconti P, Baisero D, Boitani L, Rondinini C (2013) Ecological correlates of dispersal distance in terrestrial mammals. Hystrix, Italian Journal of Mammology 24, 181-186.
| Google Scholar |
Schunter C, Pascual M, Garza JC, Raventós N, Macpherson E (2014) Kinship analyses identify fish dispersal events on a temperate coastline. Proceedings of the Royal Society B: Biological Sciences 281, 20140556.
| Crossref | Google Scholar |
Smouse PE, Peakall R (1999) Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82, 561-573.
| Crossref | Google Scholar |
Stoffel MA, Esser M, Kardos M, Humble E, Nichols H, David P, Hoffman JI (2016) inbreedR: an R package for the analysis of inbreeding based on genetic markers. Methods in Ecology and Evolution 7, 1331-1339.
| Crossref | Google Scholar |
Vangestel C, Mergeay J, Dawson DA, Vandomme V, Lens L (2011) Spatial heterogeneity in genetic relatedness among house sparrows along an urban–rural gradient as revealed by individual-based analysis. Molecular Ecology 20, 4643-4653.
| Crossref | Google Scholar |
Wahlström LK (1994) The significance of male-male aggression for yearling dispersal in roe deer (Capreolus capreolus). Behavioural Ecology and Sociobiology 35, 409-412.
| Google Scholar |


