A framework phylogeny of the diverse guinea-flowers (Hibbertia, Dilleniaceae) using high-throughput sequence data
Timothy A. Hammer A B * , Ed Biffin
A B * , Ed Biffin  B , Kor-jent van Dijk
B , Kor-jent van Dijk  A , Kevin R. Thiele
A , Kevin R. Thiele  C and Michelle Waycott
C and Michelle Waycott  A B
A B
A
B
C
Abstract
Hibbertia is the largest genus in Dilleniaceae and one of the largest Australian plant genera, with ~350 current and more than 100 known undescribed species in Australia. We present the first published phylogeny based on rigorous sampling of Hibbertia. As part of Genomics for Australian Plants Stage II, 95 Hibbertia species were newly sequenced using Angiosperm353, OzBaits nuclear and OzBaits plastid bait sets, resulting in 402 nuclear and 79 plastid loci that were subsampled to retain the most phylogenetically useful 300 and 60 loci respectively. Nuclear and plastid phylogenies were reconstructed using concatenation and coalescent approaches, and further analysed using Quartet Sampling. We found that Hibbertia and the four subgenera within the genus are robustly supported as monophyletic and recovered 14 major clades, supported in both datasets, within the two largest subgenera (subg. Hemistemma and subg. Hibbertia). However, many relationships between these major clades are unresolved and discordant. Some incongruence was also detected between the plastid and nuclear trees. Discordance was particularly high in the largest eastern Australian clade of subg. Hemistemma. Possible causes of this discordance, and relationships between and within these major clades, are discussed.
Keywords: Angiosperm353, Dilleniaceae, discordance, Genomics for Australian Plants, Hibbertia, hybrid-capture, incongruence, phylogenomics.
Introduction
Dilleniaceae Salisb. is a morphologically diverse family of 11 genera and ~600 species with a largely Gondwanan distribution (Plants of the World Online, see http://www.plantsoftheworldonline.org/, accessed 1 April 2024). The phylogenetic placement of Dilleniaceae within the angiosperms is currently uncertain, alternatively being placed in the core eudicots sister to the superrosid clade (Ruhfel et al. 2014), sister to the superasterid clade (Soltis et al. 2011) or in an unsupported position sister to the Gunnerales (Baker et al. 2022; Zuntini et al. 2024). The family is recognised as the only member of the order Dilleniales in APG IV (Angiosperm Phylogeny Group 2016). The floral morphology of the family has classically been described as ‘primitive’ based on generally having numerous stamens and carpels. Morphological characteristics of the androecium and gynoecium vary considerably within most genera (Stebbins and Hoogland 1976; Horn 2007).
Hibbertia Andrews is the largest genus in Dilleniaceae, comprising ~375 species of Southern Hemisphere, mostly temperate to semi-arid shrubs. Most of the diversity occurs in Australia, with ~350 species (Hammer and Thiele 2022), several of which extend to Papua New Guinea. A distinct radiation of ~23 species occurs in New Caledonia, 1 species of which also occurs in Fiji and 1 species is endemic to Madagascar.
Hibbertia has been the focus of intense taxonomic effort in recent decades, with species-level work ongoing. More than half of all Australian Hibbertia species have been described since 1990, principally by H. R. Toelken (focusing on eastern and northern Australian taxa), and J. R. Wheeler and K. R. Thiele (focusing on Western Australian taxa). These efforts have produced a mix of large revisions of ‘species groups’ that have been informally delimited based on shared morphological traits (e.g. Toelken 2010, 2023, 2024; Thiele 2019a, 2019b) and shorter papers describing or revising individual species (e.g. Hammer 2022, 2023a, 2023b; Hammer and Thiele 2024). A few large species group revisions have not yet been published and these are expected to add more than 100 new species to the genus, resulting in Hibbertia being one of the largest plant genera in Australia. In parallel with these revisions, treatments for all Hibbertia species within Australia are being finalised for the Flora of Australia online platform (Hammer and Thiele 2022).
Despite Hibbertia being species-rich and the family having an interesting evolutionary placement in the angiosperms, the evolution of Hibbertia has been poorly studied. Stebbins and Hoogland (1976) investigated chromosome numbers, and ecological, geographical and morphological characteristics of Hibbertia (~130 spp. were recognised at the time), noting that the floral morphological traits of Hibbertia are highly diverse. The floral diversity is often overlooked by non-experts due to most species having five similar-looking bright yellow petals (Fig. 1). The number of parts in the gynoecium ((1)2–5(–15) carpels) and androecium (1 to >200 stamens; sometimes with few to many staminodes) varies considerably, whereas the androecium shows diverse orientations, symmetries and degrees of fusion (Tucker and Bernhardt 2000). These characters are typically fixed with few, minor changes within angiosperm genera and more likely varying at supergeneric levels within families, if at all (Tucker 1997). Stebbins and Hoogland (1976) concluded that ‘with respect to leaf size, structure and venation, floral symmetry and chromosome numbers, the diversity found among the species of Hibbertia exceeds that found in all but a few genera of Angiosperms and is greater than that in any other exclusively woody genus’ (p. 10). These differences are often diagnostic for species groups and occasionally for species within Hibbertia (e.g. Toelken 2013; Thiele and Cockerton 2015).
Representative floral diversity of Hibbertia spp. sampled in the current study. (a) H. gracilipes. (b) H. exasperata. (c) H. polystachya. (d) H. drummondii. (e) H. platyphylla subsp. platyphylla. (f) H. exutiacies. (g) H. cinerea. (h) H. banksii. (i) H. conspicua. (j) H. depressa. (k) H. cunninghamii. (l) H. fasciculata. Photographs: T. Hammer (a–g, j–l), A. Chapman (h) and R. Davis (i).

Preliminary phylogenies of Dilleniaceae and Hibbertia were completed by Horn (2005, 2009), comprising (1) an analysis using four plastid loci (rbcL, infA, rps4, rpl16 intron) of 10 genera in Dilleniaceae (including 22 Hibbertia species), and (2) an analysis of a combined nuclear and plastid dataset (ITS and rpl16 intron) of 117 Hibbertia species (including putative species). Horn (2009) resolved three major clades within Hibbertia as recognised at that time and found that the genus Pachynema R.Br. ex DC. was nested within Hibbertia, forming a fourth major clade. Backbone topologies within the two largest clades were poorly resolved. Based on this, Horn (2009) synonymised Pachynema within Hibbertia and erected subgenera based on the four clades: subg. Adrastaea (DC.) J.W.Horn, subg. Hemistemma (Juss. ex Thouars) J.W.Horn, subg. Hibbertia and subg. Pachynema (R.Br. ex DC.) J.W.Horn. An unpublished phylogeny in Horn’s dissertation (Horn 2005, p. 145) recovered the same four major clades and provided greater resolution within the two largest clades (subg. Hibbertia and Hemistemma) but also lacked good resolution of the backbone for the major groups.
Raheem (2012) focused on eastern Australian species of subg. Hemistemma, assumed to be a monophyletic clade, and conducted morphological and molecular (ITS and trnL-F) analyses for 93 taxa (including putative taxa). The molecular and morphological trees were deeply incongruent and failed to recover many of the informal species groups established by H. Toelken, however the methods employed were ultimately limited in utility to resolve the group due to limited variation detected (Raheem 2012).
We revisit the phylogeny and systematics of Hibbertia as part of the Genomics for Australian Plants (GAP) Stage II initiative, with the goal of reconstructing the backbone topology of Hibbertia by sampling among the currently recognised Australian species groups. We generated an extensive DNA sequence dataset using a hybrid-capture approach targeting low copy nuclear and plastid orthologs captured using two ‘universal’ angiosperm probe sets, Angiosperms353 (Johnson et al. 2019) and OzBaits (Waycott et al. 2021). Our aim was to test (1) the performance of these marker sets for the Dilleniaceae, (2) congruence between nuclear and plastid topologies, and (3) the species and cladistic relationships recovered by previous phylogenies and expected based on morphology. This dataset will provide a springboard for further research investigating the biogeography and evolution of Hibbertia including patterns of speciation.
Materials and methods
Taxon sampling
Representatives were sampled from all four subgenera of Hibbertia, with 95 species selected to encompass the likely backbone of the genus based on morphological characteristics and prior phylogenetic studies (Table 1). Species were assigned to informal species groups by two of the authors (T.A. Hammer and K.R. Thiele) to aid ongoing taxonomic revisions within the genus. Subgenus Hemistemma (~250 spp.) was represented by 59 species sampled from 26 informal species groups and subg. Hibbertia (~100 spp.) by 32 species sampled from 15 informal species groups. The two smaller subgenera, subg. Pachynema (11 spp.) and subg. Adrastaea (1 sp.) were represented by three and one species respectively. Approximately 27% of the currently accepted and formally named species in Australia were included. As sequencing for this study was part of the GAP initiative, Hibbertia species outside of Australia (e.g. Madagascar and New Caledonia) were excluded and are planned to be included in a forthcoming study with expanded sampling instead. Four publicly available accessions of Dilleniaceae were included as outgroups (Table 1): Dillenia alata (R.Br. ex DC.) Banks ex Martelli, Tetracera daemeliana F.Muell., Curatella americana L. and Acrotrema sp. (the latter two having A353 data only, and new sequence generated for a Dillenia alata sample). In total, 100 (99 spp.) and 98 (97 spp.) genomic samples were included for nuclear and plastid markers respectively.
| Taxon | European Nucleotide Archive (ENA) or BioPlatforms Australia (GAP) accession ID | GAP Library ID | Subgenus | Species group | Clade | Voucher specimen; provenance | |
|---|---|---|---|---|---|---|---|
| Acrotrema sp. | PAFTOL; ERR4180187 | – | – | – | – | M.W. Chase 17398 (K) | |
| Curatella americana L. | PAFTOL; ERR5006125 | – | – | – | – | M.W. Chase 973 (K) | |
| Dillenia alata (R.Br. ex DC.) Banks ex Martelli | GAP; 376560 | 380926 (A353); 381022 (OzBaits) | – | – | – | J.O. Westaway 3132 (DNA D0195012); NT, Australia | |
| Dillenia alata (R.Br. ex DC.) Banks ex Martelli | GAP; 80947 | 83151 (A353) | – | – | – | C. Costion 3217 (CNS 136612); QLD, Australia | |
| Hibbertia acerosa (R.Br. ex DC.) Benth. | GAP; 376561 | 380927 (A353); 381023 (OzBaits) | Hemistemma | Acerosa | C | K.R. Thiele 5287 (PERTH 8838119); WA, Australia | |
| Hibbertia alopecota Toelken | GAP; 376562 | 380928 (A353); 381024 (OzBaits) | Hemistemma | Tomentosa | H | G.J. Leach 2785 & I.D. Cowie (AD 99037095); NT, Australia | |
| Hibbertia ancistrophylla J.R.Wheeler | GAP; 376563 | 380929 (A353); 381025 (OzBaits) | Hemistemma | Stowardii | E | K.R. Thiele 5702 (PERTH 9187618); WA, Australia | |
| Hibbertia ancistrotricha J.R.Wheeler | GAP; 376564 | 380930 (A353); 381026 (OzBaits) | Hemistemma | Axillibarba | E | J.R. Wheeler 4150 (PERTH 6315208); WA, Australia | |
| Hibbertia arcuata J.R.Wheeler | GAP; 376565 | 380931 (A353); 381027 (OzBaits) | Hemistemma | Arcuata | A | K.R. Thiele 5668 (PERTH 9187642); WA, Australia | |
| Hibbertia aspera DC. subsp. aspera | GAP; 376566 | 380932 (A353); 381028 (OzBaits) | Hemistemma | Aspera | G | R. Jensen 4313 & J.E. Kemp (BRI AQ1018953); QLD, Australia | |
| Hibbertia aurea Steud. | GAP; 376567 | 380933 (A353); 381029 (OzBaits) | Hemistemma | Aurea | C | K.R. Thiele 5134 (PERTH 8850208); WA, Australia | |
| Hibbertia australis N.A.Wakef. | GAP; 376568 | 380934 (A353); 381030 (OzBaits) | Hemistemma | Stricta | F | T.A. Hammer 145 & A.H. Thornhill (AD); SA, Australia | |
| Hibbertia axillibarba J.R.Wheeler | GAP; 376569 | 380935 (A353); 381031 (OzBaits) | Hemistemma | Axillibarba | E | K. Kershaw KK 2146 (PERTH 8013608); WA, Australia | |
| Hibbertia banksii subsp. sparsidentata Toelken | GAP; 376570 | 380936 (A353); 381032 (OzBaits) | Hemistemma | Banksii | I | K.R. McDonald KRM1797 (AD 184151); QLD, Australia | |
| Hibbertia bistrata (J.R.Wheeler) K.R.Thiele & T.Hammer | GAP; 376598 | 380964 (A353); 381060 (OzBaits) | Hibbertia | Glomerosa | J | K.R. Thiele 5671b (PERTH 9187669); WA, Australia | |
| Hibbertia cactifolia Toelken | GAP; 376571 | 380937 (A353); 381033 (OzBaits) | Hemistemma | Tomentosa | H | J.L. Egan 4722 (AD 169057); NT, Australia | |
| Hibbertia candicans (Hook.f.) Benth. | GAP; 376572 | 380938 (A353); 381034 (OzBaits) | Hemistemma | Banksii | I | R. Jensen 3183 (BRI AQ0856789); QLD, Australia | |
| Hibbertia capensis K.R.Thiele | GAP; 376573 | 380939 (A353); 381035 (OzBaits) | Hemistemma | Polystachya | C | J. English 58 (PERTH 7842228); WA, Australia | |
| Hibbertia carinata J.R.Wheeler | GAP; 376574 | 380940 (A353); 381036 (OzBaits) | Hemistemma | Carinata | E | W. Archer 806132 (PERTH 8472068); WA, Australia | |
| Hibbertia charlesii J.R.Wheeler | GAP; 376575 | 380941 (A353); 381037 (OzBaits) | Hemistemma | Charlesii | F | S. Barrett 608 (PERTH 4273710); WA, Australia | |
| Hibbertia cinerea (R.Br. ex DC.) Toelken | GAP; 376576 | 381038 (A353); 380942 (OzBaits) | Hemistemma | Aspera | G | T.A. Hammer 161 & A.E. McDougall (AD); SA, Australia | |
| Hibbertia coloensis Toelken | GAP; 376577 | 380943 (A353); 381039 (OzBaits) | Hemistemma | Vestita | F | P.H. Weston 3392 (AD 238034); NSW, Australia | |
| Hibbertia conspicua (J.Drumm. ex Harv.) Gilg | GAP; 376578 | 380944 (A353); 381040 (OzBaits) | Pachynema | Conspicua | Pachynema | K.R. Thiele 5180 (PERTH 8906319); WA, Australia | |
| Hibbertia crinita Toelken | GAP; 376579 | 380945 (A353); 381041 (OzBaits) | Hemistemma | Sericea | F | T.A. Hammer 194 & A.E. McDougall (AD); SA, Australia | |
| Hibbertia crispula J.M.Black | GAP; 376580 | 380946 (A353); 381042 (OzBaits) | Hibbertia | Virgata | N | S. Reiffer SRE 032 (PERTH 8656819); WA, Australia | |
| Hibbertia cuneiformis (Labill.) Sm. | GAP; 376581 | 380947 (A353); 381043 (OzBaits) | Hibbertia | Cuneiformis | J | G.J. Keighery 17104 (PERTH 7854234); WA, Australia | |
| Hibbertia cunninghamii Aiton ex Hook. | GAP; 376623 | 380989 (A353); 381085 (OzBaits) | Hibbertia | Cunninghamii | L | K.R. Thiele 5290 (PERTH 8838151); WA, Australia | |
| Hibbertia dentata R.Br. ex DC. | GAP; 376582 | 380948 (A353); 381044 (OzBaits) | Hibbertia | Dentata | – | N. Schultz 67 (AD 159136); NSW, Australia | |
| Hibbertia depressa Steud. | GAP; 376583 | 380949 (A353); 381045 (OzBaits) | Hibbertia | Huegelii | J | R.J. Cranfield WFM 178 & B.G. Ward (PERTH 7099975); WA, Australia | |
| Hibbertia dispar Toelken | GAP; 376584 | 380950 (A353); 381046 (OzBaits) | Hemistemma | Vestita | F | J. Whinray 13243 (AD 246589); TAS, Australia | |
| Hibbertia drummondii (Turcz.) Gilg | GAP; 376585 | 380951 (A353); 381047 (OzBaits) | Hemistemma | Drummondii | E | K.R. Thiele 5701 (PERTH 9187634); WA, Australia | |
| Hibbertia eatoniae Diels | GAP; 376586 | 380952 (A353); 381048 (OzBaits) | Hemistemma | Eatoniae | A | K.R. Thiele 5678 (PERTH 9187626); WA, Australia | |
| Hibbertia echiifolia subsp. oligantha Toelken | GAP; 376587 | 380953 (A353); 381049 (OzBaits) | Hemistemma | Melhanioides | H | A.V. Slee 2538 & L.A. Craven (AD 99401211); NT, Australia | |
| Hibbertia ericifolia Hook.f. subsp. ericifolia | GAP; 376589 | 380955 (A353); 381051 (OzBaits) | Hemistemma | Vestita | F | A. Moscal 12316 (HO 402224); TAS, Australia | |
| Hibbertia exasperata (Steud.) Briq. | GAP; 376590 | 380956 (A353); 381052 (OzBaits) | Hemistemma | Exasperata | D | G.J. Keighery 6790 & N. Gibson (PERTH 6823688); WA, Australia | |
| Hibbertia exutiacies N.A.Wakef. | GAP; 376591 | 380957 (A353); 381053 (OzBaits) | Hemistemma | Acicularis | F | T.A. Hammer 140 (AD); SA, Australia | |
| Hibbertia fasciculata R.Br. ex DC. | GAP; 376592 | 380958 (A353); 381054 (OzBaits) | Hibbertia | Fasciculata | N | T.A. Hammer 195 & A.E. McDougall (AD); SA, Australia | |
| Hibbertia fasciculiflora K.R.Thiele | GAP; 376593 | 380959 (A353); 381055 (OzBaits) | Hemistemma | Fasciculiflora | – | M.A. Langley MAL 2106 & P.M. Smith (PERTH 6391729); WA, Australia | |
| Hibbertia ferox Jackes | GAP; 376594 | 380960 (A353); 381056 (OzBaits) | Hemistemma | Drummondii | E | R. Jensen 4137 & J.E. Kemp (BRI AQ1007961); QLD, Australia | |
| Hibbertia ferruginea J.R.Wheeler | GAP; 376595 | 380961 (A353); 381057 (OzBaits) | Hibbertia | Huegelii | J | K.R. Thiele 5592 (PERTH 9187340); WA, Australia | |
| Hibbertia glaberrima F.Muell. | GAP; 376596 | 380962 (A353); 381058 (OzBaits) | Hibbertia | Glaberrima | L | J.E. Wajon 1089 (PERTH 7358407); WA, Australia | |
| Hibbertia glaucophylla (Steud.) K.R.Thiele & T.Hammer | GAP; 376635 | 381001 (A353); 381097 (OzBaits) | Hibbertia | Hemignosta | J | G. Byrne 5356 (PERTH 8787603); WA, Australia | |
| Hibbertia glomerata Benth. var. glomerata | GAP; 376597 | 380963 (A353); 381059 (OzBaits) | Hibbertia | Vaginata | J | G. Byrne 5805 (PERTH 8936463); WA, Australia | |
| Hibbertia gracilipes Benth. | GAP; 376599 | 380965 (A353); 381061 (OzBaits) | Hemistemma | Gracilipes | A | G. Byrne 5314 (PERTH 8771006); WA, Australia | |
| Hibbertia hemignosta (Steud.) J.R.Wheeler | GAP; 376600 | 380966 (A353); 381062 (OzBaits) | Hibbertia | Hemignosta | J | K.R. Thiele 5300 (PERTH 8838011); WA, Australia | |
| Hibbertia hermanniifolia subsp. recondita Toelken | GAP; 376601 | 380967 (A353); 381063 (OzBaits) | Hemistemma | Hermanniifolia | – | P. Gilmour 8373 (AD 163940); NSW, Australia | |
| Hibbertia hirsuta (Hook.) Benth. | GAP; 376602 | 380968 (A353); 381064 (OzBaits) | Hemistemma | Vestita | F | T.A. Hammer 136 (AD); SA, Australia | |
| Hibbertia hooglandii J.R.Wheeler | GAP; 376603 | 380969 (A353); 381065 (OzBaits) | Hemistemma | Banksii | I | G. Sankowsky 2222 & N. Sankowsky (PERTH 6868576); WA, Australia | |
| Hibbertia huegelii (Endl.) F.Muell. | GAP; 376604 | 380970 (A353); 381066 (OzBaits) | Hibbertia | Huegelii | J | D. Coultas DCLFOpp10 & L. Firth (PERTH 9275150); WA, Australia | |
| Hibbertia hypericoides (DC.) Benth. subsp. hypericoides | GAP; 376605 | 380971 (A353); 381067 (OzBaits) | Hemistemma | Hypericoides | H | G. Cassis L 22 (PERTH 7620020); WA, Australia | |
| Hibbertia inclusa Benth. | GAP; 376606 | 380972 (A353); 381068 (OzBaits) | Hibbertia | Virgata | N | J.M. Collins 476 (PERTH 8080623); WA, Australia | |
| Hibbertia lasiopus Benth. | GAP; 376607 | 380973 (A353); 381069 (OzBaits) | Hibbertia | Commutata | K | K.R. Thiele 5296 (PERTH 8838062); WA, Australia | |
| Hibbertia ledifolia Benth. | GAP; 376608 | 380974 (A353); 381070 (OzBaits) | Hemistemma | Banksii | I | K. Coate 757 (PERTH 7457847); WA, Australia | |
| Hibbertia lepidota R.Br. ex DC. | GAP; 376609 | 380975 (A353); 381071 (OzBaits) | Hemistemma | Tomentosa | H | T. Handasyde 6189 (AD 248078); WA, Australia | |
| Hibbertia leucocrossa K.R.Thiele | GAP; 376610 | 380976 (A353); 381072 (OzBaits) | Hibbertia | Huegelii | J | K.R. Thiele 5202 (PERTH 8828113); WA, Australia | |
| Hibbertia lineata Steud. | GAP; 376611 | 380977 (A353); 381073 (OzBaits) | Hemistemma | Lineata | A | M. Hislop 4766 (PERTH 9169636); WA, Australia | |
| Hibbertia longifolia F.Muell. | GAP; 376612 | 380978 (A353); 381074 (OzBaits) | Hibbertia | Glaberrima | L | Y. Baba 438 (CNS 134382.1); QLD, Australia | |
| Hibbertia malacophylla Toelken | GAP; 376613 | 380979 (A353); 381075 (OzBaits) | Hemistemma | Melhanioides | H | J.E. Kemp JEK20240 & R. Jensen (BRI AQ1020187); QLD, Australia | |
| Hibbertia montana Steud. | GAP; 376614 | 380980 (A353); 381076 (OzBaits) | Hibbertia | Commutata | K | K.R. Thiele 5304 (PERTH 8850143); WA, Australia | |
| Hibbertia nymphaea Diels | GAP; 376615 | 380981 (A353); 381077 (OzBaits) | Hibbertia | Cunninghamii | L | K.R. Thiele 5138 (PERTH 8850291); WA, Australia | |
| Hibbertia oblongata subsp. brevifolia (Benth.) Toelken | GAP; 376616 | 380982 (A353); 381078 (OzBaits) | Hemistemma | Tomentosa | H | T. Handasyde 4611 (AD 248081); WA, Australia | |
| Hibbertia obtusibracteata Toelken | GAP; 376617 | 380983 (A353); 381079 (OzBaits) | Hemistemma | Acicularis | F | D.J. Duval 814 & D. Hinchecliff, C. Dickson & R.J. Johnson (AD 220410); SA, Australia | |
| Hibbertia obtusifolia DC. | GAP; 376618 | 380984 (A353); 381080 (OzBaits) | Hibbertia | Linearis | M | D.M. Crayn 979 & R.G. Coveny (NSW 744242); NSW, Australia | |
| Hibbertia oligantha J.R.Wheeler | GAP; 376619 | 380985 (A353); 381081 (OzBaits) | Hemistemma | Psilocarpa | B | G. Byrne 6495 (PERTH 8995982); WA, Australia | |
| Hibbertia ovata Steud. | GAP; 376620 | 380986 (A353); 381082 (OzBaits) | Hibbertia | Commutata | K | K.R. Thiele 5135 (PERTH 8850283); WA, Australia | |
| Hibbertia pachyphylla J.R.Wheeler | GAP; 376621 | 380987 (A353); 381083 (OzBaits) | Hemistemma | Charlesii | F | P.G. Armstrong PA 11/622 (PERTH 8800650); WA, Australia | |
| Hibbertia paranthera K.R.Thiele | GAP; 376622 | 380988 (A353); 381084 (OzBaits) | Pachynema | Paranthera | Pachynema | R.L. Barrett RLB 6287 & M. Maier & P. Kendrick (PERTH 8674523); WA, Australia | |
| Hibbertia pilosa Steud. | GAP; 376624 | 380990 (A353); 381086 (OzBaits) | Hibbertia | Commutata | K | K.R. Thiele 5293 (PERTH 8838089); WA, Australia | |
| Hibbertia platyphylla Toelken subsp. platyphylla | GAP; 376625 | 380991 (A353); 381087 (OzBaits) | Hemistemma | Sericea | F | T.A. Hammer 149 & A.E. McDougall (AD); SA, Australia | |
| Hibbertia polyancistra K.R.Thiele | GAP; 376626 | 380992 (A353); 381088 (OzBaits) | Hemistemma | Lineata | A | K.R. Thiele 5139 (PERTH 8775222); WA, Australia | |
| Hibbertia polystachya Benth. | GAP; 376627 | 380993 (A353); 381089 (OzBaits) | Hemistemma | Polystachya | C | M. Hislop & H. Mills WW 209–38 (PERTH 7694830); WA, Australia | |
| Hibbertia proberae K.R.Thiele | GAP; 376628 | 380994 (A353); 381090 (OzBaits) | Hemistemma | Stenophylla | A | K.R. Thiele 5476 (PERTH 9083413); WA, Australia | |
| Hibbertia procumbens (Labill.) DC. | GAP; 376629 | 380995 (A353); 381091 (OzBaits) | Hibbertia | Fasciculata | N | D.E. Murfet 5858 & A. Lowie (AD 216770); TAS, Australia | |
| Hibbertia psilocarpa J.R.Wheeler | GAP; 376630 | 380996 (A353); 381092 (OzBaits) | Hemistemma | Psilocarpa | B | A. Coates AC 5732 (PERTH 8367329); WA, Australia | |
| Hibbertia puberula Toelken | GAP; 376642 | 381104 (A353); 381008 (OzBaits) | Hemistemma | Sericea | F | D.M. Crayn 1109 & R.G. Coveny & M.R. Whitehead (NSW 760126); NSW, Australia | |
| Hibbertia pulchra Ostenf. var. pulchra | GAP; 376631 | 380997 (A353); 381093 (OzBaits) | Hibbertia | Hemignosta | J | R. Meissner 4927 & C. McCormack & M. Langley (PERTH 8993866); WA, Australia | |
| Hibbertia racemosa (Endl.) Gilg | GAP; 376632 | 380998 (A353); 381094 (OzBaits) | Hibbertia | Vaginata | J | G.J. Keighery 147 & B.J. Keighery (PERTH 6330010); WA, Australia | |
| Hibbertia radians (Toelken) T.Hammer | GAP; 376588 | 380954 (A353); 381050 (OzBaits) | Hemistemma | Aspera | G | T.A. Hammer 127 (AD); SA, Australia | |
| Hibbertia riparia (R.Br. ex DC.) Hoogland | GAP; 376633 | 380999 (A353); 381095 (OzBaits) | Hemistemma | Sericea | F | T.A. Hammer 103 (AD); SA, Australia | |
| Hibbertia robur K.R.Thiele | GAP; 376634 | 381000 (A353); 381096 (OzBaits) | Hemistemma | Aurea | C | K.R. Thiele 5091 (PERTH 8594058); WA, Australia | |
| Hibbertia salicifolia (DC.) F.Muell. | GAP; 376636 | 381002 (A353); 381098 (OzBaits) | Adrastaea | Adrastaea | Adrastaea | P.I. Forster PIF30223 & G. Leiper (AD 196618); QLD, Australia | |
| Hibbertia saligna R.Br. ex DC. | GAP; 376637 | 381003 (A353); 381099 (OzBaits) | Hibbertia | Saligna | M | R. Johnstone 1966 & A.E. Orme (AD 212551); NSW, Australia | |
| Hibbertia sandifordiae K.R.Thiele | GAP; 376638 | 381004 (A353); 381100 (OzBaits) | Hibbertia | Commutata | K | K.R. Thiele 5283 (PERTH 8838194); WA, Australia | |
| Hibbertia scabra R.Br. ex Benth. | GAP; 376639 | 381005 (A353); 381101 (OzBaits) | Hemistemma | Melhanioides | H | I.D. Cowie 11841 (AD 246345); NT, Australia | |
| Hibbertia scandens (Willd.) Gilg | GAP; 376640 | 381006 (A353); 381102 (OzBaits) | Hibbertia | Scandens | – | C. Costion 1671 (CNS 131429.1); QLD, Australia | |
| Hibbertia sericea (R.Br. ex DC.) Benth. | GAP; 376641 | 381007 (A353); 381103 (OzBaits) | Hemistemma | Sericea | F | T.A. Hammer 205 (AD); SA, Australia | |
| Hibbertia silvestris Diels | GAP; 376643 | 381009 (A353); 381105 (OzBaits) | Hemistemma | Hypericoides | H | K.R. Thiele 5292 (PERTH 8838143); WA, Australia | |
| Hibbertia sphenandra (F.Muell. & Tate) J.W.Horn | GAP; 376644 | 381010 (A353); 381106 (OzBaits) | Pachynema | Sphenandra | Pachynema | R.L. Barrett RLB 4920 (PERTH 8044082); WA, Australia | |
| Hibbertia squarrosa K.R.Thiele | GAP; 376645 | 381011 (A353); 381107 (OzBaits) | Hibbertia | Glomerosa | J | L. Atkins 140 (PERTH 8270724); WA, Australia | |
| Hibbertia stellaris Endl. | GAP; 376646 | 381012 (A353); 381108 (OzBaits) | Hibbertia | Stellaris | J | G. Byrne 6574 (PERTH 8996423); WA, Australia | |
| Hibbertia stelligera (C.T.White) Toelken | GAP; 376647 | 381013 (A353); 381109 (OzBaits) | Hemistemma | Tomentosa | H | J.E. Kemp JEK20248 & R. Jensen (BRI AQ1022082); QLD, Australia | |
| Hibbertia stenophylla J.R.Wheeler | GAP; 376648 | 381014 (A353); 381110 (OzBaits) | Hemistemma | Stenophylla | A | K.R. Thiele 5670b (PERTH 9187677); WA, Australia | |
| Hibbertia stowardii S.Moore | GAP; 376649 | 381015 (A353); 381111 (OzBaits) | Hemistemma | Stowardii | N | K.R. Thiele 5721 (PERTH 9187685); WA, Australia | |
| Hibbertia subvaginata (Steud.) F.Muell. | GAP; 376650 | 381016 (A353); 381112 (OzBaits) | Hibbertia | Vaginata | J | K.R. Thiele 5712 (PERTH 9187650); WA, Australia | |
| Hibbertia tomentosa R.Br. ex DC. | GAP; 376651 | 381017 (A353); 381113 (OzBaits) | Hemistemma | Tomentosa | H | I.D. Cowie 9680 (AD 169068); NT, Australia | |
| Hibbertia uncinata F.Muell. | GAP; 376652 | 381018 (A353); 381114 (OzBaits) | Hemistemma | Exasperata | D | K.R. Thiele 5085 (PERTH 8804761); WA, Australia | |
| Hibbertia verrucosa Benth. | GAP; 376653 | 381019 (A353); 381115 (OzBaits) | Hemistemma | Lineata | A | K.R. Thiele 5316 (PERTH 8850127); WA, Australia | |
| Hibbertia vestita A.Cunn. ex Benth. var. vestita | GAP; 376654 | 381020 (A353); 381116 (OzBaits) | Hemistemma | Vestita | F | R. Johnstone 2451 & A.E. Orme (AD 246106); NSW, Australia | |
| Hibbertia virgata R.Br. ex DC. | GAP; 376655 | 381021 (A353); 381117 (OzBaits) | Hibbertia | Virgata | N | T.A. Hammer 131 (AD); SA, Australia | |
| Tetracera daemeliana F.Muell. | GAP; 80948 | 83152 (A353) | – | – | – | C. Costion 2160 (CNS 134905); QLD, Australia |
GAP Samples 376560–376655 were newly sequenced. Phylogenetic clades correspond to those in Fig. 2. Hibbertia species without designated clades are singletons (i.e. H. scandens and H. hermaniifolia subsp. recondita) or failed samples (i.e. H. dentata and H. fasciculiflora).
DNA extraction, library preparation and sequencing
DNA extraction, library preparation and sequencing were performed at the Australian Genome Research Facility (Melbourne, Australia) as part of the GAP initiative by Bioplatforms Australia (Sydney, Australia). Dried plant tissue (20–30 mg) was ground using a TissueLyser II (Qiagen) with tungsten carbide beads for simultaneous disruption and homogenisation of the sample, as per the manufacturer’s instructions. Genomic DNA was extracted using the DNeasy Plant mini kit (Qiagen) as per the manufacturer’s instructions on a QIAcube Connect (Qiagen). DNA quantity and quality were assessed using 1% E-gel with Sybr Safe dye (ThermoFisher) and concentrations assessed using Quantifluor dsDNA assay (Promega). DNA samples were fragmented enzymatically as part of the NEBNext Ultra II FS library preparation workflow. Angiosperms353 and OzBaits nuclear libraries were prepared using the NEBNext Ultra II FS Library Prep Kit (New England Biolabs, Ipswich, MA, USA), following the manufacturer’s instructions with inserts of ~350 bp. Leftover DNA was sent back to the University of Adelaide and OzBaits plastid libraries were prepared again based on similar conditions to the GAP libraries but following the two-step library preparation protocol from Waycott et al. (2021). This was done to perform an additional chloroplast bait enrichment and genome skim to capture a wider range of genomic components (Zeng et al. 2018).
Libraries prepared by AGRF were enriched using both the Angiosperms353 (A353; Johnson et al. 2019) and OzBaits (Waycott et al. 2021) probe kits. Pooled libraries (12–16 plex) were enriched using the A353 probe kit by hybridising at 65°C with the Arbor Biosciences MyBaits Expert Plant A353 v1 bait set with V5 chemistry (catalogue number 308108.v5) and Arbor Biosciences MyBaits custom OzBaits_NR set (catalogue number 300496R.V5) set with V5 chemistry and 64°C hybridisation following the manufacturer’s instructions. The libraries prepared in the University of Adelaide laboratory were enriched (16 plex) with the OzBaits_CP (Waycott et al. 2021) chloroplast probe kit (catalogue number 300196R.v5) at 65°C using the V5 chemistry. Post-capture libraries were amplified with indexed primers and cleaned up with Ampure (1.0×). Additionally, the unenriched library was amplified (eight cycles) with indexed p5 and p7 primers to generate a genome skim with the intention to sequence high copy loci such as 16S, 18S and ITS (e.g. Weitemier et al. 2014). Both Cp and genome skim libraries were quantified using a 4150 TapeStation System (Agilent) with High Sensitivity DNA Screen tapes. Libraries were pooled equimolar and size-selected (300–700 bp) on a Pippin Prep (Sage Sciences) 1.5% agarose 250–1.5-kb gel cassette. All sequencing was performed on a NovaSeq. 6000 (Illumina Inc., San Diego, CA, USA) with v1.5 chemistry and 150-bp paired-end reads.
Data processing
High-throughput 150-bp paired-end reads were imported into CLC Genomics Workbench (ver. 20.0.2, QIAGEN, see https://digitalinsights.qiagen.com/) for demultiplexing and trimming using a quality score limit of 0.05 (Phred score ~13). Reads for each individual were randomly sampled to 2.5 million paired-end reads. The sampled reads were de novo assembled by MEGAHIT (ver. 1.2.9, see https://github.com/voutcn/megahit; Li et al. 2015) using the ‘assemble’ function in the CAPTUS bioinformatics pipeline (ver. 1.0.0, see https://github.com/edgardomortiz/Captus; Raza et al. 2023) with the CAPSKIM default –k-list within CAPTUS (see https://edgardomortiz.github.io/captus.docs/assembly/assemble/) and with ‘–min-count’ and ‘–prune-level’ both set to three.
We used the CAPTUS ‘extract’ function to extract target gene regions using custom nuclear and plastid references. Dilleniaceae reference sequences of OzBaits gene targets were derived from the transcriptome assembly of Dillenia indica L. and Hibbertia grossulariifolia (Salisb.) Salisb. (sample codes EHNF and OKEF respectively that were generated as part of the OneKP project; Leebens-Mack et al. 2019) accessed by local BLAST searches against the transcriptome sequences using the OzBaits target gene region from Arabidopsis thaliana (Araport11, see https://phytozome-next.jgi.doe.gov/info/Athaliana_Araport11; Cheng et al. 2017). References for the A353 gene targets were sourced from H. grossulariifolia (as for OzBaits) and the Kew Tree of Life Explorer (Baker et al. 2022), consisting of Acrotrema sp. (ERR4180187), Dillenia alata (GAP 80947) and Tetracera daemeliana (ERR7599814). Plastid references were sourced from GenBank and included coding regions extracted from the plastid genome sequences of D. indica (NC_042740), D. turbinata Finet & Gagnep. (NC_062798) and Tetracera sarmentosa (L.) Vahl (NC_065058). We used a minimum score of 0.2, minimum identity of 75 and minimum coverage of 30 (for definitions, see https://edgardomortiz.github.io/captus.docs/) for both the nuclear and plastid data to match the contigs to the target references using Scipio (ver. 1.4, see http://www.webscipio.org; Keller et al. 2008). Dillenia alata (GAP 80947) and Tetracera daemeliana (GAP 80948) were previously sequenced by GAP for only A353 and were represented in the present plastid dataset by extracting off-target plastid genes.
We used the CAPTUS ‘align’ function to generate multiple sequence alignments (MSAs) for the extracted nuclear and plastid gene data. All extracted markers were aligned with MAFFT (ver. 7.511, see https://mafft.cbrc.jp/alignment/software/; Katoh and Standley 2013) using the ‘mafft_auto’ algorithm. We specified a maximum of three paralogs per sample for paralogy inference implemented through CAPTUS and used the ‘informed’ filter method to generate alignments of putative orthologs for each gene region, using ‘–tolerance 2’. Alignments were trimmed with ClipKIT (ver. 1.3.0, see https://github.com/JLSteenwyk/ClipKIT; Steenwyk et al. 2020) using default settings in CAPTUS. Subsequently, we used the Paragone (ver. 1.0.0, see https://github.com/chrisjackson-pellicle/ParaGone) pipeline to only refine alignment quality. Paragone uses HmmCleaner (ver. 0.243280, see https://metacpan.org/dist/Bio-MUST-Apps-HmmCleaner; Di Franco et al. 2019), TreeShrink (ver. 1.3.9, see https://github.com/uym2/TreeShrink; Mai and Mirarab 2018) and TrimAl (ver. 1.5.0, see http://trimal.cgenomics.org/; Capella-Gutiérrez et al. 2009) to remove sequencing and alignment errors, and rogue taxa from MSAs. Alignments with fewer than 20 sequences (≥80% missing samples) were removed from downstream analyses.
As output from CAPTUS, ‘NT’ (coding sequences in nucleotides) output was used for A353 and ‘genes-flanked’ (complete gene sequences, including exons and introns along with flanking regions) was used for OzBaits, noting that the OzBaits baits are generally designed on a single exon per gene and thus the flanking regions are potentially introns. Genes recovered by both the A353 and OzBaits bait sets were identified, and only one version of the gene was retained that was selected based on the largest number of included sequences. The resulting genes from both bait sets were subsequently combined to create the full nuclear dataset. We used the ‘NT’ sequences output from CAPTUS for the plastid dataset.
Phylogenetic analyses
The nuclear and plastid alignments were each partitioned and analysed using IQ-TREE (ver. 2.2.0, see http://www.iqtree.org/; Nguyen et al. 2015; Chernomor et al. 2016) to generate a concatenated maximum likelihood tree. ModelFinder, as implemented in IQ-TREE (MF + MERGE, see http://www.iqtree.org/ModelFinder/), was used to find the best nucleotide substitution model for each partition and merge like partitions (Kalyaanamoorthy et al. 2017). To save computational time, only the top 10% of partition merging schemes was examined by using the relaxed hierarchical clustering algorithm (Lanfear et al. 2014). Support for the topology was assessed using 1000 ultrafast bootstrap replicates (UFBS; Minh et al. 2013) and the approximate likelihood ratio test (ALRT; Guindon et al. 2010) as implemented in IQ-TREE. Interpretation of support values follows Minh et al. (2013) and recommendations in the IQ-TREE manual (see http://www.iqtree.org/doc/iqtree-doc.pdf). Tetracera daemeliana F.Muell. was used as the outgroup for all phylogenetic reconstructions given the resolution as sister to the remaining Dilleniaceae (Horn 2009). Individual locus trees were produced in IQ-TREE from the concatenated alignment with the approximate Bayes test to generate branch support values (Anisimova et al. 2011). The initial phylogenies were used as input for genesortR (see https://github.com/mongiardino/genesortR, accessed 30 January 2024; Mongiardino Koch 2021), an R script that sorts and subsamples phylogenomic datasets based on properties that quantify phylogenetic usefulness. The sorted top 300 and 60 loci were retained for downstream analyses for the nuclear and plastid datasets respectively. The previous steps generating the concatenated and loci trees using IQ-TREE were re-run using the newly subsampled datasets.
Locus trees were analysed using wASTRAL hybrid in Accurate Species Tree Estimator (ASTER, ver. 1.4.2.3, see https://github.com/chaoszhang/ASTER) that reimplements ASTRAL-III and accounts for phylogenetic uncertainty by considering signals derived from branch length and branch support in gene trees (Zhang et al. 2018; Zhang and Mirarab 2022) for both genomic datasets. We followed the author’s recommendations (see https://github.com/chaoszhang/ASTER/blob/90f475fbdc79151b86c27f0881c549a1c6d230a4/tutorial/astral-hybrid.md) for maximum and minimum values for local Bayesian supports. The concatenated and coalescent (ASTRAL) trees were both analysed using Quartet Sampling (QS) (see https://github.com/fephyfofum/quartetsampling; Pease et al. 2018). QS indicates how well the data support the tree topology of the phylogeny. Branch support for the QS analyses was assessed by the Quartet Concordance (QC), Quartet Differential (QD) and Quartet Informativeness (QI) values that were described in Pease et al. (2018). Branches were considered concordant with the displayed topology if QC ≥ 0.5, indicating that half or more of all quartet trees are concordant with the focal branch. High QD means that neither of the discordant topologies are favoured versus a skew to one discordant topology at low QD values. QD can therefore provide evidence as to whether discordance is due to incomplete lineage sorting (ILS) or introgression, with the former indicated by high QD and the latter by low QD. QI reports the proportion of the replicates that are informative for the quartet and can be used to distinguish lack of support from conflict (low to high QI respectively). Additionally, terminal nodes (taxa) were assessed by Quartet Fidelity (QF) values. QF reports the proportion of replicates that included the taxon and resulted in a concordant topology (Pease et al. 2018). The suitability of using coalescent methods on the plastome has been questionable (e.g. Doyle 2022) but we have nevertheless included this analysis for comparison.
Results
Dataset creation
Assembly of the nuclear dataset recovered 351 loci using the A353 bait set and 66 loci using OzBaits, of which 5 and 10 were respectively removed due to having fewer than 20 sequences or being overlapping loci. Before subsampling with genesortR, the nuclear dataset contained 346 A353 loci and 56 OzBaits loci, resulting in the combined alignment containing 402 loci and an overall concatenated length of 227,848 bp. Assembly of the plastid dataset recovered 79 loci that were all retained for the genesortR step and totalled 61,776 bp in length. Hibbertia dentata R.Br. ex DC. and Hibbertia fasciculiflora K.R.Thiele sequenced poorly for all bait sets and were excluded from all subsequent analyses. The H. dentata and H. fasciculiflora samples were respectively represented in the nuclear dataset by only 3 and 16% of loci and 1 and 7% of the total alignment length, and in the plastid dataset by 5 and 3% of loci and 1 and 0.6% of total alignment length.
The subsampled nuclear alignment of 300 loci and 98 sequenced samples had a concatenated length of 188,421 bp, and the subsampled plastid alignment of 60 loci and 96 sequenced samples had a concatenated length of 58,370 bp. The nuclear dataset, after subsampling, consisted of 258 loci derived from A353 and 42 loci derived from OzBaits. Output figures from genesortR are available as Supplementary Fig. S1 and S2. Gene recovery for samples in the nuclear dataset was reasonably high, with 81/98 samples represented in 80% or more of the loci (mean 82%), whereas 90/98 samples had an ungapped length of ≥50% of the total alignment (≥94,210 bp; mean 75%). In the plastid dataset, 71/96 samples were represented in 80% or more of the loci (mean 85%) and 82/96 samples had an ungapped length of ≥50% of the total alignment (≥29,185 bp; mean 69%). Coverage information for the genomic datasets is available in Supplementary Table S1.
Phylogenetic analyses
Topologies from all phylogenetic reconstructions were highly similar but not perfectly congruent, especially with relationships that were poorly supported. The IQ-TREE concatenated nuclear and chloroplast datasets exhibited overall high congruence in the phylogenies, with most incongruences observed in a reasonably small number of branches lacking robust support (Fig. 2). Likewise, the phylogenies reconstructed through a coalescent approach implemented by ASTRAL were mostly congruent with concatenated trees of the corresponding dataset (Supplementary Fig. S3, S4).
IQ-TREE analysis of the concatenated datasets for nrDNA (a) and cpDNA (b). Nodes are coloured based on values from approximate likelihood ratio test (ALRT) and ultrafast bootstrap (UFBS) supports. Nodes with red circles indicate that both values fail to support the node (ALRT < 80 and UFBS < 95) and orange nodes indicate that only one value supports the node. Unlabelled nodes are supported with ALRT ≥ 80 and UFBS ≥ 95. Taxa are coloured based on the geographic distribution within Australia, with regions indicated on the inset map. Hibbertia scandens (dark green) occupies the northern and eastern Australian regions. Branches of the four subgenera of Hibbertia are labelled i–iv. Major monophyletic clades of subg. Hibbertia and subg. Hemistemma represented in all analyses for both datasets are labelled clades A–N; finer dashed, unlabelled lines are species without clade relationships.
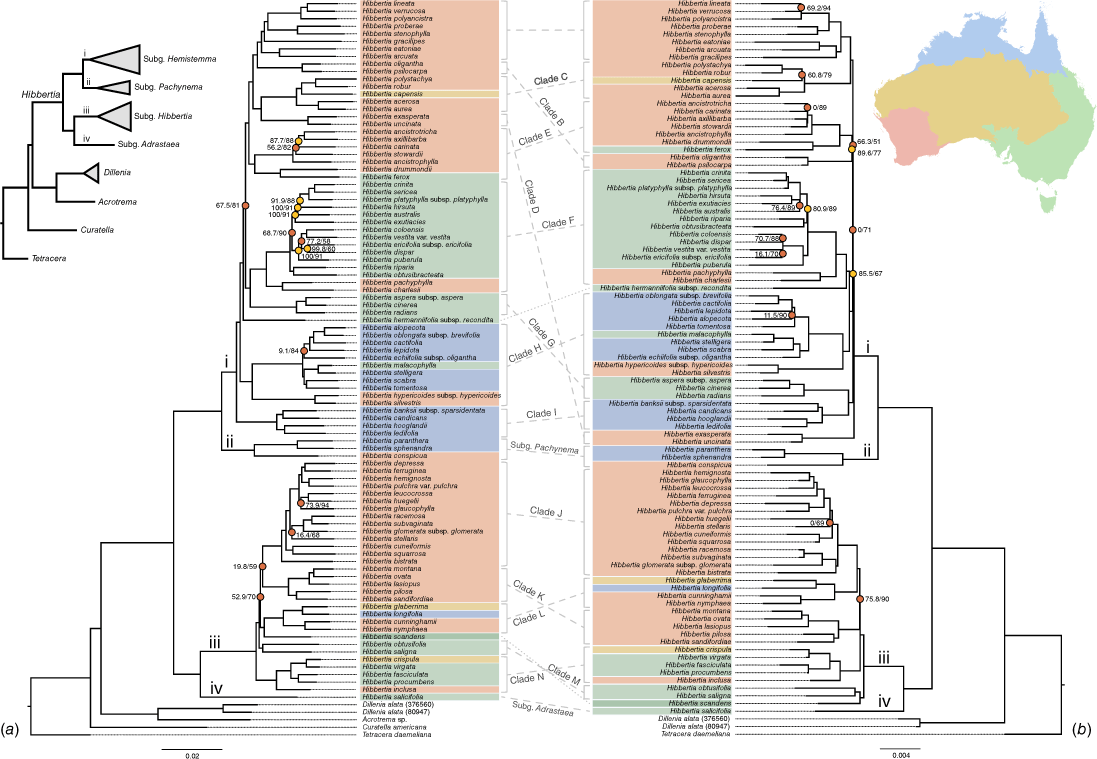
In all analyses of both genomic datasets, Hibbertia and the four subgenera were monophyletic with very high support, with subg. Adrastaea and subg. Hibbertia placed as sisters, and subg. Hemistemma and subg. Pachynema as sisters. The concatenated nuclear phylogeny was mostly well supported along the backbone apart from one node in subg. Hemistemma and two nodes in subg. Hibbertia (Fig. 2a). The concatenated plastid phylogeny was particularly poorly resolved on the backbone of subg. Hemistemma with very short branch lengths and had one poorly supported node on the backbone of subg. Hibbertia (Fig. 2b). Nevertheless, we were able to use these phylogenies to identify clades that were consistently monophyletic in every analysis to aid in the discussion of the relationships for which we can be reasonably certain. In subg. Hemistemma and subg. Hibbertia, ‘major clades’ have been labelled as A–I and J–N respectively, with the two smaller subgenera labelled as ‘subg. Adrastaea’ and ‘subg. Pachynema’ (Fig. 2). Hibbertia hermaniifolia subsp. recondita Toelken and H. scandens (Willd.) Gilg were consistently resolved outside of the other clades. Several of the relationships between the major clades were incongruent between the two genomic datasets, especially where the trees were poorly supported. This poor support was reflected in both concatenated phylogenies in many of the same areas, such as in subg. Hibbertia with the relationships between clades J, K and L, and along the whole backbone of subg. Hemistemma due to the many unresolved nodes in the plastid trees (Fig. 2, Supplementary Fig. S3, S4). There was genuine incongruence between the trees in some well-supported relationships, such as the different placements of clades D and M, H. scandens, H. hermanniifolia subsp. recondita, and placement of H. gracilipes Benth. within clade A. The clade having the highest number of nodes with poor support was consistently clade F, which included most of the species of subg. Hemistemma from eastern Australia (Fig. 2, Supplementary Fig. S3, S4).
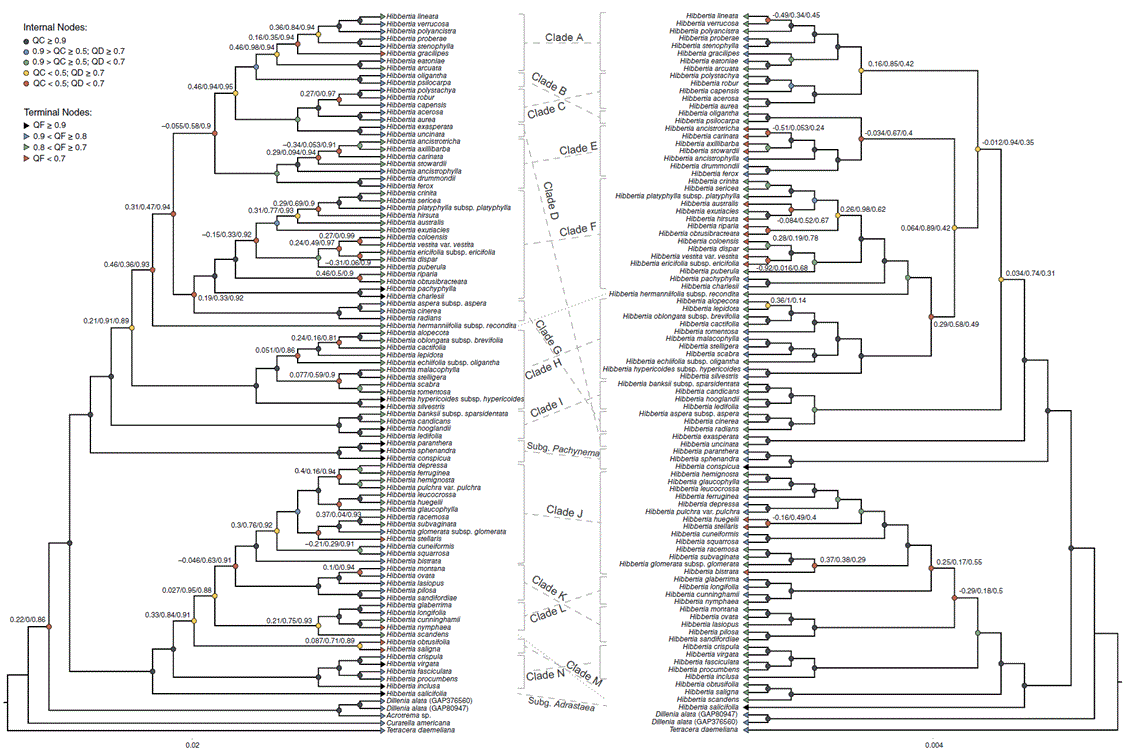
The QS analyses of the concatenated phylogenies of Fig. 2 revealed more discordant nodes than those found poorly supported with UFBS and ALRT methods (Fig. 3). The QS analyses of the ASTRAL trees (Supplementary Fig. S5, S6) were similar to the other analyses. Hibbertia and the subgenera were well supported in the QS analyses. Although all labelled clades were well supported in the prior analyses, clades A and M on the nuclear tree were discordant in the QS analysis (Fig. 3a), with QC values of 0.46 and 0.087 respectively; the QD scores for these nodes were reasonably high (0.98 and 0.71 respectively), indicating that no particular topology was favoured. All other major clades in the nuclear tree were concordant in the QS analyses and all major clades were concordant on the plastid tree. In both genomic datasets, the same general patterns were observed in the QS analyses as for the IQ-TREE and ASTRAL analyses, with the backbones of the two largest subgenera being poorly supported or discordant, leading to uncertainty in the relationships between the major clades within subgenera. In both genomic datasets, there were a few discordant nodes in clades E, F and J, and also within clade H for the nuclear tree. Clade F had the most discordant nodes of any clade, with six discordant nodes on the nuclear tree (Fig. 3a) and four discordant nodes on the plastid tree (Fig. 3b). In total, 22 of the 32 discordant nodes on the nuclear tree had low QD scores (<0.7) indicating that an alternate topology is favoured for these relationships. In general, there were more discordant nodes for the nuclear tree than the plastid tree, possibly indicating more conflict between genes in the former dataset. Reasonably low QF scores (<0.7) were found for five terminals on the nuclear tree (Fig. 3). QF scores were lower in the plastid tree (14 terminals with <0.7) that could indicate more uncertainty in taxon placement due to lower variation relative to the nuclear data. In general, terminals with lower QF scores (<0.8) corresponded to relationships that show some degree of discordance.
Quartet Sampling analysis on the nuclear (left) and plastid (right) phylogenies of the IQ-TREE concatenated analysis (shown in Fig. 2). Phylogenies are presented as cladograms for readability. Internal nodes are coloured based on their Quartet Concordance (QC) and Quartet Differential (QD) values; nodes with <0.5 QC have the QC, QD and Quartet Informativeness (QI) values reported respectively. Terminal nodes are coloured based on Quartet Fidelity (QF) values. Clades are labelled as for Fig. 2.
Discussion
Performance of the methods
The phylogenies presented here are the first published that focus on Hibbertia and the first to implement phylogenomic methods within Dilleniaceae, being based on 300 nuclear and 60 plastid loci for ~100 species. Implementation of the Genomics for Australian Plants (GAP) Phylogenomics protocols for the family was successful in generating a significant dataset with high quality phylogenetic resolution. Only two samples (H. dentata and H. fasciculiflora) failed among the 96 that were newly sequenced for this study, with most of the remaining samples having high recovery of loci for all three bait sets (Supplementary Table S1). Both nuclear and plastid genomic data were successful in reconstructing phylogenies that are largely congruent (Fig. 2), yet we have identified areas where both datasets consistently fail to satisfactorily resolve relationships due to a lack of phylogenetic signal or from conflicting loci. All four subgenera and all 14 of the identified major clades of subg. Hibbertia and subg. Hemistemma have been found to be consistently monophyletic with robust support in both datasets.
Subgenera and diagnostic features
The phylogenies presented here agree in large part with those of Horn (2005, 2009). As in Horn (2005, 2009), the two largest subgenera, subg. Hemistemma and subg. Hibbertia, are each sister to the smaller subgenera, subg. Pachynema and subg. Adrastaea respectively (Fig. 2). These relationships are interesting in that the two largest subgenera are more similar in most morphological characters to each other than either is to the smaller subgenera and vice versa for the small subgenera.
All species in Hibbertia subg. Pachynema are distinct in the genus in having mature plants that are seemingly leafless, the stems, that in some species are distinctly flattened, being the primary photosynthetic organ at maturity. Broad leaves are only present at the base of these plants at the juvenile or regrowth stages and are absent at maturity, the mature stems of the plants having scale-like leaves that are similar in appearance to floral bracts (Hammer 2022). Hibbertia subg. Adrastaea comprises the single species H. salicifolia (DC.) F.Muell. that despite the leafy habit is reasonably similar in floral morphology to the H. conspicua (J.Drumm. ex Harv.) Gilg species group of H. subg. Pachynema that consists of H. conspicua, H. goyderi F.Muell. and H. triquetra T.Hammer (represented here by H. conspicua). These four species have a cone-like androecium comprising two or three whorls of stamens enclosing the two-carpellate gynoecium at anthesis (Fig. 1i), occasionally with a ring of staminodes on the outside in some species. Other members of subg. Pachynema usually have two distinct whorls of stamens (occasionally the inner reduced to staminodes). Petal colour in subg. Pachynema is the most varied in Hibbertia, ranging from yellow or white to pink or dark red; besides three orange-flowered species in south-western Australian subg. Hibbertia, all other Hibbertia species have yellow (sometimes very pale) petals.
Floral morphology in subg. Hibbertia and subg. Hemistemma is varied (especially in the latter) but both subgenera contain species that have the stamens arranged all around the carpels (but not in distinct whorls as in the smaller subgenera); the carpels usually number three or five in subg. Hibbertia and two, three or five in subg. Hemistemma (Fig. 1b, d, j–l). In subg. Hibbertia, stamens may be free in a single or multi-layered ring around the carpels (Fig. 1k), free in gaps between the carpels (Fig. 1l) or fused in bundles between the carpels (Fig. 1j). In subg. Hemistemma, stamens are most commonly placed on one side of two carpels, where these are either more or less erect with the styles erect to spreading (Fig. 1e, f) or curved over the carpels as a hand of bananas with the styles short and curved beneath the stamens (Fig. 1a, g, h). Most Hibbertia species have solitary flowers but a few distinct groups of species in subg. Hemistemma have multi-flowered inflorescences as pseudocincinnae (Hammer and Thiele 2022), including the H. polystachya Benth. species group in south-western Australia (Fig. 1c; Thiele 2019b), the H. banksii (R.Br. ex DC.) Benth. species group (Fig. 1h; Toelken 2023) in the Australian monsoon tropics, and the non-Australian groups in Madagascar and New Caledonia (e.g. Veillon 1990). The two former groups are not closely related here (Fig. 2), indicating that the multi-flowered inflorescence is independently derived. Vestiture is another key feature that varies between these otherwise similar subgenera, with species in subg. Hibbertia having only simple hairs, whereas those in subg. Hemistemma have hairs that range from simple to hooked or fasciculate, the latter often being star-like to scale-like in appearance (Toelken 2010).
Clades and species groups
All informal taxonomic species groups represented by more than one sample were found to occur in the same major clade (Fig. 2); examples of these include H. lineata Steud. and H. verrucosa (Turcz.) Benth. from the H. lineata species group in clade A (Thiele 2017) and H. hypericoides (DC.) Benth. and H. silvestris Diels from the H. hypericoides species group in clade H (Thiele and Cockerton 2015). In some cases, major clades comprise a single species group; examples of these include the H. aspera DC. group (e.g. H. cinerea Toelken and H. radians (Toelken) T. Hammer; Hammer 2023b) corresponding to clade G, the H. banksii group (e.g. H. candicans (Hook.f.) Benth., H. hooglandii J.R.Wheeler and H. ledifolia Benth.; Toelken 2023) corresponding to clade I, and the H. commutata species group (e.g. H. montana Steud., H. ovata Steud. and H. pilosa Steud.; Thiele 2019a) corresponding to clade K. Some of these clades (e.g. J, K, L and N) are also reflected in the relationships recovered by Horn (2005).
Revisionary work in Hibbertia has until recently been split between eastern and northern Australia on the one hand, and south-western Australia on the other, due to there being little overlap between the species that occur in these regions. We show, however, that species or clades of Hibbertia are often closely related or nested within clades that occur elsewhere on the continent. A noteworthy example is H. ferox (from eastern Queensland) that is robustly supported as sister to H. drummondii (Turcz.) Gilg (from south-western Western Australia; Fig. 2). An emphasis on vegetative rather than floral characters and geographic bias has led previous authors to assume that the sharply pointed leaves of H. ferox indicate a close relationship to other pungent-leaved species (such as H. exutiacies and other members of the H. acicularis group from eastern Australia; Jackes 2018; Toelken 2024). Hibbertia drummondii and H. ferox, although differing markedly in morphological leaf characteristics, share important and overlooked morphological floral characteristics, with two glabrous carpels surrounded by a whorl of stamens (Fig. 1d; Hammer et al. 2019, p. 5), an unusual combination within subg. Hemistemma. Although H. drummondii does not have sharply pointed leaves, this trait is characteristic of some species in clades A and C–F, and has clearly evolved multiple times in subg. Hemistemma.
None of the a priori species groups are found to cross two or more major clades; however, not all species groups are monophyletic. For example, clade F (Fig. 2) includes a large, taxonomically challenging group of species from eastern Australia that have been informally classified by H. Toelken into the H. acicularis (Labill.) F.Muell., H. sericea (R.Br. ex DC.) Benth., H. stricta (DC.) R.Br. ex F.Muell. and H. vestita A.Cunn. ex Benth. species groups (e.g. Toelken 2000, 2013, 2024). However, the H. acicularis species group (represented by H. exutiacies N.A.Wakef. and H. obtusibracteata Toelken) is clearly not monophyletic. Clade F also contains a small group of Western Australia species (i.e. H. charlesii J.R.Wheeler and H. pachyphylla J.R.Wheeler) that are well resolved as sister to the eastern Australian species in every analysis.
Species in clade F are morphologically diverse, covering a broad diversity of floral and vegetative character states in Hibbertia. The eastern component of clade F, although represented by only 13 species in this study (Fig. 2), is likely to include ~100 current species and ~100 putative new species (H. R. Toelken, pers. comm.), making this the most morphologically diverse and species-rich radiation of Hibbertia in eastern Australia. The study by Raheem (2012) treated the eastern Australian species of subg. Hemistemma as monophyletic but we resolve some subg. Hemistemma species from subtropical and temperate eastern Australia in clades other than clade F (e.g. H. aspera group, H. hermaniifolia subsp. recondita and H. ferox Jackes). The difficulties encountered by Raheem (2012) in finding support for morphological species groups and resolving trees based on ITS and trnL-F alone are not surprising given our results using far more data.
Possible causes of discordance
Biological factors affecting phylogenetic signal within genomic datasets is a subject of intense study, with many groups of organisms having recalcitrant relationships even with large datasets (e.g. Roycroft et al. 2019; Wikström et al. 2020; Nge et al. 2021; Yang et al. 2021; McLay et al. 2023). This may also be the case within Hibbertia given that both genomic datasets show poor support and discordance along the backbones of the two largest subgenera and within a few complex groups (e.g. clade F; Fig. 2 and 3). The QS analysis indicates that some of this discordance could be due to non-tree-like evolution (e.g. due to ILS and introgression) associated with short branches (e.g. Paetzold et al. 2019; Kong et al. 2022). Internal nodes with low QC and high QD (coloured red on Fig. 3) show discordant topologies with a low or no skew towards a particular topology that can indicate ILS (Pease et al. 2018; Paetzold et al. 2019). Internal nodes with low QC and low QD (coloured red on Fig. 3) show discordant topologies with high skew towards a particular topology that can indicate the presence of a secondary evolutionary history (e.g. due to introgression; Pease et al. 2018; Paetzold et al. 2019). Clade F, for example, contains nodes showing both of these patterns, that in combination could significantly blur the evolutionary relationships and account for discordance and poor support values in this clade (e.g. McLay et al. 2023). A complex evolutionary history of clade F, possibly including introgression and rapid radiation, could also account for the difficulties encountered with working on the complex taxonomic classification of the group. In comparison, species groups in subg. Hemistemma in south-western Australia, although very diverse, have not had the same taxonomic issues and are reasonably well defined with readily identifiable characters. These groups are also represented in clades of the phylogeny with usually longer branches and higher support values (Fig. 2 and 3). The difference between clade F and these clades from south-western Australia could be due to differing diversification dynamics. Many other plant groups also show this pattern between south-eastern and south-western Australia that may be attributed to the historically more stable environment in the south-west (Nge et al. 2020). Future work will attempt to elucidate biological factors influencing relationships recovered by phylogenies and explore the historical complexities of the evolution of Hibbertia.
Conclusion
As part of GAP Stage II, we used genomic data to reconstruct nuclear and plastid phylogenies of Hibbertia, the largest genus in Dilleniaceae and one of the largest flowering plant genera in Australia. Methods employed were largely successful and identified key areas of incongruence within and between datasets, with the backbones of two of the largest subgenera incompletely resolved. Nevertheless, several robustly supported major clades were resolved on the phylogeny and these correspond well to hypothesised relationships based on morphological groupings. Interestingly, several species and clades on the phylogeny were found to have close transcontinental relationships. Future work will expand the taxon sampling here to investigate historical biogeography, character evolution and relationships within poorly resolved groups.
Data availability
GAP and ENA accession numbers for the data used in this study can be found in Table 1. Raw GAP data are available through the BioPlatforms Australia Data Portal (see https://data.bioplatforms.com/organization/bpa-plants). The final datasets generated and analysed are available on FigShare repository (see https://figshare.com/s/69f59d19c0a3c738605e).
Conflicts of interest
K. R. Thiele and M. Waycott are Associate Editors of Australian Systematic Botany but did not at any stage have editor-level access to this manuscript while in peer review, as is the standard practice when handling manuscripts submitted by an editor to this journal. Australian Systematic Botany encourages its editors to publish in the journal and they are kept totally separate from the decision-making processes for their manuscripts. The authors have no further conflicts of interest to declare.
Declaration of funding
The Initiative is supported by funding from Bioplatforms Australia (enabled by NCRIS), the Ian Potter Foundation, Royal Botanic Gardens Foundation (Victoria), Royal Botanic Gardens Victoria, the Royal Botanic Gardens and Domain Trust, the Council of Heads of Australasian Herbaria, CSIRO, Centre for Australian National Biodiversity Research and the Department of Biodiversity, Conservation and Attractions, Western Australia. Through the GAP consortium, co-funding for generation of the nuclear data was provided by Bioplatforms Australia (see https://bioplatforms.com/) as part of Genomics for Australian Plants Phylogenomics Stage II (see https://www.genomicsforaustralianplants.com/). The remaining data generation was funded through a Marlies Eichler Postdoctoral Fellowship (Australasian Systematic Botany Society, Inc.; see https://asbs.org.au/) awarded to T. A. Hammer in 2021. During this study, T. A. Hammer was supported through a Postdoctoral Fellowship to complete the project ‘Delineating the diversity of Dilleniaceae: a revisionary synthesis of Hibbertia for the Flora of Australia and investigations into its taxonomy, systematics, evolution and biogeography’ that was funded by the Australian Government’s Australian Biological Resources Study (ABRS) National Taxonomy Research Grant Program from 2020 to 2023 (grant number 4-EHOJ11Y); M. Waycott is the supervisor and E. Biffin, K. van Dijk and K. R. Thiele are collaborators on this project.
Acknowledgements
We acknowledge the contribution of the Genomics for Australian Plants Framework Initiative consortium (see https://www.genomicsforaustralianplants.com/consortium/) in the generation of data used in this publication. Herbarium material sampled for this study was sourced from the State Herbarium of South Australia (AD), Australian Tropical Herbarium (CNS), Tasmanian Herbarium (HO) and Western Austraian Herbarium (PERTH); we thank the directors and staff of these herbaria for granting access and permissions for use. We additionally thank directors and staff of CNS and PERTH for sending silica material used in this study. T. A. Hammer acknowledges A. E. McDougall and A. H. Thornhill for assistance with field work, and H. R. Toelken for useful discussions regarding ongoing taxonomic work.
References
Angiosperm Phylogeny Group (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical Journal of the Linnean Society 181, 1-20.
| Crossref | Google Scholar |
Anisimova M, Gil M, Dufayard J-F, Dessimoz C, Gascuel O (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Systematic Biology 60, 685-699.
| Crossref | Google Scholar | PubMed |
Baker WJ, Bailey P, Barber V, Barker A, Bellot S, Bishop D, Botigué LR, Brewer G, Carruthers T, Clarkson JJ, Cook J, Cowan RS, Dodsworth S, Epitawalage N, Françoso E, Gallego B, Johnson MG, Kim JT, Leempoel K, Maurin O, McGinnie C, Pokorny L, Roy S, Stone M, Toledo E, Wickett NJ, Zuntini AR, Eiserhardt WL, Kersey PJ, Leitch IJ, Forest F (2022) A comprehensive phylogenomic platform for exploring the angiosperm tree of life. Systematic Biology 71, 301-319.
| Crossref | Google Scholar | PubMed |
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972-1973.
| Crossref | Google Scholar | PubMed |
Cheng C, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. The Plant Journal 89, 789-804.
| Crossref | Google Scholar | PubMed |
Chernomor O, von Haeseler A, Minh BQ (2016) Terrace aware data structure for phylogenomic inference from supermatrices. Systematic Biology 65, 997-1008.
| Crossref | Google Scholar | PubMed |
Di Franco A, Poujol R, Baurain D, Philippe H (2019) Evaluating the usefulness of alignment filtering methods to reduce the impact of errors on evolutionary inferences. BMC Evolutionary Biology 19, 21.
| Crossref | Google Scholar | PubMed |
Doyle JJ (2022) Defining coalescent genes: theory meets practice in organelle phylogenomics. Systematic Biology 71, 476-489.
| Crossref | Google Scholar | PubMed |
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59, 307-321.
| Crossref | Google Scholar | PubMed |
Hammer TA (2022) Updated nomenclature and an identification key for Hibbertia subg. Pachynema (Dilleniaceae), and description of a new species from the Northern Territory. Swainsona 36, 153-160.
| Google Scholar |
Hammer TA (2023a) Hibbertia fulva (Dilleniaceae), a new species from the Northern Territory in the H. banksii species group. Swainsona 37, 71-74.
| Google Scholar |
Hammer TA (2023b) Hibbertia radians (Dilleniaceae), a new combination from South Australia. Swainsona 37, 63-70.
| Google Scholar |
Hammer TA, Thiele KR (2022) Hibbertia Andrews. In ‘Flora of Australia’. (Ed. PG Kodela) (Australian Biological Resources Study, Department of Climate Change, the Environment and Water: Canberra, ACT, Australia) Available at https://profiles.ala.org.au/opus/foa/profile/Hibbertia [Verified 1 April 2024]
Hammer TA, Thiele KR (2024) New combinations of Hibbertia (Dilleniaceae) segregated from H. hibbertioides and H. glomerata. Nuytsia 35, 31-45.
| Crossref | Google Scholar |
Hammer TA, Toelken HR, Thiele KR (2019) Hibbertia advena (Dilleniaceae), a new and rare species from Queensland with transcontinental affinities. Australian Journal of Taxonomy 9, 1-5.
| Crossref | Google Scholar |
Horn JW (2009) Phylogenetics of Dilleniaceae using sequence data from four plastid loci (rbcL, infA, rps4, rpl16 intron). International Journal of Plant Sciences 170, 794-813.
| Crossref | Google Scholar |
Jackes BR (2018) Hibbertia ferox Jackes (Dilleniaceae), a new species from the White Mountains area of north Queensland. Austrobaileya 10, 282-285.
| Crossref | Google Scholar |
Johnson MG, Pokorny L, Dodsworth S, Botigué LR, Cowan RS, Devault A, Eiserhardt WL, Epitawalage N, Forest F, Kim JT, Leebens-Mack JH, Leitch IJ, Maurin O, Soltis DE, Soltis PS, Wong GK, Baker WJ, Wickett NJ (2019) A universal probe set for targeted sequencing of 353 nuclear genes from any flowering plant designed using k-medoids clustering. Systematic Biology 68, 594-606.
| Crossref | Google Scholar | PubMed |
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587-589.
| Crossref | Google Scholar | PubMed |
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772-780.
| Crossref | Google Scholar | PubMed |
Keller O, Odronitz F, Stanke M, Kollmar M, Waack S (2008) Scipio: using protein sequences to determine the precise exon/intron structures of genes and their orthologs in closely related species. BMC Bioinformatics 9, 278.
| Crossref | Google Scholar | PubMed |
Kong H, Condamine FL, Yang L, Harris AJ, Feng C, Wen F, Kang M (2022) Phylogenomic and macroevolutionary evidence for an explosive radiation of a plant genus in the Miocene. Systematic Biology 71, 589-609.
| Crossref | Google Scholar | PubMed |
Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A (2014) Selecting optimal partitioning schemes for phylogenomic datasets. BMC Ecology and Evolution 14, 82.
| Crossref | Google Scholar | PubMed |
Leebens-Mack JH, Barker MS, Carpenter EJ, Deyholos MK, Gitzendanner MA, Graham SW, Grosse I, Li Z, Melkonian M, Mirarab S, et al. (2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574, 679-68.
| Crossref | Google Scholar |
Li D, Liu CM, Luo R, Sadakane K, Lam TW (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674-1676.
| Crossref | Google Scholar | PubMed |
Mai U, Mirarab S (2018) TreeShrink: fast and accurate detection of outlier long branches in collections of phylogenetic trees. BMC Genomics 19(Suppl 5), 272.
| Crossref | Google Scholar | PubMed |
McLay TGB, Fowler RM, Fahey PS, Murphy DJ, Udovicic F, Cantrill DJ, Bayly MJ (2023) Phylogenomics reveals extreme gene tree discordance in a lineage of dominant trees: hybridization, introgression, and incomplete lineage sorting blur deep evolutionary relationships despite clear species groupings in Eucalyptus subgenus Eudesmia. Molecular Phylogenetics and Evolution 187, 107869.
| Crossref | Google Scholar | PubMed |
Minh BQ, Nguyen MAT, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30, 1188-1195.
| Crossref | Google Scholar | PubMed |
Mongiardino Koch N (2021) Phylogenomic subsampling and the search for phylogenetically reliable loci. Molecular Biology and Evolution 38, 4025-4038.
| Crossref | Google Scholar | PubMed |
Nge FJ, Biffin E, Thiele KR, Waycott M (2020) Extinction pulse at Eocene–Oligocene boundary drives diversification dynamics of two Australian temperate floras. Proceedings of the Royal Society B 287, 20192546.
| Crossref | Google Scholar | PubMed |
Nge FJ, Biffin E, Thiele KR, Waycott M (2021) Reticulate evolution, ancient chloroplast haplotypes, and rapid radiation of the Australian plant genus Adenanthos (Proteaceae). Frontiers in Ecology and Evolution 8, 616741.
| Crossref | Google Scholar |
Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Molecular Biology and Evolution 32, 268-274.
| Crossref | Google Scholar | PubMed |
Paetzold C, Wood KR, Eaton DA, Wagner WL, Appelhans MS (2019) Phylogeny of Hawaiian Melicope (Rutaceae): RAD-seq resolves species relationships and reveals ancient introgression. Frontiers in Plant Science 10, 1074.
| Crossref | Google Scholar | PubMed |
Pease JB, Brown JW, Walker JF, Hinchliff CE, Smith SA (2018) Quartet Sampling distinguishes lack of support from conflicting support in the green plant tree of life. American Journal of Botany 105, 385-403.
| Crossref | Google Scholar | PubMed |
Raza M, Ortiz EM, Schwung L, Shigita G, Schaefer H (2023) Resolving the phylogeny of Thladiantha (Cucurbitaceae) with three different target capture pipelines. BMC Ecology and Evolution 23, 75.
| Crossref | Google Scholar | PubMed |
Roycroft EJ, Moussalli A, Rowe KC (2019) Phylogenomics uncovers confidence and conflict in the rapid radiation of Australo-Papuan rodents. Systematic Biology 69, 431-444.
| Crossref | Google Scholar |
Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG (2014) From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evolutionary Biology 14, 23.
| Crossref | Google Scholar | PubMed |
Soltis DE, Smith SA, Cellinese N, Wurdack KJ, Tank DC, Brockington SF, Refulio-Rodriguez NF, Walker JB, Moore MJ, Carlsward BS, Bell CD, Latvis M, Crawley S, Black C, Diouf D, Xi Z, Rushworth CA, Gitzendanner MA, Sytsma KJ, Qiu Y, Hilu KW, Davis CC, Sanderson MJ, Beaman RS, Olmstead RG, Judd WS, Donoghue MJ, Soltis PS (2011) Angiosperm phylogeny: 17 genes, 640 taxa. American Journal of Botany 98, 704-730.
| Crossref | Google Scholar | PubMed |
Stebbins GL, Hoogland RD (1976) Species diversity, ecology and evolution in a primitive angiosperm genus: Hibbertia (Dilleniaceae). Plant Systematics and Evolution 125, 139-154.
| Crossref | Google Scholar |
Steenwyk JL, Buida TJ, Li Y, Shen X-X, Rokas A (2020) ClipKIT: a multiple sequence alignment trimming software for accurate phylogenomic inference. PLoS Biology 18, e3001007.
| Crossref | Google Scholar | PubMed |
Thiele KR (2017) A revision of the Hibbertia lineata (Dilleniaceae) species group. Nuytsia 28, 173-191.
| Crossref | Google Scholar |
Thiele KR (2019a) A revision of the Hibbertia commutata (Dilleniaceae) species group. Australian Systematic Botany 32, 71-109.
| Crossref | Google Scholar |
Thiele KR (2019b) The Hibbertia polystachya–H. spicata (Dilleniaceae) species group in Western Australia. Nuytsia 30, 291-308.
| Crossref | Google Scholar |
Thiele KR, Cockerton G (2015) A revision of the Hibbertia hypericoides species group (Dilleniaceae). Nuytsia 25, 285-300.
| Crossref | Google Scholar |
Toelken HR (2000) Notes on Hibbertia (Dilleniaceae) 3. H. sericea and associated species. Journal of the Adelaide Botanic Gardens 19, 1-54.
| Google Scholar |
Toelken HR (2010) Notes on Hibbertia (Dilleniaceae) 5. H. melhanioides and H. tomentosa groups from tropical Australia. Journal of the Adelaide Botanic Gardens 23, 1-117.
| Google Scholar |
Toelken HR (2013) Notes on Hibbertia subg. Hemistemma (Dilleniaceae) 9. The eastern Australian H. vestita group, including H. pedunculata and H. serpyllifolia. Journal of the Adelaide Botanic Gardens 26, 31-69.
| Google Scholar |
Toelken HR (2023) Notes on Hibbertia (Dilleniaceae: subgen. Hemistemma) — 12. The northern Australian species of the H. banksii group. Swainsona 37, 7-34.
| Google Scholar |
Toelken HR (2024) Notes on Hibbertia subgen. Hemistemma (Dilleniaceae) – 13. The eastern Australian H. acicularis and H. perhamata groups. Swainsona 38, 79-124.
| Google Scholar |
Tucker SC (1997) Floral evolution, development, and convergence: the hierarchical-significance hypothesis. International Journal of Plant Sciences 158(S6), S143-S161.
| Crossref | Google Scholar |
Tucker SC, Bernhardt P (2000) Floral ontogeny, pattern formation, and evolution in Hibbertia and Adrastaea (Dilleniaceae). American Journal of Botany 87, 1915-1936.
| Google Scholar | PubMed |
Waycott M, van Dijk K, Biffin E (2021) A hybrid capture RNA bait set for resolving genetic and evolutionary relationships in angiosperms from deep phylogeny to intraspecific lineage hybridization. BioRxiv
| Crossref | Google Scholar |
Weitemier K, Straub SCK, Cronn RC, Fishbein M, Schmickl R, McDonnell A, Liston A (2014) Hyb-Seq: combining target enrichment and genome skimming for plant phylogenomics. Applications in Plant Sciences 2, 1400042.
| Crossref | Google Scholar | PubMed |
Wikström N, Bremer B, Rydin C (2020) Conflicting phylogenetic signals in genomic data of the coffee family (Rubiaceae). Journal of Systematics and Evolution 58, 440-460.
| Crossref | Google Scholar |
Yang Y-Y, Qu Z-J, Zhang R, Stull GW, Yi T-S (2021) Plastid phylogenomic analyses of Fagales reveal signatures of conflict and ancient chloroplast capture. Molecular Phylogenetics and Evolution 163, 107232.
| Crossref | Google Scholar | PubMed |
Zeng CX, Hollingsworth PM, Yang J, He ZS, Zhang ZR, Li DZ, Yang JB (2018) Genome skimming herbarium specimens for DNA barcoding and phylogenomics. Plant Methods 14, 43.
| Crossref | Google Scholar | PubMed |
Zhang C, Mirarab S (2022) Weighting by gene tree uncertainty improves accuracy of quartet-based species trees. Molecular Biology and Evolution 39, msac215.
| Crossref | Google Scholar | PubMed |
Zhang C, Rabiee M, Sayyari E, Mirarab S (2018) ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics 19(S6), 153.
| Crossref | Google Scholar | PubMed |
Zuntini AR, Carruthers T, Maurin O, et al. (2024) Phylogenomics and the rise of the angiosperms. Nature 629, 843-850.
| Crossref | Google Scholar | PubMed |


